ABSTRACT
Streptococcus mutans is known to possess rhamnose-glucose polysaccharide (RGP), a major cell wall antigen. S. mutans strains deficient in rgpG, encoding the first enzyme of the RGP biosynthesis pathway, were constructed by allelic exchange. The rgpG deficiency had no effect on growth rate but caused major defects in cell division and altered cell morphology. Unlike the coccoid wild type, the rgpG mutant existed primarily in chains of swollen, “squarish” dividing cells. Deficiency of rgpG also causes significant reduction in biofilm formation (P < 0.01). Double and triple mutants with deficiency in brpA and/or psr, genes coding for the LytR-CpsA-Psr family proteins BrpA and Psr, which were previously shown to play important roles in cell envelope biogenesis, were constructed using the rgpG mutant. There were no major differences in growth rates between the wild-type strain and the rgpG brpA and rgpG psr double mutants, but the growth rate of the rgpG brpA psr triple mutant was reduced drastically (P < 0.001). Under transmission electron microscopy, both double mutants resembled the rgpG mutant, while the triple mutant existed as giant cells with multiple asymmetric septa. When analyzed by immunoblotting, the rgpG mutant displayed major reductions in cell wall antigens compared to the wild type, while little or no signal was detected with the double and triple mutants and the brpA and psr single mutants. These results suggest that RgpG in S. mutans plays a critical role in cell division and biofilm formation and that BrpA and Psr may be responsible for attachment of cell wall antigens to the cell envelope.
IMPORTANCE Streptococcus mutans, a major etiological agent of human dental caries, produces rhamnose-glucose polysaccharide (RGP) as the major cell wall antigen. This study provides direct evidence that deficiency of RgpG, the first enzyme of the RGP biosynthesis pathway, caused major defects in cell division and morphology and reduced biofilm formation by S. mutans, indicative of a significant role of RGP in cell division and biofilm formation in S. mutans. These results are novel not only in S. mutans, but also other streptococci that produce RGP. This study also shows that the LytR-CpsA-Psr family proteins BrpA and Psr in S. mutans are involved in attachment of RGP and probably other cell wall glycopolymers to the peptidoglycan. In addition, the results also suggest that BrpA and Psr may play a direct role in cell division and biofilm formation in S. mutans. This study reveals new potential targets to develop anticaries therapeutics.
KEYWORDS: BrpA, LCP proteins, Psr, Streptococcus mutans, TEM analysis, biofilm formation, cell division, cell wall antigens, dental caries, rhamnose-glucose polysaccharides
INTRODUCTION
Streptococcus mutans, a key etiological agent of human dental caries, lives almost exclusively on the tooth surface in high-density, high-diversity plaque biofilms. In the oral cavity, S. mutans and other bacteria encounter frequent and sometimes drastic and detrimental insults, including feast and famine in nutrients, toxic metabolites such as lactic acid and other weak organic acids, reactive oxygen species, and antimicrobial agents of bacterial origin and from oral care products. S. mutans is well known to have evolved multiple mechanisms to colonize the tooth surface, survive various environmental insults, persist in the plaque community, and under certain conditions, become numerically significant, thus causing carious lesions on the tooth surface (1–3).
The cell envelope is essential in maintenance of bacterial cell shape, cell growth, and cell division and plays a fundamental role in protection against various environmental insults. In addition, the cell envelope is directly involved in environmental signaling and bacterial cell-surface and cell-cell interactions and, thus, bacterial adherence and biofilm formation. The cell envelope of Gram-positive bacteria is characterized by the presence of a thick layer of peptidoglycan and peptidoglycan-attached anionic cell wall polymers, including capsular polysaccharides and (lipo)teichoic acids (4, 5). S. mutans is not known to have copious capsules, but does produce rhamnose-glucose polysaccharide (RGP) and lipoteichoic acid (6–8), although the role of RGP in this and other streptococci remains unclear (9). Our recent studies have shown that BrpA and Psr, two members of the LytR-CpsA-Psr (LCP) family of proteins (10–13), in S. mutans play critical roles in cell envelope homeostasis, stress tolerance response, and biofilm formation and regulation of genes known to be involved in these processes (14–16). BrpA deficiency leads to major defects in acid and oxidative stress tolerance responses, defects in cell division and alterations in cell envelope morphology, and reduction in biofilm formation. BrpA deficiency also alters the expression of a number of genes, including those known to play a critical role in cell envelope biogenesis and cell division and biofilm formation (8, 16, 17). Paralogue Psr also plays a major role in acid tolerance and biofilm formation, but unlike BrpA, has no major effect on oxidative and cell envelope stress and cell morphology under the conditions studied (17). Psr also influences gene expression, although differences exist between these two LCP proteins in the scope and effect of their gene regulation (16, 17). Like Bacillus subtilis but different from Staphylococcus aureus, a functional BrpA or Psr appears to be required for viability in S. mutans (13, 18).
In S. mutans, RgpG is reported as the first enzyme of the RGP biosynthesis pathway (19). When cloned in Escherichia coli strains deficient of the enzyme for rhamnose synthesis, the rgpG gene was able to rescue the ability of the E. coli mutants to produce rhamnans on the surface (19, 20). Our recent studies have shown that RgpG production is part of the BrpA-mediated regulation of cell envelope biogenesis (16). In this study, an rgpG mutant was constructed and characterized for the impact of the RgpG deficiency on cell morphology, stress tolerance response, and biofilm formation. Our results consistently showed that RgpG deficiency alone had little effect on growth and stress tolerance response but significantly reduced biofilm formation, regardless of carbohydrate source. Deficiency of RgpG also caused major defects in cell division, leading to drastic alterations in cell morphology. Further characterization of the rgpG, brpA, and/or psr mutants also suggests that brpA and psr in S. mutans are required for attachment of surface antigens to the cell wall.
RESULTS
RgpG deficiency reduces growth rates when grown in semidefined medium.
Consistent with previous findings (16), no major differences in growth rate were observed between the rgpG mutant, TW322, and the wild-type strain, UA159, during growth in regular brain heart infusion (BHI) broth (Fig. 1A), although compared to UA159, TW322 did show a higher tendency to form aggregates in the bottom of the test tube. When grown in semidefined biofilm medium (BM) with glucose (BMG), TW322 displayed an extended lag phase, although no significant differences in growth rate were measured between the mutant and its parent strain, UA159 (P > 0.05). Similar results were also seen when strains were grown in BM containing sucrose (Fig. 1A). When grown in BHI broth adjusted to pH 6.0, however, no major differences were observed in growth rates between UA159 and the rgpG mutants, TW393 and TW322 (Fig. 1B). In addition, no major differences were measured between UA159 and TW322 when grown in regular BHI broth in the presence of methyl viologen, a reagent commonly used to induce oxidative stress (data not shown). When subjected to acid killing or hydrogen peroxide killing assays, no significant differences in survival rates were observed between UA159 and TW322 (data not shown).
FIG 1.
