ABSTRACT
Target-gene amplicon sequencing is the most exploited high-throughput sequencing application in microbial ecology. The targets are taxonomically relevant genes, with 16S rRNA being the gold standard for bacteria. As for fungi, the most commonly used target is the internal transcribed spacer (ITS). However, the uneven ITS length among species may promote preferential amplification and sequencing and incorrect estimation of their abundance. Therefore, the use of different targets is desirable. We evaluated the use of three different target amplicons for the characterization of fungal diversity. After an in silico primer evaluation, we compared three amplicons (the ITS1-ITS2 region [ITS1-2], 18S ribosomal small subunit RNA, and the D1/D2 domain of the 26S ribosomal large subunit RNA), using biological samples and a mock community of common fungal species. All three targets allowed for accurate identification of the species present. Nevertheless, high heterogeneity in ITS1-2 length was found, and this caused an overestimation of the abundance of species with a shorter ITS, while both 18S and 26S amplicons allowed for more reliable quantification. We demonstrated that ITS1-2 amplicon sequencing, although widely used, may lead to an incorrect evaluation of fungal communities, and efforts should be made to promote the use of different targets in sequencing-based microbial ecology studies.
IMPORTANCE Amplicon-sequencing approaches for fungi may rely on different targets affecting the diversity and abundance of the fungal species. An increasing number of studies will address fungal diversity by high-throughput amplicon sequencing. The description of the communities must be accurate and reliable in order to draw useful insights and to address both ecological and biological questions. By analyzing a mock community and several biological samples, we demonstrate that using different amplicon targets may change the results of fungal microbiota analysis, and we highlight how a careful choice of the target is fundamental for a thorough description of the fungal communities.
KEYWORDS: fungal diversity, primer bias, amplicon sequencing, high-throughput sequencing, microbial ecology
INTRODUCTION
Communities of fungi are of utmost importance in many different environments (1–3), and an accurate and detailed description of their structure is desirable in order to address fundamental biological questions in microbial ecology. The analysis of environmental samples based on high-throughput sequencing (HTS) technologies allowed for study of the abundance and richness of microbial species directly in the matrix of interest, overcoming the limitations imposed by methods based on cultivation yet incurring biases introduced in both “wet-lab” procedures and bioinformatics data analysis (4–7). Studies based on shotgun sequencing of total DNA provide a description of the microbial community free from PCR biases. Nevertheless, the shotgun approach is still not routinely used due to the higher costs and intensive computational analysis involved compared to those of amplicon-based sequencing (1). On the contrary, public databases are rich in studies investigating the composition of microbial populations in different environments using the sequencing of taxonomically relevant genes (1). The 16S rRNA gene is considered the gold standard as the target in HTS studies aiming at characterization of bacterial communities. As for fungi, more variability in chosen targets is found in the literature (1). Most studies employed the internal transcribed spacer 1–2 (ITS1-2) region of ribosomal DNA. This target was proposed as the official primary barcode for fungi thanks to the presence of a rich and up-to-date database, which is necessary for data analysis (8). However, the length of the ITS1-2 region is extremely variable among different fungal genera and species (9); therefore, the amplification of this gene, starting from environmental DNA, can give rise to preferential amplification and a biased assessment of the abundance of the taxonomic units in the matrix (1, 10). For this reason, in recent years, some efforts have been focused on promoting the use of alternative target genes in HTS studies, such as portions of the genes coding for 26S (11–13) and 18S (14–16) rRNA. In addition, Stielow et al. (17) showed that translation elongation factor 1-α (TEF1α) gene sequencing has superior resolution compared to that of ITS amplicon sequencing and proposed it as a secondary DNA barcode for the Fungi kingdom.
We aimed to evaluate how the choice of the target gene in HTS studies may influence the description of fungal diversity. Three different targets (ITS1-2, 18S rRNA, and 26S rRNA genes) were compared in this study. A “mock community” of known composition, including common fungal species as well as environmental samples, was amplified using the three targets, and HTS experiments were carried out in order to compare the results achieved.
RESULTS
In silico evaluation of primer efficiency.
The efficacy of the three primer sets used in this study was evaluated in silico using Primer Prospector. We computationally predicted and compared the primer coverage of known fungal sequences. First, all the primers were tested for overall database matches using the “analyze primers” module. The 18S targeting set showed the lowest (best) weighted score, indicating higher coverage across the database sequences and lower number of mismatches (Table 1; see also Fig. S1A in the supplemental material). The worst weighted score was reported for the ITS1 forward primer, which also showed the highest number of mismatches (Table 1; see also Fig. S1A). Next, we used the “taxonomic coverage” module to predict the coverage of the different fungal clades for each primer. All of the primer sets showed wide coverage across fungal lineages (see Fig. S1B). However, the lowest percentage of coverage was observed for the ITS1 forward primer; only Glomeromycota was covered >80% with this primer, while the values were around 40% for all the other clades. Also in this case, the 18S primer set showed the best results in terms of taxa coverage, with values close to 90% for all the fungal clades. Finally, the 26S primer pair showed lower coverage for the forward primer (see Fig. S1B). The two most abundant fungal phyla (Ascomycota and Basidiomycota) reported a predicted coverage of about 60% and >80% with the forward and reverse 26S primers, respectively (see Fig. S1B).
