ABSTRACT
The Bacillus cereus group comprises nine species, several of which are pathogenic. Differentiating between isolates that may cause disease and those that do not is a matter of public health and economic importance, but it can be particularly challenging due to the high genomic similarity within the group. To this end, we have developed BTyper, a computational tool that employs a combination of (i) virulence gene-based typing, (ii) multilocus sequence typing (MLST), (iii) panC clade typing, and (iv) rpoB allelic typing to rapidly classify B. cereus group isolates using nucleotide sequencing data. BTyper was applied to a set of 662 B. cereus group genome assemblies to (i) identify anthrax-associated genes in non-B. anthracis members of the B. cereus group, and (ii) identify assemblies from B. cereus group strains with emetic potential. With BTyper, the anthrax toxin genes cya, lef, and pagA were detected in 8 genomes classified by the NCBI as B. cereus that clustered into two distinct groups using k-medoids clustering, while either the B. anthracis poly-γ-d-glutamate capsule biosynthesis genes capABCDE or the hyaluronic acid capsule hasA gene was detected in an additional 16 assemblies classified as either B. cereus or Bacillus thuringiensis isolated from clinical, environmental, and food sources. The emetic toxin genes cesABCD were detected in 24 assemblies belonging to panC clades III and VI that had been isolated from food, clinical, and environmental settings. The command line version of BTyper is available at https://github.com/lmc297/BTyper. In addition, BMiner, a companion application for analyzing multiple BTyper output files in aggregate, can be found at https://github.com/lmc297/BMiner.
IMPORTANCE Bacillus cereus is a foodborne pathogen that is estimated to cause tens of thousands of illnesses each year in the United States alone. Even with molecular methods, it can be difficult to distinguish nonpathogenic B. cereus group isolates from their pathogenic counterparts, including the human pathogen Bacillus anthracis, which is responsible for anthrax, as well as the insect pathogen B. thuringiensis. By using the variety of typing schemes employed by BTyper, users can rapidly classify, characterize, and assess the virulence potential of any isolate using its nucleotide sequencing data.
KEYWORDS: Bacillus cereus group, taxonomy, virulence genes, whole-genome sequencing
INTRODUCTION
The Bacillus cereus group, also known as Bacillus cereus sensu lato (s.l.), consists of nine closely related bacterial species: B. anthracis (1), B. cereus sensu stricto (s.s.), B. cytotoxicus (2), B. mycoides (3), B. pseudomycoides (4), B. thuringiensis, B. toyonensis (5), B. weihenstephanensis (3), and B. wiedmannii (6). The pathogenic potentials of members of the B. cereus group vary widely; while some isolates are capable of causing anthrax or anthrax-like disease (7), foodborne illness (8), or food spoilage issues (9–11), others are used in industrial settings as probiotics (5, 12–14), insecticides and pest control agents (15), agents in environmental pollutant bioremediation (15–17), plant growth promoters (15, 18), and even as producers of bacteriocins (19, 20) or parasporins with anticancer activities (15, 21, 22). As the industrial and agricultural applications of these microorganisms expand, differentiating between isolates that can cause anthrax or gastrointestinal illness and those that can be used as beneficial microbes in industrial or agricultural settings becomes critical. Relying strictly on taxonomic classification at the species level can lead not only to isolate misclassification, but also to an inaccurate assessment of a given isolate's virulence potential. There have been numerous cases in which probiotics containing B. cereus group isolates sold for human and/or animal consumption were found to possess strains capable of producing toxins Nhe and/or Hbl (12, 14, 23), or the species they contained were incorrectly identified (12, 14, 24). Additionally, B. thuringiensis, a biopesticide, can possess B. cereus s.s. toxin genes and potentially infect humans via the food chain (25), a notable example being a foodborne outbreak associated with salad that was potentially caused by B. thuringiensis serovar aizawai that had been sprayed on a produce field (26).