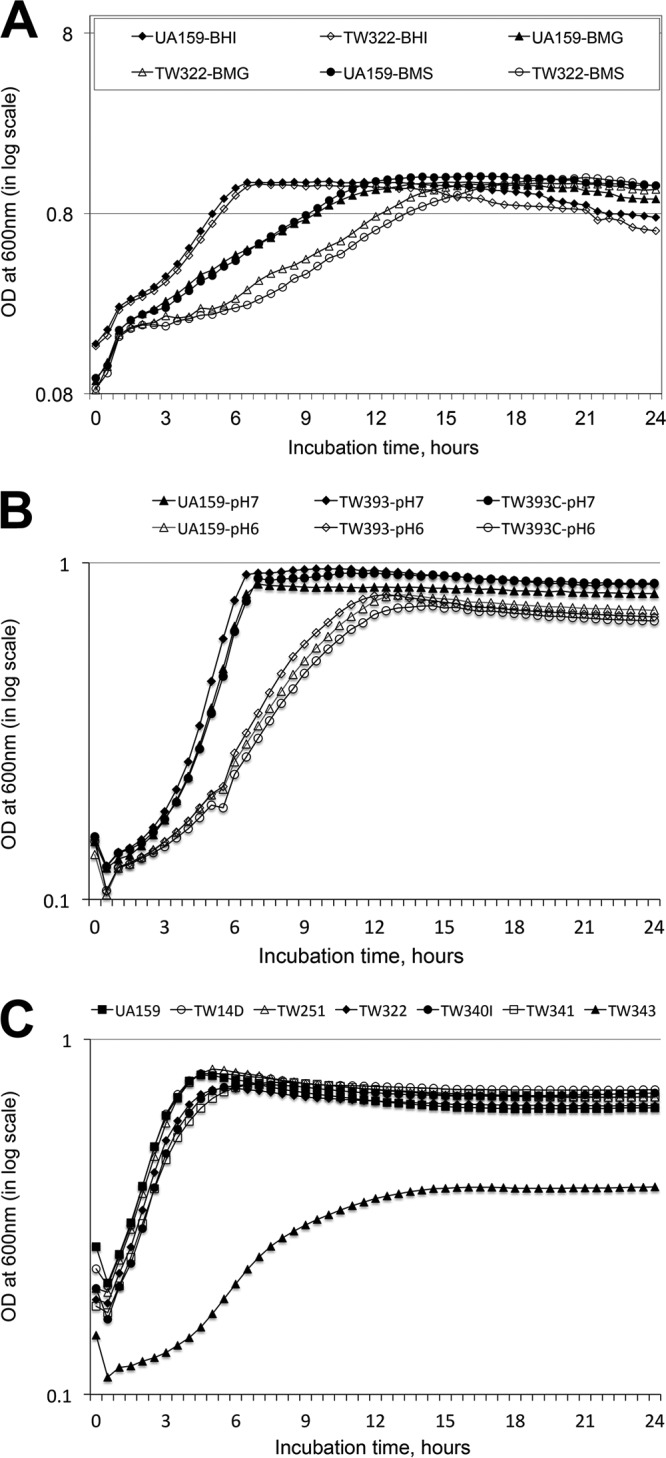
Growth study. The S. mutans wild-type strain (UA159), its rgpG (TW322 and TW393), brpA (TW14D), psr (TW251), rgpG brpA (TW340), rgpG psr (TW341), and rgpG brpA psr (TW343) mutants, and the rgpG complement strain (TW393C) were grown in BHI (A and C), semidefined biofilm medium (A) with glucose (BMG) and sucrose (BMS) as the carbohydrate sources, and BHI adjusted to pH 7.0 and 6.0 (B). The optical density at 600 nm (OD600) of the cultures was monitored continuously using Bioscreen C with a sterile mineral oil overlay. The data presented here are representative of more than three separate experiments.
In an effort to evaluate the role of BrpA and Psr in RgpG-mediated biosynthesis of secondary cell wall polymers, brpA rgpG and psr rgpG double mutants were constructed by allelic exchange. Previously, multiple attempts to make brpA psr double mutants failed (17). In this study, a triple mutant deficient in both brpA and psr was also constructed using the rgpG mutant. Like UA159 and TW322 (Fig. 1A and B) and the brpA and psr single mutants, TW14D and TW251, respectively (14, 15, 17), both the brpA rgpG (TW340) and psr rgpG (TW341) double mutants showed some moderate reductions in growth rates when grown in regular BHI broth, although such reductions were not statistically significant (P > 0.05) (Fig. 1C). In contrast, the brpA psr rgpG triple mutant, TW343, displayed a dramatic reduction in growth rate (with a doubling time of 157 ± 17 min for TW343 versus 93 ± 5.7 min for UA159) (P < 0.001) (Fig. 1C) and the culture density after 24 h (just under 0.4 for TW343 versus 0.79 for UA159) (P < 0.001). When grown in BM with glucose and/or sucrose, TW343 yielded little or no growth overnight (data not shown).
RgpG deficiency causes major defects in cell division, and such defects are exacerbated by BrpA and Psr deficiency.
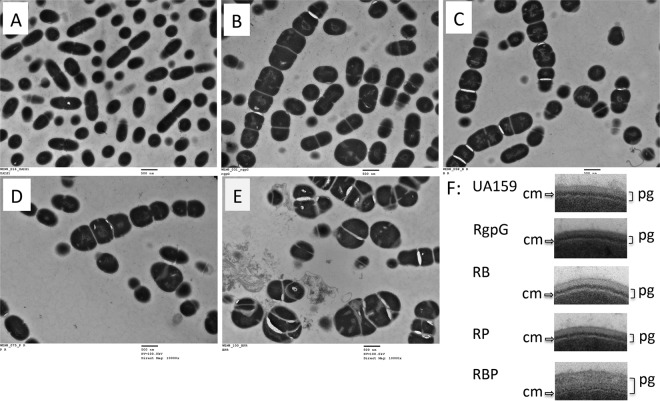
When analyzed under transmission electron microscopy (TEM), UA159 generally appeared in a regular coccoid shape and displayed short chains of dividing cells with septa placed at the end. UA159 cells appeared to be slightly elongated during septum formation and cell division. In contrast, TW322 did not seem to elongate prior to cell division but rather became distended and swollen, the cells being larger and “squarish.” Most of the TW322 cells failed to separate completely and displayed aberrant septal placement (Fig. 2). Both TW340 and TW341 existed primarily in long chains of enlarged squarish cells resembling the rgpG single mutant, TW322 (Fig. 2). However, unlike UA159, TW322, TW340, and TW341, the triple mutant, TW343, existed predominantly in giant bovine kidney-like cells with multiple asymmetric septa, illustrative of severe defects in cell division (Fig. 2). In addition, TW343 also displayed a significantly thicker peptidoglycan (PG) layer and a thinner/fainter cytoplasmic membrane (Fig. 2F), both of which were different from those of UA159, TW340, and TW341.
FIG 2.
TEM analysis. The S. mutans wild-type strain (UA159) (A) and its rgpG (TW322) (B), rgpG brpA (TW340) (C), rgpG psr (TW341) (D), and rgpG brpA psr (TW343) (E) mutants were grown in BHI until the mid-exponential phase (OD600 of ≅0.4). The images presented here were taken at a magnification of 10,000×, and scale bars are 500 nm. Panel F shows inserts of the magnified cell envelope of the different strains with the triple mutant TW343 showing a significantly thicker peptidoglycan (pg) layer and a thinner/fainter cytoplasmic membrane (cm).
RgpG deficiency reduces biofilm formation.