TABLE 1.
Primers used in this study and results from Primer Prospector in silico analysis
| rDNA | Primer | Sequence (5′ → 3′) | Amplicon length (bp) | Reference no. | Non-3′ mismatcha | 3′ mismatcha | Non-3′ gapsa | 3′ gapsa | Weighted scorea |
|---|---|---|---|---|---|---|---|---|---|
| SSU | 18Sf | ATTCCAKCTCCAAKAGCG | 436 | 17 | 0.137 | 0.055 | 0.002 | 0.000 | 0.156 |
| 18Sr | GATNAGATACCRTCGTAGTC | 17 | 0.445 | 0.012 | 0.009 | 0.000 | 0.245 | ||
| LSU | 26Sf | GCATATCAATAAGCGGAGGAAAAG | 579 | 29 | 2.977 | 0.339 | 0.018 | 0.000 | 1.720 |
| 26Sr | CCGTCTTGAAACACGGACCG | 29 | 1.556 | 0.158 | 0.012 | 0.001 | 0.904 | ||
| ITS1-2 | ITS1f | TCCGTAGGTGAACCTGCGG | Variable | 23 | 3.503 | 1.024 | 0.059 | 0.002 | 3.050 |
| ITS2r | GCATCGATGAAGAACGCAGC | 23 | 0.527 | 0.164 | 0.021 | 0.001 | 0.593 |
Weighted score = (non-3′ mismatches × 0.4) + 3′ mismatches + non-3′ gaps + (3′ gaps × 3.0) + final 3′ base mismatch × 3.0).
Mock community analysis.
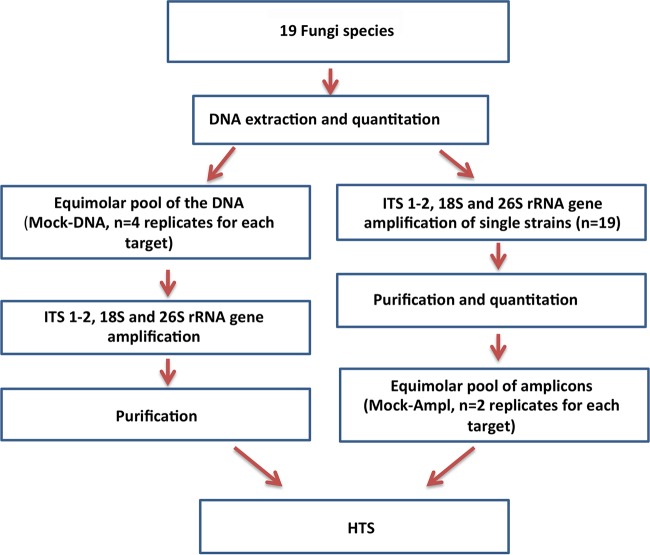
A mock community containing 19 fungal species was prepared to test the real performances of the three primer sets. In order to evaluate the effect of coamplification biases and to ensure that the primer sets used were able to detect all the species included, two different protocols were followed (Fig. 1). In the first case (Mock-Ampl), amplification of single strains was carried out, and amplicons were pooled at equimolar concentration. In the second case (Mock-DNA), the mock community was prepared by pooling the DNA of the 19 strains at the same concentration, and the PCR was performed afterward.
FIG 1.
Schematic representation of the study design.