Differentiating between pathogenic and nonpathogenic B. cereus group isolates is a matter of public health and economic importance but can be a challenging task. Phenotypic and biochemical methods (27), as well as many commonly used molecular methods, such as 16S rRNA gene sequencing, may not have sufficient discriminatory power to differentiate between members of the B. cereus group (28, 29). In addition, the ability of a particular B. cereus group isolate to cause disease in humans is not species dependent, and taxonomic classification can often be a poor predictor of an isolate's virulence potential (30); for example, genes encoding diarrheal toxins have been found in B. cereus, B. mycoides, B. pseudomycoides, B. thuringiensis, and B. weihenstephanensis (30–32). For these reasons, better tools are needed to classify B. cereus isolates, from both taxonomical and food safety risk perspectives (33).
A number of genetic loci have been proposed as markers that can be used to taxonomically classify and/or differentiate between pathogenic and nonpathogenic B. cereus group isolates at greater resolution than phenotypic methods and 16S rRNA gene sequencing (30). Some examples of taxonomic markers include the housekeeping gene rpoB (6, 30, 34–38), the pantoate-beta-alanine ligase gene panC (39–43), and multiple loci used in a 7-gene multilocus sequence typing (MLST) scheme (i.e., glp, gmk, ilv, pta, pur, pyc, and tpi) (30, 44–49) (https://pubmlst.org/bcereus/). Each of these methods alone provides greater resolution than its predecessors, and the methods may be implemented in combination with each other and/or with phenotypic methods (30, 33, 40, 49).
The presence and absence of virulence and toxin genes have also served as indicators in a method by which B. cereus group isolates can be classified as pathogenic or nonpathogenic (28, 30, 50). These methods are beneficial from a clinical perspective, as genes associated with many medically relevant phenotypes are plasmid carried (51), including anthrax toxin and capsule genes (52), and ces genes, which encode cereulide synthetase (53). This can be contrasted with the fact that many genes that encode phenotypic traits used to distinguish members of the B. cereus group using biochemical and microbiological tests are contained on the chromosome (motility, hemolysis, etc.) (51). As a result, a disease phenotype, such as the ability to cause anthrax-like symptoms in a particular host (52), may not be confined to a single B. cereus group species, making species-level taxonomy a poor indicator of an isolate's pathogenic potential.
Molecular typing methods using housekeeping and virulence genes found in members of the B. cereus group have been essential for classifying isolates from both a taxonomical and a public health perspective. However, as whole-genome sequencing (WGS) becomes cheaper, faster, and more accessible, the ability to perform molecular typing methods in silico becomes even more attractive. With the goal of creating a readily accessible open-source pipeline that can be easily used by B. cereus researchers and public health officials, we have created BTyper, a computational tool to perform (i) virulence gene detection, (ii) MLST, (iii) panC clade typing, and (iv) rpoB allelic typing using B. cereus group nucleotide sequencing data in either FASTA, SRA, or gzipped FASTQ format. Additionally, we applied BTyper and BMiner, a companion application for analyzing BTyper's output files in aggregate, to a set of 662 B. cereus group genome assemblies, with the goal of identifying (i) anthrax-associated genes in non-anthracis Bacillus members of the B. cereus group, and (ii) assemblies from B. cereus group strains with emetic potential.
RESULTS
Construction and validation of BTyper using in vitro methods.
BTyper was used to perform in silico (i) virulence gene detection, (ii) MLST, (iii) panC clade typing, and (iv) rpoB allelic typing using the default settings described in Materials and Methods. Both assembled genomes and Illumina paired-end reads from 46 B. cereus group genomes were used (Fig. 1). BTyper was successfully able to predict rpoB allelic types and whole-genome phylogenetic clade using panC for all B. cereus group genomes tested (n = 46; Table 1). For in silico MLST, it was successful at predicting the sequence type in all but one isolate (45 out of 46; Table 1); isolate FSL M8-0091 was the only isolate for which in silico prediction of sequence type did not match the sequence type obtained by Sanger sequencing. For this isolate, the only allele that differed between the two methods was the tpi allele: Sanger sequencing yielded a tpi allelic type of 20, while BTyper's in silico prediction was tpi allelic type 175, which was a perfect match and differed from tpi 20 by a single nucleotide at position 284. However, SRST2 (54) also obtained a tpi allelic type of 175, making it likely that (i) the colony selected to undergo WGS had a different tpi allele than the colony selected to undergo Sanger sequencing, or (ii) there was an error in either WGS or Sanger sequencing.