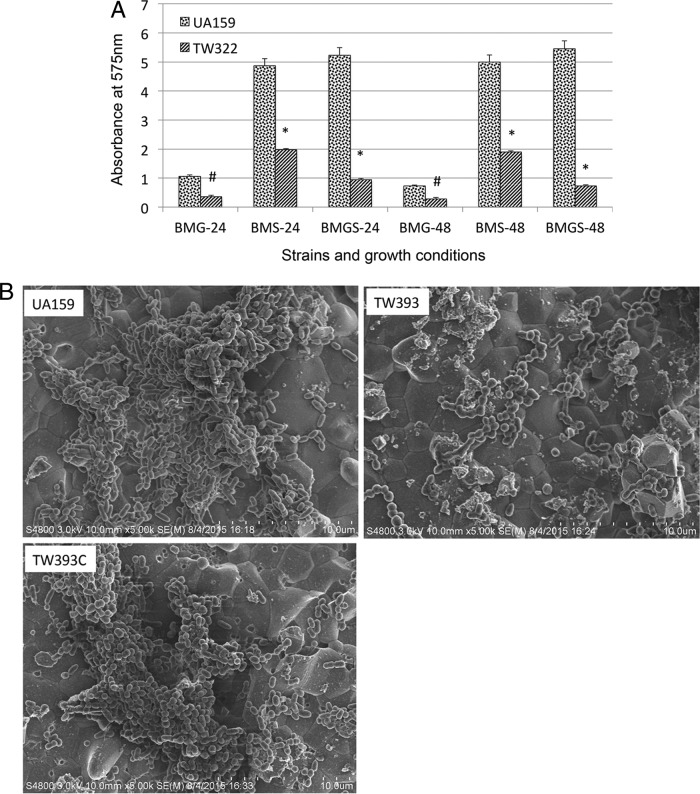
Compared to UA159, biofilm formation in 96-well plates by TW322 was reduced by >60% when grown in semidefined biofilm medium with glucose as the supplemental carbohydrate source (BMG; P < 0.01) (Fig. 3A). Similar observations were also made with cultures grown in BM containing sucrose (BMS) and BM containing glucose and sucrose (BMGS) (P < 0.001, respectively). When analyzed using scanning electron microscopy (SEM), UA159 accumulated a substantial amount of biofilms after 24 h during growth on hydroxylapatite (HA) discs, especially when sucrose was present in the growth medium (Fig. 3B). TW322 and TW393 appeared to be able to bind and colonize the surface, but only limited accumulation was observed under the same conditions. The complement strain of the deficient mutant, TW393C, which carries a coding sequence of rgpG plus its cognate promoter region in a multicopy shuttle vector, developed biofilms similar to that of the wild-type strain, UA159 (Fig. 3B).
FIG 3.
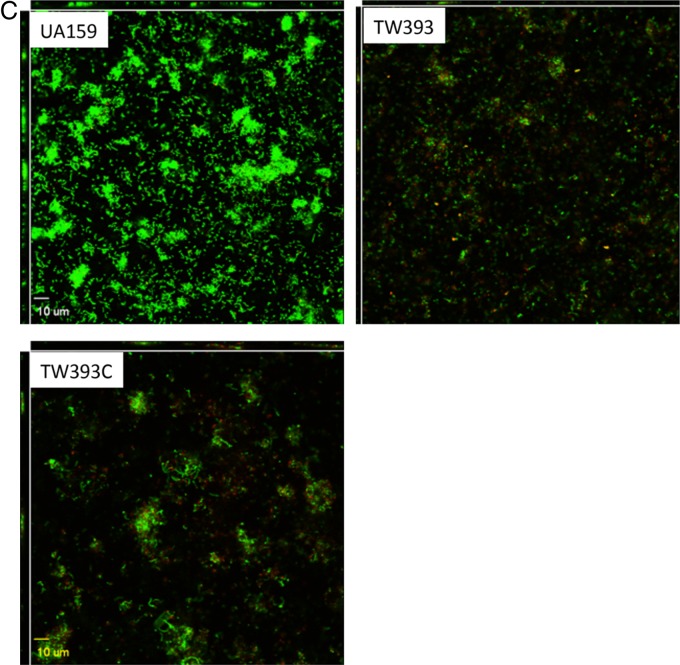
Biofilm formation. The S. mutans wild-type strain (UA159), its rgpG mutants (TW322 for the 96-well plate model and TW393 for HA discs), and the rgpG complement strain (TW393C) were grown in BM supplemented with glucose (BMG), sucrose (BMS), and glucose plus sucrose (BMGS). Biofilms were grown in 96-well plates (A) and HA discs vertically placed in 12-well plates (B and C) for 24 and 48 h. By the end of the experiments, biofilms on 96-well plates were stained with crystal violet and analyzed using a spectrophotometer (A); biofilms on HA discs were fixed overnight and processed for SEM analysis (B), or stained using the LIVE/DEAD fluorescent staining kit and examined by confocal microscopy (C). The results presented in panel A are averages (±standard deviation [SD] shown by error bars) of biofilms after 24 and 48 h from more than three independent experiments. * and # indicate statistically significant difference at P < 0.001 and P < 0.05, respectively, compared to the wild type. Panel B shows representative SEM images (at 5,000×) of 24-h biofilms grown in BM plus glucose and sucrose. Panel C shows representative compressed xyz, xz, and yz confocal microscopic images (512 by 512) of biofilms grown in BM with glucose and sucrose.
To further evaluate the impact of rgpG deficiency on biofilm structure and accumulation, 24-h biofilms grown on the HA discs were stained using LIVE/DEAD fluorescent dye (Invitrogen) and then optically dissected using a laser scanning confocal microscope (Fig. 3C). Following postacquisition analysis using COMSTAT, UA159 biofilms were shown to have an average thickness of 4.2 ± 1.5 μm with a biovolume of 2.68 ± 0.9 μm3/μm2. In contrast, TW393 biofilms were significantly thinner and had significantly less biovolume, with an average thickness of just 0.83 ± 0.19 μm (P = 0.016) and a biovolume of 0.69 ± 0.19 μm3/μm2 (P = 0.020), respectively. Biofilm formation by TW393C was partially restored, with a thickness of 1.62 ± 1.75 μm (P = 0.119) and biovolume of 1.06 ± 1.02 μm3/μm2 (P = 0.11), respectively.
When grown in BM with glucose and sucrose using the 96-well plate model and crystal violet staining, both TW340 and TW341 accumulated much less biofilm than UA159 (P < 0.001) and TW322 (P < 0.05) (Fig. 4), but not significantly different from those of the brpA and psr single mutants, TW14D and TW251, respectively. Unlike UA159 and all other single and double mutants, however, the triple mutant TW343 accumulated little or no biofilm after 24 h (P < 0.001) (Fig. 4).
FIG 4.
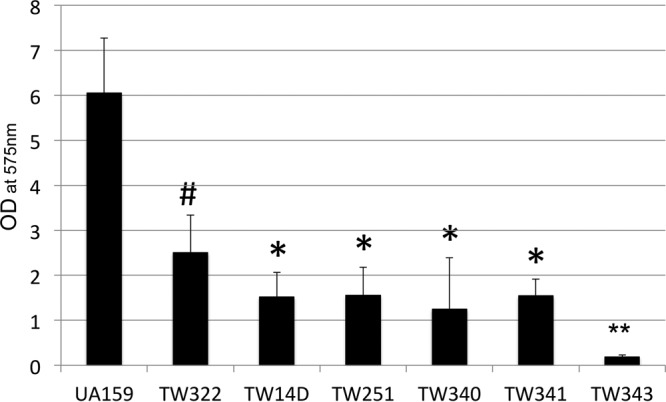
Biofilm formation. The S. mutans wild-type strain (UA159) and its rgpG (TW322), brpA (TW14D), psr (TW251), rpgG brpA (TW340), rgpG psr (TW341), and rgpG brpA psr (TW343) mutants were grown in BM containing glucose, sucrose, and glucose plus sucrose. Biofilms were grown in 96-well plates and measured using a spectrophotometer following crystal violet staining. ANOVA and Tukey's pairwise comparison were used to analyze the differences between different strains. The data presented here represent the average (±SD shown by error bars) from more than three separate sets of experiments with cultures grown in BM plus glucose and sucrose. #, P < 0.001 versus UA159 and TW343 and <0.05 versus TW14D, TW251, TW340, and TW341; *, P < 0.001 versus UA159 and TW343 and <0.05 versus TW322; **, P < 0.001 versus all others.
RgpG deficiency caused reduction of cell wall antigens, while deficiency of BrpA resulted in accumulation of cell wall antigens in the cell-free culture medium.