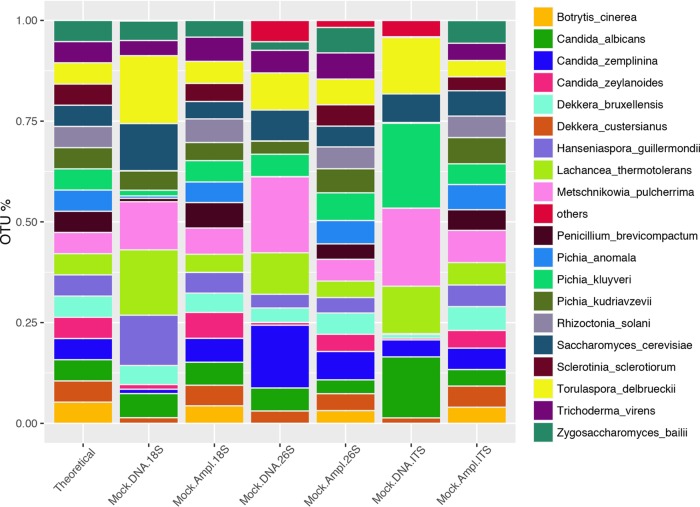
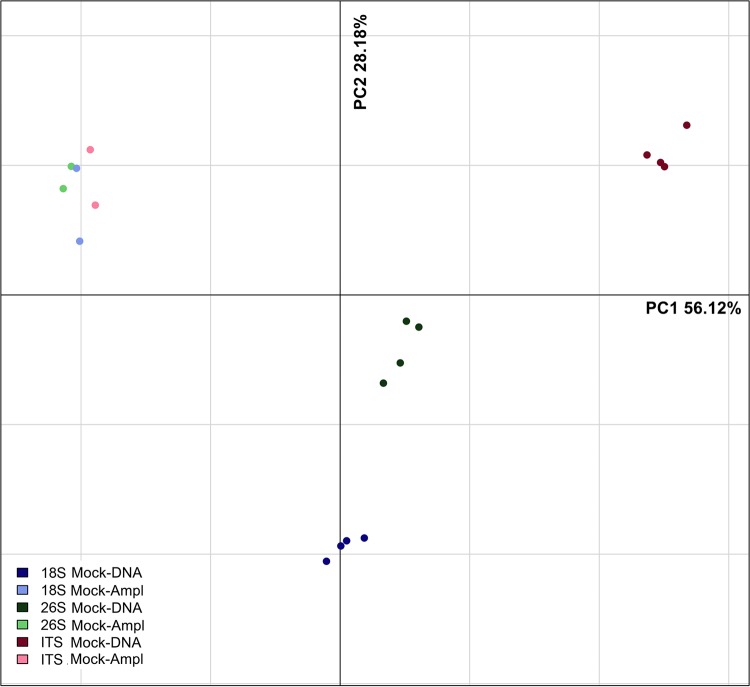
Mock-Ampl samples did not show significant differences in mycobiotal composition (P > 0.05), and all the target genes were able to detect the 19 species used (Fig. 2). The strains used showed an ITS1-2 length ranging from about 180 to 420 bp (see Fig. S2 and Table S1 in the supplemental material). Interestingly, the yeast strains Metschnikowia pulcherrima and Pichia kluyveri, with ITS1-2 lengths of about 180 and 190 bp, respectively, were overrepresented in ITS Mock-DNA samples, while Zygosacharomyces bailii and Hanseniaspora guillermondii (both with an ITS1-2 length of about 400 bp) were clearly underestimated in ITS Mock-DNA samples (Fig. 2). The Mock-Ampl samples showed a community composition similar to the theoretical, regardless of the target gene used. On the contrary, multivariate analysis of variance (MANOVA) based on Jaccard, Euclidean, and Bray Curtis distances detected differences in the overall fungal community composition in Mock-DNA samples according to the target used (P < 0.05). Spearman's correlation between the abundance matrices of Mock-DNA samples was never higher than 0.61 (ρ18S|versus|26S 0.61; ρ18S|versus|ITS 0.47; ρITS|versus|26S 0.58). Accordingly, the average Bray Curtis distance between 26S and 18S was lower than those of the other targets, while ITS gave the most different results (see Fig. S3 in the supplemental material). Moreover, PCA analysis showed a clear clustering of Mock-DNA samples according to the target gene, while Mock-Ampl samples grouped together, regardless of the target used (Fig. 3). On the contrary, the target gene chosen did not seem to influence alpha-diversity parameters (P > 0.05, data not shown).
FIG 2.
Bar chart showing the relative abundance of the fungal species identified in the mock community amplified using three different target genes. The expected concentration (theoretical) is shown in the first bar. Average abundance for 2 (Mock-Ampl) or 4 (Mock-DNA) replicates is reported.
FIG 3.
Principal component analysis based on the species-level microbiota.
In all cases, some of the species included in the mock community were not identified in the Mock-DNA or were found at abundance levels lower than expected. Taxa showing a significantly different abundance than expected are reported in Table 2. Metschnikowia pulcherrima was overestimated by 26S rRNA and ITS sequencing, while the abundance of Candida zeylanoides and Pichia anomala was underestimated with all the target genes. In addition, Candida zemplinina was detected only in Mock-DNA samples amplified with the 26S rRNA gene (Fig. 2 and Table 2).
TABLE 2.