RGP is a major component of the cell wall antigens in streptococci. To assess the role of RgpG and the LCP proteins BrpA and Psr in cell envelope biogenesis, cell wall-associated antigens were prepared from murein sacculi of the rgpG, brpA, and/or psr single, double, and triple mutants, blotted onto a nitrocellulose membrane, and then analyzed using properly adsorbed whole-cell antiserum as a probe. When probed with S. mutans whole-cell antiserum that was adsorbed with the wild-type whole cells, little or no signal was detected with any strains analyzed (data not shown). In contrast, when probed with the antiserum adsorbed with the triple mutant whole cells, UA159 displayed the strongest signal, followed by TW322, but little or no signal was detected with TW340, TW341, TW343, TW14D, and TW251, while TW322C displayed a signal similar to UA159 (Fig. 5a). Similar results were also observed when probed with antiserum adsorbed with whole cells of the rgpG mutant (data not shown). When the preparations of the cell-free culture medium were analyzed, little or no signal was detected when probed with the antiserum adsorbed with the wild-type whole cells (data not shown). When probed with the antiserum adsorbed with the triple mutant, little or no signal was detected with UA159 and TW322C (Fig. 5b). On the other hand, strong signals were detected with TW14D, TW341, TW340, and TW343, but only limited signal with TW322 and TW251.
FIG 5.
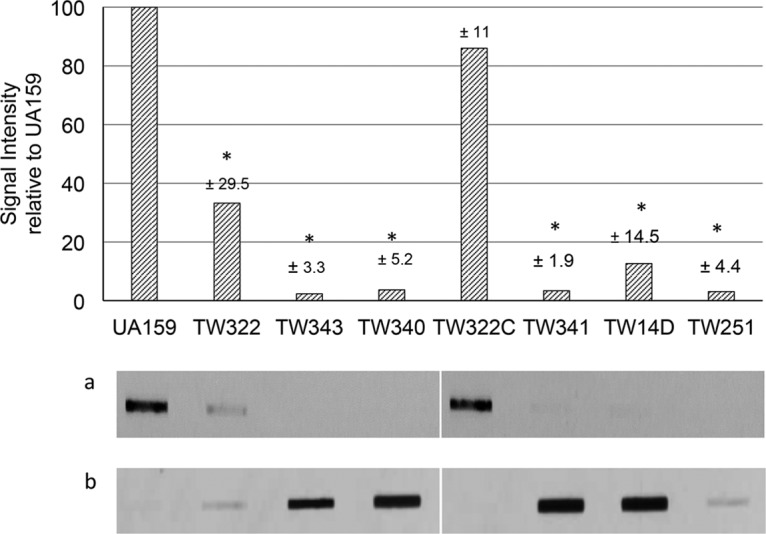
Immunoblot analysis. Cell wall antigens were isolated from murein sacculi of the S. mutans wild-type strain (UA159), its rgpG (TW322), rpgG brpA (TW340), rgpG psr (TW341), and rgpG brpA psr (TW343) mutants, and the rgpG complement strain (TW322C) and were probed with whole-cell antiserum adsorbed with UA159 or TW343. The bar graph shows results when probed with antiserum adsorbed with the triple mutant, TW343, and expressed as mean of the percentage of signal intensities relative to UA159 (±SD represented in numbers above the individual bars) from two separate experiments. *, significant difference at P < 0.001 compared to the wild type. Images show the slot blots of cell wall antigens (a) and cell-free culture medium preparations (b) probed with the TW343 adsorbed antiserum. Samples were loaded as indicated in the bar graph above. The two images presented in panels a and b were taken from the same immunoblots with samples loaded at different rows.
DISCUSSION
This study has shown that deficiency of RgpG in S. mutans resulted in significant reduction of cell wall antigens. Relative to the wild-type strain UA159, the RgpG-deficient mutants also displayed major defects in cell division and alterations in cell morphology and attenuated biofilm formation regardless of the carbohydrate sources. The triple mutant deficient in RgpG and the LCP proteins BrpA and Psr displayed exacerbated defects in cell division, morphology, and biofilm formation. Like B. subtilis and Streptococcus pneumoniae, deficiency of the LCP protein BrpA and/or Psr led to major reduction of cell wall-associated antigens and their concurrent extracellular accumulations in the spent culture medium.
S. mutans is well known to produce RGP. The genes involved in the biosynthesis of the RGP polymers have been well characterized (19, 20). Of the RGP synthetic pathway, RgpG is an E. coli WecA homologue that is believed to function in initiation of the synthesis of the rhamnose-glucose polysaccharides of the cell wall and was shown to be able to complement a WecA-deficient E. coli mutant, restoring rhamnan production by the deficient mutant (19, 20). It is therefore consistent with the observation that RgpG deficiency in S. mutans results in major reduction of cell wall-associated antigens.
In S. mutans as well as many other streptococci, the rhamnose-containing cell wall polysaccharides are the major anionic cell wall polymer (9, 19). However, other than being commonly used as surface antigens in serotyping and classification, little is known about the role of RGP and other rhamnose-containing cell wall polymers in pathophysiology of these streptococci. As shown by the results of the allelic exchange mutagenesis of rgpG, RGP in S. mutans is not essential but plays a critical role in cell division and cell morphology, as featured by the long chains of swollen, squarish cells by the deficient mutants. Defects in cell division will certainly have an impact on biofilm formation, although it is noteworthy that the rgpG mutant does not show major differences in growth rate compared to the parent strain during growth in rich medium like BHI. However, when grown in semidefined BM, the rgpG-deficient mutant displayed an extended lag phase, but again did not show any significant differences in growth rate compared to the parent strain, UA159. Determination of what factors contributed to the extended lag phase when grown in the BM awaits further investigation.
It is also worth noting that by BLAST search, RgpG was identified as a B. subtilis TagO homologue (41% identity and 62% similarity), which is the enzyme responsible for catalyzing the first step in the synthesis of all anionic cell wall polymers, including cell wall teichoic acid and the secondary cell wall polysaccharides (21–24). Like S. mutans, TagO deficiency in Bacillus also causes severe compromises in cell growth and leads to loss of the ability to elongate, forming round and swollen cells (25, 26). However, unlike B. subtilis (27, 28), S. mutans is not known to possess cell wall teichoic acid (9, 29), and key members of the Bacillus teichoic acid biosynthesis pathway cannot be identified in this oral pathogen. Whether S. mutans produces cell wall teichoic acid and what role RgpG plays in the biosynthesis pathway await further investigation.
Biofilm formation is a sequential process that is initiated with bacterial cell-substratum interaction and bacterial adherence and is followed by cell-cell interactions and accumulation of multicellular clusters. The bacterial cell envelope is believed to play a crucial role in biofilm initiation and biofilm accumulation. Both cell aggregation and growth defects in extended lag phase when grown in semidefined biofilm medium of the RgpG-deficient mutant can contribute to the reduced ability to form biofilms. As a major component of the cell envelope, RGP deficiency as a result of the loss of the initiation enzyme will likely result in alterations in the composition as well as the configuration of the cell envelope, which in turn will affect cell surface-associated functions, including cell-cell interaction and biofilm development. Similar results have also been reported in Staphylococcus epidermidis, whose TagO deficiency was shown to cause major defects in biofilm development (30). Like S. mutans, the TagO-deficient S. epidermidis mutant also displayed a higher tendency of aggregation, especially during growth in the BHI with glucose. Different from S. mutans, however, TagO deficiency in S. epidermidis causes major defects in polystyrene surface attachment, and it was in part attributed to the increased cell surface hydrophobicity, which is a major contributing factor to bacterial cell surface adherence. However, no major differences in cell surface hydrophobicity were detected between the S. mutans wild-type strain and the RgpG-deficient mutants (data not shown). Also unlike S. epidermidis, the impact of RgpG deficiency on biofilm formation in S. mutans appears to be mostly on biofilm development rather than initial attachment: as shown by confocal microscopy, the RgpG-deficient mutant still binds to the HA surface similarly to the parent strain. Likely, the lack of RGP as a result of rgpG deficiency caused some major alterations in structure and function of the cell envelope, which is crucial in intercellular interactions and biofilm development.