Significantly different species detected in the mock community analyzed with three different target genesb
| Species | Target genea |
||
|---|---|---|---|
| 18S | 26S | ITS | |
| Candida albicans | 5.98 B | 5.71 B | 15.13 A |
| Candida zemplinina | 1.09 C | 15.59 A | 4.22 B |
| Dekkera bruxellensis | 4.73 A | 3.58 A | 0.89 B |
| Dekkera custersianus | 1.40 B | 3.07 A | 1.35 B |
| Hanseniaspora guillermondi | 12.49 A | 3.41 B | 0.22 C |
| Metschnikowia pulcherrima | 11.94 C | 18.82 B | 19.35 A |
| Penicillium brevicompactum | 0.85 A | 0.00 B | 0.02 C |
| Pichia anomala | 0.59 A | 0.09 B | 0.02 B |
| Pichia kluyveri | 1.41 A | 5.54 B | 21.05 A |
| Pichia kudriavzevii | 4.80 A | 3.24 A | 0.09 B |
| Saccharomyces cerevisiae | 11.68 A | 7.67 B | 7.09 B |
| Torulaspora delbrueckii | 16.85 A | 9.20 C | 13.96 B |
| Trichoderma virens | 3.77 A | 5.62 A | 0.00 B |
Different letters indicate values significantly different (P < 0.05) after correction for multiple comparisons, as determined by pairwise Wilcoxon test, among the three target genes.
Only Mock-DNA samples were considered.
Analysis of mycobiota in biological samples.
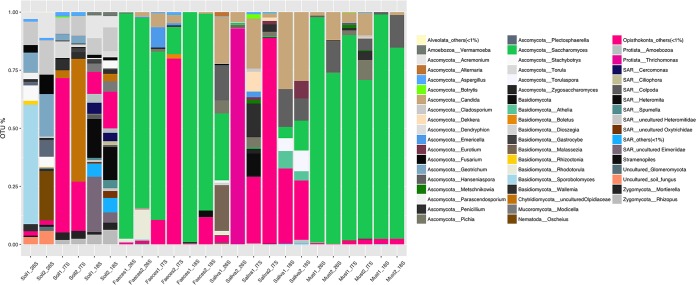
We used the same protocol to analyze biological samples. Samples sequenced via 26S rRNA showed higher alpha-diversity than samples sequenced via the other targets (P < 0.05). In addition, in all cases, we observed striking differences in fungal community composition according to the type of amplicon used (Fig. 4). Sporobolomyces (Basidiomycota), dominant in soil samples sequenced by 26S rRNA, was absent when using both of the other targets, while Hanseniaspora was underestimated by ITS sequencing (Fig. 4). In addition, besides fungi, a high abundance of other eukarya belonging to the SAR supergroup (e.g., Heteromita, Cercomonas) were found in soil samples only when using the 18S rRNA gene as the target.
FIG 4.
Bar chart showing the relative abundance of the eukaryotic species identified in the biological samples analyzed using three different target genes.
DISCUSSION
In this past decade, HTS has revolutionized the approach to microbial ecology. In particular, amplicon sequencing of taxonomically relevant genes has become a routinely used approach in most microbiology laboratories. Nevertheless, the existence of potential biases was previously emphasized (4–7), and PCR can be considered the most important source of biases due to primer preferential amplification (10, 18). The genes coding for the small subunit (SSU) (18S) or the large subunit (LSU) (28S) of rRNA and the internal transcribed spacer (ITS) region are the targets commonly used for fungal identification (19). However, the ITS region was proposed as the primary target for fungi (8), and it has been widely used in HTS studies (20, 21). Although having higher resolution in species identification compared to the other targets (19), the uneven lengths of ITS fragments are known to promote preferential amplification of shorter sequences, leading to a biased quantification of taxon relative abundance (22). Nevertheless, its use in HTS is often preferred thanks to the existence of a more curated and rich database compared to that available for SSU/LSU rRNA genes. Indeed, after filtering of existing databases, we recovered 67,073, 21,099, and 3,015 sequences, for ITS, SSU and LSU, respectively.
We assessed the effect of the target gene used in describing fungal communities, employing three previously designed primer sets. We are not aware of other reports in which the three targets were compared with the use of a mock community. In silico analysis demonstrated that the traditionally used ITS1 forward primer (23) yielded the worst results in terms of taxonomic coverage and number of mismatches with database sequences. In addition, we sequenced a mock community containing 19 fungal species in order to evaluate how they would potentially perform in real conditions. Mock communities are a useful tool for validating and optimizing experimental procedures, although their use to evaluate potential experimental biases is still limited (18, 24, 25). Indeed, when DNA of all the strains was amplified in the same PCR, resembling the typical condition of an HTS analysis, primer biases were evident, and clear preferential amplification phenomena were detected with all the primer sets. A further confirmation of such evidence was obtained by analyzing real biological samples using the three different amplicon targets. Indeed, fungal community structures of feces, saliva, soil, and musts differed depending on the PCR target used. An additional bias resulted from ITS region sequencing. In fact, those species with a longer ITS1-2 region (e.g., Zygosacharomyces bailii and Hanseniaspora guillermondii) were underestimated in Mock-DNA, while an overamplification of shorter fragments from Metschnikowia pulcherrima and Pichia kluyveri occurred. Accordingly, Hanseniaspora was also found at lower concentrations in biological samples sequenced with ITS1-2 primers. This bias may be particularly important in some environments, such as in oenological fermentations, where non-Saccharomyces yeasts dominate in the first steps of fermentation, while Saccharomyces species take over afterward (16). Indeed, when comparing the results obtained from ITS amplicon sequencing and TRFLP, Hanseniaspora was found to be underestimated by HTS (18). Therefore, although all three primer sets led to PCR biases, the results obtained highlighted that the use of ITS1-2 sequencing may further distort the community description due to preferential amplification of shorter fragments. Finally, it was only when 18S rRNA was used that we detected higher organisms belonging to the SAR supergroup. This may be particularly important in some environments, such as soil, where these organisms may play an important biological role (14).
Although based on pyrosequencing technology that is not available anymore, the results show insights of very general interest. Indeed, the primer used and the results obtained can be easily transposed to the sequencing technologies currently in use and employed in short amplicon sequencing for the description of the fungal communities. Our study suggests that results from ITS1-2 sequencing should be considered carefully, and efforts should be made to improve available primers targeting different loci and related databases to promote their fruitful and reliable use in HTS studies.
MATERIALS AND METHODS
In silico primer analysis.
The primer analysis module of Primer Prospector (26) was used with default settings to compare primer specificity and taxonomic coverage. The reference database was created by filtering the Silva SSU/LSU rRNA genes, release 128 (27) and the UNITE ITS, release 7.1 (28) databases. Hits in the taxonomy file containing the terms “environmental sample,“ “mycorrhizal,” “mycorrhizae,” “unidentified,” or “unclassified” were filtered out. Moreover, only sequences identified as fungi were kept. The final database contained a total of 67,073 (ITS), 21,099 (SSU), and 3,015 (LSU) reads, covering all fungal lineages. A matching taxonomy file was created, and the taxonomy coverage module of Primer Prospector was used to predict the coverage for each fungal phylum.
Strain culturing and DNA extraction.
Strains of fungi used for the preparation of the mock community were previously isolated and identified through complete ITS sequencing. Strain names and ITS1-2 region lengths are reported in Table 3. We chose 19 strains, including the two largest fungal clades (Ascomycota and Basidiomycota). In order to avoid biases due to the DNA extraction procedure, DNA extraction was carried out on each strain individually. Yeasts were grown in yeast extract-peptone-dextrose (YPD) broth (10 g/liter yeast extract, 20 g/liter peptone, 20 g/liter dextrose, 0.1 mg of chloramphenicol/100 ml [all from Oxoid, Milan, Italy]) incubated for 3 days at 28°C. Five milliliters was centrifuged at 12,000 × g, and the pellet was used for DNA extraction. Filamentous fungi were grown on malt extract agar (17 g/liter malt extract, 3 g/liter peptone, agar n.1, 1.5% [Oxoid]) and incubated at 28°C for 7 to 10 days. Mycelium was scraped from the plates with a sterile scalpel, and 250 mg was used for DNA extraction. Extraction was carried out with the PowerSoil DNA extraction kit (MoBio Laboratories, Inc., Carlsbad, CA). DNA was quantified using the Qubit fluorimeter (Life Technologies, Carlsbad, CA).
TABLE 3.
Fungal species used to prepare the mock communities
| Species | ITS1-2 fragment length (bp) | Phylum |
|---|---|---|
| Botrytis cinerea | 290 | Ascomycota |
| Candida albicans | 260 | Ascomycota |
| Candida zemplinina | 220 | Ascomycota |
| Candida zeylanoides | 300 | Ascomycota |
| Dekkera bruxellensis (Brettanomyces bruxellensis) | 200 | Ascomycota |
| Dekkera custersianus (Brettanomyces custersianus) | 210 | Ascomycota |
| Hanseniaspora guillermondii | 400 | Ascomycota |
| Issatchenkia orientalis | 210 | Ascomycota |
| Lachancea thermotolerans | 350 | Ascomycota |
| Metschnikowia pulcherrima | 180 | Ascomycota |
| Penicillium brevicompactum | 300 | Ascomycota |
| Pichia anomala | 320 | Ascomycota |
| Pichia kluyveri | 190 | Ascomycota |
| Saccharomyces cerevisiae | 420 | Ascomycota |
| Sclerotinia sclerotiorum | 300 | Ascomycota |
| Torulaspora delbrueckii | 400 | Ascomycota |
| Trichoderma virens | 340 | Ascomycota |
| Zygosaccharomyces bailii | 390 | Ascomycota |
| Rhizoctonia solani | 350 | Basidiomycota |
Mock community preparation and sequencing of amplicon libraries.