B. subtilis possesses three LCP proteins, but deficiency of all three corresponding genes (tagTUV) is lethal (13, 18) unless TagO or TagA is also inactivated, which blocks the earliest steps of the teichoic acid (TA) biosynthesis pathway preventing substrate sequestration of lipid II (13). S. mutans possesses only two LCP homologues, BrpA and Psr. Previously, repeated attempts failed to yield a brpA psr double mutant (17), indicating that one functional protein is required for viability, which is like B. subtilis but different from S. aureus (13, 18). In this study, we were able to generate a mutant with both brpA and psr inactivated, but it was achieved only in the rgpG mutant background, which is again similar to B. subtilis. These results further suggest that like LCP in B. subtilis, BrpA and/or Psr in S. mutans is required for downstream processes of the RgpG products.
The LCP proteins in B. subtilis and several others that have been studied are shown to function as ligases that attach the synthesized anionic cell wall polymers, such as teichoic acids and capsule polysaccharides to the peptidoglycan (11–13). In this study, we have provided further evidence that BrpA plays a similar role in S. mutans. Like B. subtilis and S. pneumoniae, deficiency of BrpA significantly reduced cell wall surface-associated antigens, which RGP is expected to be a major component of, and led to concurrent accumulation of cell wall antigens in the cell-free culture medium. However, the results presented here also suggest that BrpA is involved in attachment of additional cell wall antigens (other than RGP), which can be attributed to the strong signal of the rgpG brpA mutant, TW340. Similar results were also obtained with the Psr-deficient mutants when the murein sacculus preparations were analyzed. With the spent culture medium preparations, however, the rgpG psr double mutant displayed strong signal, while little or no signal was detected with the psr single mutant. Currently, what factor(s) contributed to the discrepancy remains unclear. What other surface antigens remain in the rgpG mutant also await further investigation.
The LCP proteins are highly conserved in Gram-positive bacteria, although differences in both structure and function exist between different species (11–13, 15, 17, 31–33). The essentiality of the LCP proteins in B. subtilis but not in S. aureus indicates differences in functionalities of these proteins between these two bacteria (13, 18). Different functions of LCP proteins have also been reported recently in Actinomyces oris, Streptococcus agalactiae, and by us in S. mutans (17, 31–33). In A. oris, an LCP-like protein acts as a glycosyltransferase on the cell surface transferring glycan macromolecules to the surface-associated protein GspA (33). In S. agalactiae, CpsA can bind to the promoters and modulate the expression of selected genes (32). Our recent studies have shown that like S. agalactiae (32), both BrpA and Psr in S. mutans play a role in regulation of gene expression, including those involved in cell envelope biogenesis, cell division, and biofilm formation, although differences can also be identified in both the scope of regulated genes and the effects of regulation between the two paralogues (17). Besides, differences have also been observed between the brpA mutant and the psr mutant in stress tolerance response, cell morphology (16), and cell wall-associated antigens, as described above.
Consistent with our previous studies (14, 15, 17), both the psr-deficient mutant (TW251) and especially the brpA-deficient mutant (TW14D) displayed major reductions in biofilm formation. Relatively, both brpA and psr deficiency showed a more severe impact on biofilm formation than seen in the rgpG mutants, which is consistent with the broad roles of BrpA and Psr in S. mutans physiology (16, 17). On the other hand, the effect of rgpG deficiency on cell division and morphology was a lot more significant than that of brpA and psr mutants (17). Likely, part of the defects observed with the double and triple mutants deficient in rgpG, brpA, and/or psr in growth, cell division, and biofilm formation can be attributed to the loss of cell wall polymers as a result of deficiency in biosynthesis and/or attachment. However, such effects cannot fully explain the differences between the rgpG brpA psr triple mutant, TW343, and the respective rgpG brpA and rgpG psr double mutants, TW340 and TW341. It is possible that for the double mutants, the deficiency of one LCP protein was at least partly compensated for by the other existing paralogue. This explains why the double mutants formed biofilms very similarly to the brpA and psr single mutants TW14D and TW251, respectively, while morphologically resembling the rgpG mutant. Deficiency of both BrpA and Psr, however, exacerbated the defects in cell division and biofilm formation, resulting in drastic differences in the triple mutant from the respective single and double mutants. Consistent with our previous findings (17), these results again further suggest that LCP proteins in S. mutans contribute directly to cell division and cell morphology. Similar results have also recently been reported with lcpB3 in Bacillus anthracis, which has six lcp genes (34, 35).
In summary, this study has provided evidence that RgpG in S. mutans plays an important role in cell division, cell morphology, and biofilm formation. This is novel concerning RGP in S. mutans and other streptococci. Like B. subtilis and other bacteria studied, LCP proteins BrpA and Psr in S. mutans also promote attachment of cell wall anionic polymers to the peptidoglycan, although consistent with our previous findings, differences do exist between these two paralogues and other members of the LCP proteins. In support of our previous findings, this study has also provided further evidence that BrpA and, probably, Psr play a direct role in S. mutans cell division, although the mechanisms that govern these processes await further investigation.
MATERIALS AND METHODS
Plasmids, bacterial strains, and growth conditions.
All bacterial strains and plasmids used in this study are listed in Table 1. Unless otherwise stated specifically, S. mutans UA159 and is derivatives were grown in brain heart infusion (BHI, Difco Laboratories) in a 37°C aerobic chamber containing 5% CO2. When needed, erythromycin (Erm [10 μg/ml]), kanamycin (Kan [1 mg/ml]), and/or spectinomycin (Spc [1 mg/ml]) was added to the growth medium. For agar medium, Bacto agar (Difco Laboratories) was added at a level of 1.5% (wt/vol). For growth characterization, overnight cultures were properly diluted in fresh medium and allowed to grow until the mid-exponential phase (optical density [OD] of ≅ 0.4 to 0.5), when they were further diluted by 1:100, and growth was continuously monitored using Bioscreen C (Oy Growth Curves AB, Ltd., Finland) at 37°C with and without a sterile mineral oil overlay (36, 37).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Major characteristic(s)a | Reference or source |
|---|---|---|
| Strains | ||
| S. mutans | ||
| UA159 | Wild type | 52 |
| TW322 | rgpG (ΔSMU.246) Spcr | 17 |
| TW393 | rgpG (ΔSMU.246) Kanr | This study |
| TW393C | TW393/pDL278:rgpG Kanr Spcr | This study |
| TW322C | TW322 gtfA::rgpG Kanr Spcr | This study |
| TW340 | rgpG brpA Spcr Ermr | This study |
| TW341 | rgpG psr Spcr Kanr | This study |
| TW343 | rgpG brpA psr Spcr Kanr Ermr | This study |
| E. coli DH10B | Cloning host | Invitrogen |
| Plasmids | ||
| pDL278 | Shuttle vector, Spcr | 42 |
| pBGK(3) | Integration vector | 43 |
Kanr, Spcr, and Ermr indicate kanamycin, spectinomycin, and erythromycin resistance, respectively.
DNA manipulation and construction of mutants.