A scheme of the study design is reported in Fig. 1. In order to exploit the possible bias introduced by PCR, two different paths were followed. (i) DNA from each strain was individually amplified (for all the three target genes), and amplicons were equimolarly pooled after purification and quantitation (Mock-Ampl). (ii) The DNA of each strain was quantified and diluted at 50 ng/μl. Diluted DNA was then pooled at the same concentration (Mock-DNA) and the pool was subjected to PCR (for all the three target genes).
Primer pairs targeting the V4 region of the 18S rRNA gene (16), the D1/D2 domain of the 26S rRNA gene (29), and the ITS1-2 (23) were used with PCR conditions previously described (11, 16, 23). The expected PCR product had sizes of 436 and 579 bp for 18S and 26S, respectively, while it ranged from 150 to 420 bp for ITS. Amplicons were purified with Agencourt AMPure XP beads (Beckman Coulter, Milan, IT), quantified using a Plate Reader AF2200 (Eppendorf, Milan, IT) and the Quant-iT PicoGreen double-stranded DNA assay kit (Life Technologies, Monza, IT). Amplicons of single strains were then pooled at equimolar concentration (in the case of Mock-Ampl samples). Four (Mock-DNA) or two (Mock-Ampl) independent replicates were sequenced for each target gene (Fig. 1). Sequencing was carried out on a 454 GS Junior platform using Titanium chemistry (454 Life Sciences, Roche Diagnostics, Milan, IT).
Environmental samples.
Biological samples used in this study included soil (14), human saliva (30), human feces (31), and grape must (16). Sample collection and DNA extraction were carried out as described in the original studies. Extracted DNA was amplified and sequenced as described above.
Data analysis.
Raw reads were first filtered according to the 454-processing pipeline. Sequences were then analyzed and further filtered by using QIIME 1.9.0 software (32). Reads were excluded from the analysis if they had an average quality score lower than 25, if they were shorter than 300 bp, and if there were ambiguous base calls. Sequences that passed the quality filter were denoised (33), and singletons were excluded. OTUs defined by a 97% similarity were picked using the uclust method, and the representative sequences were submitted to the RDPII classifier (34) to obtain the taxonomy assignment using the Silva SSU/LSU rRNA gene database release 128 (27) or the user-friendly Nordic ITS ectomycorrhiza database (28). Sequences were double-checked using the BLASTN search tool (http://www.ncbi.nlm.nih.gov/blast/) to confirm the taxonomy assignment. The OTU tables obtained were rarefied at the lowest number of sequences/sample.
Statistical analyses and plotting were carried out in R environment (http://www.r-project.org). Principal component analysis (PCA) was carried out on the log transformed abundance table by using dudi.pca function in made4 package. Spearman's correlation was computed between OTU abundance matrices (cor.test function). Alpha-diversity analysis was carried out in QIIME on OTU tables rarefied at 3,200 reads/sample. Pairwise Wilcoxon tests were used in order to determine significant differences in alpha diversity parameters or in OTU abundance. Permutational multivariate analysis of variance (nonparametric PERMANOVA) based on Jaccard and Bray Curtis distance matrices was carried out by using 999 permutations to detect significant differences in the overall fungal community composition, using the adonis function in the vegan package. Results were considered significant at a P value of <0.05.
Accession number(s).
The gene sequences produced in this study are available at the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI) under accession number SRP104109.
Supplementary Material
ACKNOWLEDGMENTS
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
We thank Sheridan L. Woo, Department of Pharmacy, University of Naples Federico II, Italy, for providing some of the fungus strains and Carmen Cretella for skillful technical assistance.
We declare no competing financial interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00905-17.
REFERENCES
- 1.De Filippis F, Parente E, Ercolini D. 2017. Metagenomics insights into food fermentations. Microb Biotechnol 10:1–9. doi: 10.1111/1751-7915.12421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lange L. 2014. The importance of fungi and mycology for addressing major global challenges. IMA Fungus 5:463–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Selin C, de Kievit TR, Belmonte MF, Fernando WG. 2016. Elucidating the role of effectors in plant-fungal interactions: progress and challenges. Front Microbiol 7:600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowers RM, Clum A, Tice H, Lim J, Singh K, Ciobanu D, Ngan CY, Cheng JF, Tringe SG, Woyke T. 2015. Impact of library preparation protocols and template quantity on the metagenomic reconstruction of a mock microbial community. BMC Genomics 16:856. doi: 10.1186/s12864-015-2063-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fouhy F, Clooney AG, Stanton C, Claesson MJ, Cotter PD. 2016. 16S rRNA gene sequencing of mock microbial populations-impact of DNA extraction method, primer choice and sequencing platform. BMC Microbiol 16:123. doi: 10.1186/s12866-016-0738-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.May A, Abeln S, Crielaard W, Heringa J, Brandt BW. 2014. Unraveling the outcome of 16S rDNA-based taxonomy analysis through mock data and simulations. Bioinformatics 30:1530–1538. doi: 10.1093/bioinformatics/btu085. [DOI] [PubMed] [Google Scholar]
- 7.Rideout JR, He Y, Navas-Molina JA, Walters WA, Ursell LK, Gibbons SM, Chase J, McDonald D, Gonzalez A, Robbins-Pianka A, Clemente JC, Gilbert JA, Huse SM, Zhou HW, Knight R, Caporaso JG. 2014. Subsampled open-reference clustering creates consistent, comprehensive OTU definitions and scales to billions of sequences. PeerJ 2:e545. doi: 10.7717/peerj.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Fungal Barcoding Consortium . 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc Natl Acad Sci U S A 109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esteve-Zarzoso B, Belloch C, Uruburu F, Querol A. 1999. Identification of yeasts by RFLP analysis of the 5.8S rRNA gene and the two ribosomal internal transcribed spacers. Int J Syst Bacteriol 49:329–337. doi: 10.1099/00207713-49-1-329. [DOI] [PubMed] [Google Scholar]
- 10.Ercolini D. 2013. High-throughput sequencing and metagenomics: moving forward in the culture-independent analysis of food microbial ecology. Appl Environ Microbiol 79:3148–3155. doi: 10.1128/AEM.00256-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stellato G, De Filippis F, La Storia A, Ercolini D. 2015. Coexistence of lactic acid bacteria and potential spoilage microbiota in a dairy processing environment. Appl Environ Microbiol 81:7893–7904. doi: 10.1128/AEM.02294-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garofalo C, Osimani A, Milanovic V, Aquilanti L, De Filippis F, Stellato G, Di Mauro S, Turchetti B, Buzzini P, Ercolini D, Clementi F. 2015. Bacteria and yeast microbiota in milk kefir grains from different Italian regions. Food Microbiol 49:123–133. doi: 10.1016/j.fm.2015.01.017. [DOI] [PubMed] [Google Scholar]
- 13.Wang C, García-Fernández D, Mas A, Esteve-Zarzoso B. 2015. Fungal diversity in grape must and wine fermentation assessed by massive sequencing, quantitative PCR and DGGE. Front Microbiol 6:1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonanomi G, De Filippis F, Cesarano G, La Storia A, Ercolini D, Scala F. 2016. Organic farming induces changes in soil microbiota that affect agro-ecosystem functions. Soil Biol Biochem 103:327–336. doi: 10.1016/j.soilbio.2016.09.005. [DOI] [Google Scholar]
- 15.David V, Terrat S, Herzine K, Claisse O, Rousseaux S, Tourdot-Maréchal R, Masneuf-Pomarede I, Ranjard L, Alexandre H. 2014. High-throughput sequencing of amplicons for monitoring yeast biodiversity in must and during alcoholic fermentation. J Ind Microbiol Biotechnol 41:811–821. doi: 10.1007/s10295-014-1427-2. [DOI] [PubMed] [Google Scholar]
- 16.De Filippis F, La Storia A, Blaiotta G. 2017. Monitoring the mycobiota during Greco di Tufo and Aglianico wine fermentation by 18S rRNA gene sequencing. Food Microbiol 63:117–122. doi: 10.1016/j.fm.