To construct a Kanr RgpG-deficient mutant, a PCR-ligation-mutagenesis strategy was used as described previously (38–40). Briefly, the 5′ and 3′ regions flanking rgpG were amplified by PCR using high-fidelity DNA polymerase (New England BioLabs, Ipswich, MA) with gene-specific primers (Table 2). The flanking regions were digested using proper restriction enzymes and ligated to a nonpolar Kanr element (aphA encoding aminoglycoside 3′-phosphotransferase) (37) that was digested with the same enzymes, and the resulting ligation mixtures were then used to directly transform S. mutans UA159 in the presence of synthetic competence-stimulating peptide (CSP) (41). Allelic replacement mutant TW393 with rgpG deficiency was isolated on BHI agar plates with Kan and further confirmed by colony PCR with gene-specific primers 55 and 33 and by Sanger sequencing. For complementation of the rgpG mutant, the rgpG coding sequence plus its cognate promoter region was amplified by PCR using the rgpG 55 and 33 primers (Table 1), and following sequence verification by sequencing, it was directly cloned into shuttle vector pDL278 (42). The resulting construct, pDL278:rgpG, was then used to transform TW393, generating complement strain, TW393C. For single-copy complementation, rgpG and its cognate promoter region were PCR amplified and cloned into integration vector pBGK(3), a modified version of pBGK with both the tetracycline and ampicillin resistance elements deleted and the addition of an NheI site in the multiple cloning site (43). The resulting construct was then transformed into the Spcr RgpG-deficient mutant TW322 via double crossover recombination (17), resulting in generation of TW322C. For construction of the brpA rgpG and the rgpG psr double mutants, PCRs were performed using genomic DNA from TW14D, a BrpA-deficient mutant with its coding sequence replaced by an Ermr element (ermr) (15), and TW251, a Psr-deficient mutant with its coding sequence replaced by a nonpolar Kanr marker (kanr) (17), as the respective template with gene-specific 55 and 33 primers (15, 17). The resulting amplicon that contains ermr and the flanking regions of brpA (15) as well as the amplicon with kanr and the psr flanking regions was used to transform TW322 (16) separately. Double mutants deficient in rgpG and brpA (TW340) and rgpG and psr (TW341) were isolated from BHI-Erm-Spc agar and BHI-Kan-Spc agar, respectively. For the rgpG brpA psr triple mutant TW343, the rgpG brpA double mutant TW340 was transformed with the psr amplicon similarly, and the transformation mixture was plated on BHI-Erm-Spc-Kan.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′ to 3′)a | Application |
|---|---|---|
| rgpG5 | ||
| 55 | ACTACTCGTATGAGGTAAAGAC | 5′ fragment for mutation |
| 53 | TCCGTCTCGCATCTAGATTATCAAC | |
| rgpG3 | ||
| 35 | ACTTACATTCTAGATTGCTTGCTATG | 3′ fragment for mutation |
| 33 | ACCATCCATCATAACATGAAC | |
| rgpGS | ||
| P5 | ATGCTGGTAGAGGAACAAAGACAC | Confirmation of ΔrgpG |
| P3 | TGCTGAAAGCGTTGATTTCCCAGTC | |
| rgpGC | ||
| P5 | TAAATGAAACAAGCTAGCGAAACAAC | Single-copy complementation |
| P3 | TAAGCGAGCTCGCTCATCAACTTC |
Underlined portions of sequences are restriction sites engineered for cloning.
Biofilm analysis.
For biofilms, S. mutans strains were grown in modified biofilm medium with glucose (20 mM; BMG), sucrose (20 mM; BMS), or glucose (16 mM) plus sucrose (4 mM) (BMGS) as the supplemental carbon and energy sources as described previously (16, 41, 44). Biofilms were grown on polystyrene surfaces using flat-bottom 96-well culture plates (Corning) and hydroxylapatite (HA) discs (Clarkson Chromatography Products, Inc., PA) that were vertically placed in culture medium in 24-well plates for 24 and 48 h (2, 45). When grown on 96-well plates, biofilms were stained using 0.1% crystal violet by the end of the incubation, and the quantity in absorbance at 575 nm was measured using a Synergy II multidetection reader (Synergy II; BioTek, Inc.) as described previously (14, 16). Biofilms on HA discs were analyzed using scanning electron microscopy (SEM) and confocal laser scanning microscopy (CLSM). For SEM analysis, biofilms were fixed using 2.5% glutaraldehyde (Polysciences, Warrington, PA) in phosphate-buffered saline (PBS [pH 7.4]), dehydrated using increasing concentrations of ethanol, critical point dried, carbon coated, and analyzed using a field emission-scanning electron microscope (Hitachi, Ltd., Tokyo, Japan) under 5-kV acceleration voltage (36, 38). For CLSM, biofilms were stained using LIVE/DEAD Baclight fluorescent dye, which stains live cells with green and dead cells with red fluorescence (Invitrogen, Life Technologies), and optically dissected using an upright Olympus Fluoview BX61 (15, 46). Postacquisition analyses, such as average thickness and biovolume of biofilms, were carried out using COMSTAT as described previously (17, 36). The biovolume is defined as the volume (cubic micrometers) of the biomass per square micrometer of substratum area.
Acid and hydrogen peroxide killing assays.
The impact of RgpG deficiency on S. mutans' ability to withstand acid and oxidative stress was determined by using acid killing and hydrogen peroxide challenge assays by following procedures described elsewhere (15, 38). For these assays, planktonic cultures of S. mutans strains were grown in BHI until the mid-exponential phase (OD600 of ≅0.4) (15).
TEM analysis.
For transmission electron microscopy (TEM) analysis, S. mutans strains were grown in BHI broth until an OD600 of ≅0.4, harvested by centrifugation at 3,000 × g, 4°C for 10 min, washed once with PBS, and then fixed in 2% paraformaldehyde–2.5% glutaraldehyde (Polysciences, Warrington, PA) in PBS for 1 h at room temperature. Cells were washed in PBS and postfixed in 1% osmium tetroxide (Polysciences) for 1 h. Samples were then rinsed extensively in distilled water (dH2O) prior to en bloc staining with 1% aqueous uranyl acetate (Ted Pella, Inc., Redding, CA) for 1 h. The cells were then washed with dH2O and dehydrated in a graded series of ethanol and embedded in Eponate 12 resin (Ted Pella, Inc., Redding CA). Sections of 90 to 100 nm were prepared, stained with uranyl acetate and lead citrate, and viewed under a JEOL 1200 EX transmission electron microscope (JEOL USA, Inc., Peabody, MA) equipped with an AMT 8 megapixel digital camera and AMT Image Capture Engine V602 software (Advanced Microscopy Techniques, Woburn, MA).
Cell wall antigen preparation and slot blot analysis.
Cell wall antigens were extracted from murein sacculi using the formamide technique described by Wetherell and Bleweis (47). Murein sacculi were prepared by following the protocols of Chan et al. (10, 11) from cultures grown overnight in BHI and were suspended in formamide and heated at 180°C for 30 min in an oil bath. The suspensions were then mixed with 2 volumes of 2 N HCl and absolute ethanol (1:19 [vol/vol]) and centrifuged at 350 × g for 20 min, and the supernatants were collected and mixed with 5 volumes of acetone at 4°C for 4 h. The white precipitates were spun down at 350 × g for 20 min. The pellets were dissolved in water, and following centrifugation at 21,000 × g for 10 min, the supernatants were dialyzed (molecular mass cutoff of 3.5 kDa [Fisher Scientific]) at 4°C overnight and then freeze-dried. The antigen extracts were resuspended in 1 ml of sterile deionized water.
For cell wall antigens in spent culture medium (10, 11), S. mutans strains were grown in chemically defined FMC medium (48) until mid-exponential phase (OD600 of 0.6 to 0.7). The cell-free culture medium was collected by centrifugation at 10,000 × g at 4°C for 10 min and then treated with 3 volumes of 95% ethanol at 4°C overnight. Subsequently, the precipitate was collected by centrifugation, washed once in 70% ethanol, and air dried. The pellet was resuspended in 50 mM Tris-HCl (pH 7.5) containing 5 mM CaCl2 and 25 mM MgCl2 and treated with 10 μg/ml DNase I at 37°C for 3 h. It was then extracted with chloroform-methanol, and the aqueous layer was collected, freeze-dried, and resuspended in 50 mM Tris-HCl (pH 7.5).