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 17.Stielow JB, Levesque CA, Seifert KA, Meyer W, Irinyi L, Smits D, Renfurm R, Verkley GJ, Groenewald M, Chaduli D, Lomascolo A, Welti S, Lesage-Meessen L, Favel A, Al-Hatmi AM, Damm U, Yilmaz N, Houbraken J, Lombard L, Quaedvlieg W, Binder M, Vaas LA, Vu D, Yurkov A, Begerow D, Roehl O, Guerreiro M, Fonseca A, Samerpitak K, van Diepeningen AD, Dolatabadi S, Moreno LF, Casaregola S, Mallet S, Jacques N, Roscini L, Egidi E, Bizet C, Garcia-Hermoso D, Martín MP, Deng S, Groenewald JZ, Boekhout T, de Beer ZW, Barnes I, Duong TA, Wingfield MJ, de Hoog GS, Crous PW, Lewis CT, Hambleton S, Moussa TA, Al-Zahrani HS, Almaghrabi OA, Louis-Seize G, Assabgui R, McCormick W, Omer G, Dukik K, Cardinali G, Eberhardt U, de Vries M, Robert V. 2015. One fungus, which genes? Development and assessment of universal primers for potential secondary fungal DNA barcodes. Persoonia 35:242–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bokulich NA, Mills DA. 2013. Improved selection of internal transcribed spacer-specific primers enables quantitative, ultrahigh-throughput profiling of fungal communities. Appl Environ Microbiol 79:2519–2526. doi: 10.1128/AEM.03870-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindahl BD, Nilsson RH, Tedersoo L, Abarenkov K, Carlsen T, Kjøller R, Kõljalg U, Pennanen T, Rosendahl S, Stenlid J, Kauserud H. 2013. Fungal community analysis by high-throughput sequencing of amplified markers—a user's guide. New Phytol 199:288–299. doi: 10.1111/nph.12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bokulich NA, Thorngate JH, Richardson PM, Mills DA. 2014. Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc Natl Acad Sci U S A 111:E139–E148. doi: 10.1073/pnas.1317377110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bokulich NA, Joseph CM, Allen G, Benson AK, Mills DA. 2012. Next-generation sequencing reveals significant bacterial diversity of botrytized wine. PLoS One 7:e36357. doi: 10.1371/journal.pone.0036357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bellemain E, Carlsen T, Brochmann C, Coissac E, Taberlet P, Kauserud H. 2010. ITS as an environmental DNA barcode for fungi: an in silico approach reveals potential PCR biases. BMC Microbiol 10:189. doi: 10.1186/1471-2180-10-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.White TJ, Bruns T, Lee S, Taylor JW. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 315–322. In Innis MA, Gelfand DH, Sninsky JJ, White TJ (ed), PCR protocols: a guide to methods and applications. Academic, San Diego, CA. [Google Scholar]
- 24.Taylor DL, Walters WA, Lennon NJ, Bochicchio J, Krohn A, Caporaso JG, Pennanen T. 2016. Accurate estimation of fungal diversity and abundance through improved lineage-specific primers optimized for Illumina amplicon sequencing. Appl Environ Microbiol 82:7217–7226. doi: 10.1128/AEM.02576-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tonge DP, Pashley CH, Gant TW. 2014. Amplicon-based metagenomic analysis of mixed fungal samples using proton release amplicon sequencing. PLoS One 9:e93849. doi: 10.1371/journal.pone.0093849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walters WA, Caporaso JG, Lauber CL, Berg-Lyons D, Fierer N, Knight R. 2011. PrimerProspector: de novo design and taxonomic analysis of barcoded polymerase chain reaction primers. Bioinformatics 27:1159–1161. doi: 10.1093/bioinformatics/btr087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AF, Bahram M, Bates ST, Bruns TD, Bengtsson-Palme J, Callaghan TM, Douglas B, Drenkhan T, Eberhardt U, Dueñas M, Grebenc T, Griffith GW, Hartmann M, Kirk PM, Kohout P, Larsson E, Lindahl BD, Lücking R, Martín MP, Matheny PB, Nguyen NH, Niskanen T, Oja J, Peay KG, Peintner U, Peterson M, Põldmaa K, Saag L, Saar I, Schüßler A, Scott JA, Senés C, Smith ME, Suija A, Taylor DL, Telleria MT, Weiss M, Larsson KH. 2013. Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol 22:5271–5277. doi: 10.1111/mec.12481. [DOI] [PubMed] [Google Scholar]
- 29.Kurtzman CP, Robnett CJ. 1998. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 73:331–371. doi: 10.1023/A:1001761008817. [DOI] [PubMed] [Google Scholar]
- 30.De Filippis F, Vannini L, La Storia A, Laghi L, Piombino P, Stellato G, Serrazanetti DI, Gozzi G, Turroni S, Ferrocino I, Lazzi C, Di Cagno R, Gobbetti M, Ercolini D. 2014. The same microbiota and a potentially discriminant metabolome in the saliva of omnivore, ovo-lacto-vegetarian and vegan individuals. PLoS One 9:e112373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vitaglione P, Mennella I, Ferracane R, Rivellese AA, Giacco R, Ercolini D, Gibbons SM, La Storia A, Gilbert JA, Jonnalagadda S, Thielecke F, Gallo MA, Scalfi L, Fogliano V. 2015. Whole-grain wheat consumption reduces inflammation in a randomized controlled trial on overweight and obese subjects with unhealthy dietary and lifestyle behaviors: role of polyphenols bound to cereal dietary fiber. Am J Clin Nutr 101:251–261, 2015. doi: 10.3945/ajcn.114.088120. [DOI] [PubMed] [Google Scholar]
- 32.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reeder J, Knight R. 2010. Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat Methods 7:668–669. doi: 10.1038/nmeth0910-668b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.