For immunoblot analysis, equal amounts of the above antigen preparations were blotted onto a nitrocellulose membrane using a Bio-Dot SF slot blot device (Bio-Rad) (49) and probed with S. mutans whole-cell antiserum that was generated using inactivated whole cells of the wild-type, UA159, to immunize rabbits (Lampire Biological Laboratories, Inc.) and was adsorbed with live cells of either the wild type, the rgpG mutant, or the rgpG brpA psr triple mutant (50, 51). Antigen-antibody reactions were detected using SuperSignal West Pico chemiluminescent substrate (Thermo Fisher Scientific). The signal intensities were further analyzed using Quantity One (Bio-Rad).
Statistical analysis.
All quantitative data were analyzed using the paired Student t test. In some cases as stated, analysis of variance (ANOVA) and Tukey's pairwise comparison were also used. A P value of 0.05 or less between the groups compared was considered significantly different.
ACKNOWLEDGMENTS
We thank Alexander H. Rickard at the University of Michigan for kindly providing us with the S. mutans whole-cell antiserum.
This work was supported in part by NIH/NIDCR grant DE19452 to Z.T.W. and H.W.
REFERENCES
- 1.Moye ZD, Zeng L, Burne RA. 2014. Modification of gene expression and virulence traits in Streptococcus mutans in response to carbohydrate availability. Appl Environ Microbiol 80:1–14. doi: 10.1128/AEM.03579-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liao S, Klein MI, Heim KP, Fan Y, Bitoun JP, Ahn SJ, Burne RA, Koo H, Brady LJ, Wen ZT. 2014. Streptococcus mutans extracellular DNA is upregulated during growth in biofilms, actively released via membrane vesicles, and influenced by components of the protein secretion machinery. J Bacteriol 196:2355–2366. doi: 10.1128/JB.01493-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lemos JA, Burne RA. 2008. A model of efficiency: stress tolerance by Streptococcus mutans. Microbiology 154:3247–3255. doi: 10.1099/mic.0.2008/023770-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Navarre WW, Schneewind O. 1999. Surface proteins of Gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol Mol Biol Rev 63:174–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Silhavy TJ, Kahne D, Walker S. 2010. The bacterial cell envelope. Cold Spring Harb Perspect Biol 2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hong SW, Baik JE, Kang SS, Yun CH, Seo DG, Han SH. 2014. Lipoteichoic acid of Streptococcus mutans interacts with Toll-like receptor 2 through the lipid moiety for induction of inflammatory mediators in murine macrophages. Mol Immunol 57:284–291. doi: 10.1016/j.molimm.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Hong SW, Seo DG, Baik JE, Cho K, Yun CH, Han SH. 2014. Differential profiles of salivary proteins with affinity to Streptococcus mutans lipoteichoic acid in caries-free and caries-positive human subjects. Mol Oral Microbiol 29:208–218. doi: 10.1111/omi.12057. [DOI] [PubMed] [Google Scholar]
- 8.Boyd DA, Cvitkovitch DG, Bleiweis AS, Kiriukhin MY, Debabov DV, Neuhaus FC, Hamilton IR. 2000. Defects in d-alanyl-lipoteichoic acid synthesis in Streptococcus mutans results in acid sensitivity. J Bacteriol 182:6055–6065. doi: 10.1128/JB.182.21.6055-6065.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mistou MY, Sutcliffe IC, van Sorge NM. 2016. Bacterial glycobiology: rhamnose-containing cell wall polysaccharides in Gram-positive bacteria. FEMS Microbiol Rev 40:464–479. doi: 10.1093/femsre/fuw006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan YG, Frankel MB, Dengler V, Schneewind O, Missiakas D. 2013. Staphylococcus aureus mutants lacking the LytR-CpsA-Psr family of enzymes release cell wall teichoic acids into the extracellular medium. J Bacteriol 195:4650–4659. doi: 10.1128/JB.00544-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan YG, Kim HK, Schneewind O, Missiakas D. 2014. The capsular polysaccharide of Staphylococcus aureus is attached to peptidoglycan by the LytR-CpsA-Psr (LCP) family of enzymes. J Biol Chem 289:15680–15690. doi: 10.1074/jbc.M114.567669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eberhardt A, Hoyland CN, Vollmer D, Bisle S, Cleverley RM, Johnsborg O, Havarstein LS, Lewis RJ, Vollmer W. 2012. Attachment of capsular polysaccharide to the cell wall in Streptococcus pneumoniae. Microb Drug Resist 18:240–255. doi: 10.1089/mdr.2011.0232. [DOI] [PubMed] [Google Scholar]
- 13.Kawai Y, Marles-Wright J, Cleverley RM, Emmins R, Ishikawa S, Kuwano M, Heinz N, Bui NK, Hoyland CN, Ogasawara N, Lewis RJ, Vollmer W, Daniel RA, Errington J. 2011. A widespread family of bacterial cell wall assembly proteins. EMBO J 30:4931–4941. doi: 10.1038/emboj.2011.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wen ZT, Burne RA. 2002. Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans. Appl Environ Microbiol 68:1196–1203. doi: 10.1128/AEM.68.3.1196-1203.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wen ZT, Baker HV, Burne RA. 2006. Influence of BrpA on critical virulence attributes of Streptococcus mutans. J Bacteriol 188:2983–2992. doi: 10.1128/JB.188.8.2983-2992.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bitoun JP, Liao S, Yao X, Ahn S-J, Isoda R, Nguyen AH, Brady LJ, Burne RA, Abranches A, Wen ZT. 2012. BrpA is involved in regulation of cell envelope stress responses in Streptococcus mutans. Appl Environ Microbiol 78:2914–2922. doi: 10.1128/AEM.07823-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bitoun JP, Liao S, McKey BA, Yao X, Fan Y, Abranches J, Beatty WL, Wen ZT. 2013. Psr is involved in regulation of glucan production, and double deficiency of BrpA and Psr is lethal in Streptococcus mutans. Microbiology 159:493–506. doi: 10.1099/mic.0.063032-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Over B, Heusser R, McCallum N, Schulthess B, Kupferschmied P, Gaiani JM, Sifri CD, Berger-Bachi B, Stutzmann Meier P. 2011. LytR-CpsA-Psr proteins in Staphylococcus aureus display partial functional redundancy and the deletion of all three severely impairs septum placement and cell separation. FEMS Microbiol Lett 320:142–151. doi: 10.1111/j.1574-6968.2011.02303.x. [DOI] [PubMed] [Google Scholar]
- 19.Yamashita Y, Shibata Y, Nakano Y, Tsuda H, Kido N, Ohta M, Koga T. 1999. A novel gene required for rhamnose-glucose polysaccharide synthesis in Streptococcus mutans. J Bacteriol 181:6556–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shibata Y, Yamashita Y, Ozaki K, Nakano Y, Koga T. 2002. Expression and characterization of streptococcal rgp genes required for rhamnan synthesis in Escherichia coli. Infect Immun 70:2891–2898. doi: 10.1128/IAI.70.6.2891-2898.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soldo B, Lazarevic V, Karamata D. 2002. tagO is involved in the synthesis of all anionic cell-wall polymers in Bacillus subtilis 168. Microbiology 148:2079–2087. doi: 10.1099/00221287-148-7-2079. [DOI] [PubMed] [Google Scholar]
- 22.Soldo B, Lazarevic V, Pooley HM, Karamata D. 2002. Characterization of a Bacillus subtilis thermosensitive teichoic acid-deficient mutant: gene mnaA (yvyH) encodes the UDP-N-acetylglucosamine 2-epimerase. J Bacteriol 184:4316–4320. doi: 10.1128/JB.184.15.4316-4320.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kern J, Ryan C, Faull K, Schneewind O. 2010. Bacillus anthracis surface-layer proteins assemble by binding to the secondary cell wall polysaccharide in a manner that requires csaB and tagO. J Mol Biol 401:757–775. doi: 10.1016/j.jmb.2010.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lunderberg JM, Liszewski Zilla M, Missiakas D, Schneewind O. 2015. Bacillus anthracis tagO is required for vegetative growth and secondary cell wall polysaccharide synthesis. J Bacteriol 197:3511–3520. doi: 10.1128/JB.00494-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.D'Elia MA, Henderson JA, Beveridge TJ, Heinrichs DE, Brown ED. 2009. The N-acetylmannosamine transferase catalyzes the first committed step of teichoic acid assembly in Bacillus subtilis and Staphylococcus aureus. J Bacteriol 191:4030–4034. doi: 10.1128/JB.00611-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D'Elia MA, Millar KE, Beveridge TJ, Brown ED. 2006. Wall teichoic acid polymers are dispensable for cell viability in Bacillus subtilis. J Bacteriol 188:8313–8316. doi: 10.1128/JB.01336-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Denapaite D, Bruckner R, Hakenbeck R, Vollmer W. 2012. Biosynthesis of teichoic acids in Streptococcus pneumoniae and closely related species: lessons from genomes. Microb Drug Resist 18:344–358. doi: 10.1089/mdr.2012.0026. [DOI] [PubMed] [Google Scholar]
- 28.Brown S, Santa Maria JP Jr, Walker S. 2013. Wall teichoic acids of Gram-positive bacteria. Annu Rev Microbiol 67:313–336. doi: 10.1146/annurev-micro-092412-155620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schleifer KH, Kilpper-Balz R, Kraus J, Gehring F. 1984. Relatedness and classification of Streptococcus mutans and “mutans-like” streptococci. J Dent Res 63:1047–1050. doi: 10.1177/00220345840630080701. [DOI] [PubMed] [Google Scholar]
- 30.Holland LM, Conlon B, O'Gara JP. 2011. Mutation of tagO reveals an essential role for wall teichoic acids in Staphylococcus epidermidis biofilm development. Microbiology 157:408–418. doi: 10.1099/mic.0.042234-0. [DOI] [PubMed] [Google Scholar]
- 31.Rowe HM, Hanson BR, Runft DL, Li Q, Firestine SM, Neely MN. 2015. Modification of the CpsA protein reveals a role in alteration of the Streptococcus agalactiae cell envelope. Infect Immun 83:1497–1506. doi: 10.1128/IAI.02656-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanson BR, Runft DL, Streeter C, Kumar A, Carion TW, Neely MN. 2012. Functional analysis of the CpsA protein of Streptococcus agalactiae. J Bacteriol 194:1668–1678. doi: 10.1128/JB.06373-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu C, Huang IH, Chang C, Reardon-Robinson ME, Das A, Ton-That H. 2014. Lethality of sortase depletion in Actinomyces oris caused by excessive membrane accumulation of a surface glycoprotein. Mol Microbiol 94:1227–1241. doi: 10.1111/mmi.12780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liszewski Zilla M, Chan YG, Lunderberg JM, Schneewind O, Missiakas D. 2015. LytR-CpsA-Psr enzymes as determinants of Bacillus anthracis secondary cell wall polysaccharide assembly. J Bacteriol 197:343–353. doi: 10.1128/JB.02364-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liszewski Zilla M, Lunderberg JM, Schneewind O, Missiakas D. 2015. Bacillus anthracis lcp genes support vegetative growth, envelope assembly, and spore formation. J Bacteriol 197:3731–3741. doi: 10.1128/JB.00656-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bitoun JP, Nguyen AH, Fan Y, Burne RA, Wen ZT. 2011. Transcriptional repressor Rex is involved in regulation of oxidative stress response and biofilm formation by Streptococcus mutans. FEMS Microbiol Lett 320:110–117. doi: 10.1111/j.1574-6968.2011.02293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeng L, Wen ZT, Burne RA. 2006. A novel signal transduction system and feedback loop regulate fructan hydrolase gene expression in Streptococcus mutans. Mol Microbiol 62:187–200. doi: 10.1111/j.1365-2958.2006.05359.x. [DOI] [PubMed] [Google Scholar]
- 38.Wen ZT, Burne RA. 2004. LuxS-mediated signaling in Streptococcus mutans is involved in regulation of acid and oxidative stress tolerance and biofilm formation. J Bacteriol 186:2682–2691. doi: 10.1128/JB.186.9.2682-2691.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lau PCY, Sung CK, Lee JH, Morrison DA, Cvitkovitch DG. 2002. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J Microbiol Methods 49:193–201. doi: 10.1016/S0167-7012(01)00369-4. [DOI] [PubMed] [Google Scholar]
- 40.Burne RA, Abranches J, Ahn SJ, Lemos JA, Wen ZT, Zeng L. 2011. Functional genomics of Streptococcus mutans, p 185–204. In Kolenbrander PE. (ed), Oral microbial communities: genomic inquiries and interspecies communication. ASM Press, Washington, DC. [Google Scholar]
- 41.Li YH, Tang N, Aspiras MB, Lau PC, Lee JH, Ellen RP, Cvitkovitch DG. 2002. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J Bacteriol 184:2699–2708. doi: 10.1128/JB.184.10.2699-2708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.LeBanc D, Lee L. 1991. Replication function of pVA380-1, p 235–239. In Dunny G, Cleary PP, Mckay LL (ed), Genetics and molecular biology of streptococci, lactococci, and enterococci. ASM Press, Washington, DC. [Google Scholar]
- 43.Wen TZ, Burne RA. 2001. Construction of a new integration vector for use in Streptococcus mutans. Plasmid 45:31–36. doi: 10.1006/plas.2000.1498. [DOI] [PubMed] [Google Scholar]
- 44.Loo CY, Corliss DA, Ganeshkumar N. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J Bacteriol 182:1374–1382. doi: 10.1128/JB.182.5.1374-1382.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lemos JA, Abranches J, Koo H, Marquis RE, Burne RA. 2010. Protocols to study the physiology of oral biofilms. Methods Mol Biol 666:87–102. doi: 10.1007/978-1-60761-820-1_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wen ZT, Nguyen AH, Bitoun JP, Abranches J, Baker HV, Burne RA. 2011. Transcriptome analysis of LuxS-deficient Streptococcus mutans grown in biofilms. Mol Oral Microbiol 26:2–18. doi: 10.1111/j.2041-1014.2010.00581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wetherell JR Jr, Bleiweis AS. 1975. Antigens of Streptococcus mutans: characterization of a polysaccharide antigen from walls of strain GS-5. Infect Immun 12:1341–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Terleckyj B, Willett NP, Shockman GD. 1975. Growth of several cariogenic strains of oral streptococci in a chemically defined medium. Infect Immun 11:649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wen TZ, Suntharaligham P, Cvitkovitch DG, Burne RA. 2005. Trigger factor in Streptococcus mutans is involved in stress tolerance, competence development, and biofilm formation. Infect Immun 73:219–225. doi: 10.1128/IAI.73.1.219-225.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elder BL, Boraker DK, Fives-Taylor PM. 1982. Whole-bacterial cell enzyme-linked immunosorbent assay for Streptococcus sanguis fimbrial antigens. J Clin Microbiol 16:141–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Palmer RJ Jr, Gordon SM, Cisar JO, Kolenbrander PE. 2003. Coaggregation-mediated interactions of streptococci and actinomyces detected in initial human dental plaque. J Bacteriol 185:3400–3409. doi: 10.1128/JB.185.11.3400-3409.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ajdic D, McShan WM, McLaughlin RE, Savic G, Chang J, Carson MB, Primeaux C, Tian R, Kenton S, Jia H, Lin S, Qian Y, Li S, Zhu H, Najar F, Lai H, White J, Roe BA, Ferretti JJ. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci U S A 99:14434–14439. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]