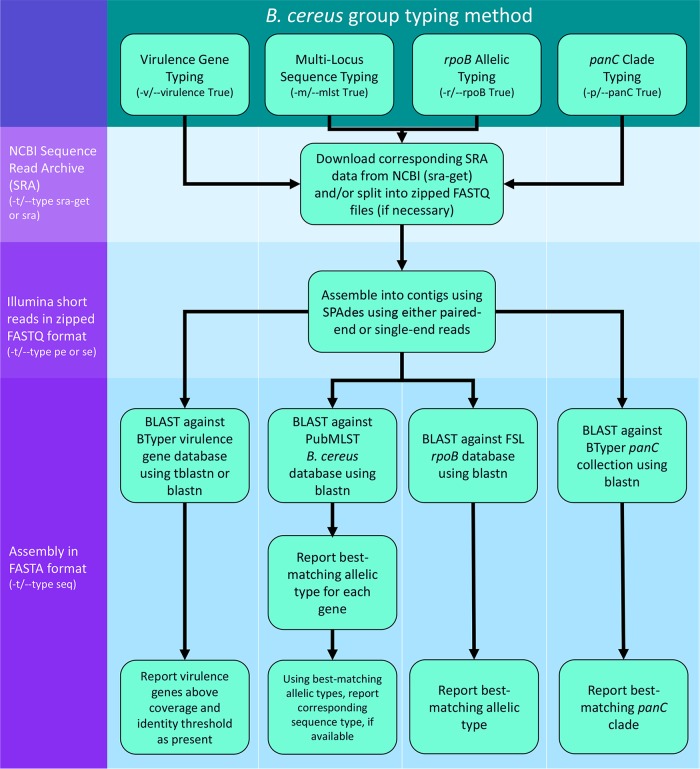
FIG 1.
BTyper command line workflow for various types of data and default typing methods. Input datum type is listed in the left margin, while typing methods are listed at the top of the chart. Command line parameters associated with a particular typing method are shown in parentheses. FSL, Food Safety Lab.
TABLE 1.
Percentage of isolates in which BTyper correctly identified the presence/absence of eight virulence genes, MLST, rpoB AT, and panC clade
| Data set | Virulence gene (%)a |
MLST ST (%)b | rpoB AT (%)c | panC clade (%)d | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| hblA | hblC | hblD | nheA | nheB | nheC | cytK | entFM | ||||
| Training (n = 22) | |||||||||||
| Assemblies | 100 | 100 | 100 | 100 | 95.5 | 100 | 90.9 | 95.5 | 100 | 100 | 100 |
| PE readse | 100 | 90.9 | 100 | 90.9 | 95.5 | 95.5 | 90.9 | 95.5 | 100 | 100 | 100 |
| Validation (n = 24) | |||||||||||
| Assemblies | 91.7 | 100 | 95.8 | 87.5 | 95.8 | 100 | 100 | 91.7 | 95.8 | 100 | 100 |
| PE reads | 91.7 | 100 | 91.7 | 87.5 | 95.8 | 100 | 100 | 91.7 | 95.8 | 100 | 100 |
| Total (n = 46) | |||||||||||
| Assemblies | 95.7 | 100 | 97.8 | 93.5 | 95.7 | 100 | 95.7 | 93.5 | 97.8 | 100 | 100 |
| PE readse | 95.7 | 95.7 | 95.7 | 89.1 | 95.7 | 97.8 | 95.7 | 93.5 | 97.8 | 100 | 100 |
Presence/absence of eight virulence genes from previously published WGS data (training set) or PCR (validation set).
Multilocus sequence typing (MLST) results from previously published WGS data (training set) or Sanger sequencing (validation set).
rpoB allelic typing (AT) results from previously published WGS data (training set) or Sanger sequencing (validation set).
panC clade typing results from previously published WGS data.
Illumina paired-end (PE) reads.
For virulence gene detection, the results obtained from BTyper matched the PCR results for eight selected virulence genes in over 89% of all isolates (n = 46; Table 1). This resulted in an overall sensitivity and specificity of 99.0% and 85.5%, respectively, when the default parameters for assembled genomes were used, and an overall sensitivity and specificity of 97.0% and 85.5%, respectively, when default parameters for Illumina paired-end reads were used.
Characteristics associated with B. cereus group phylogenetic clade III are most prevalent among genome assemblies currently available at NCBI.
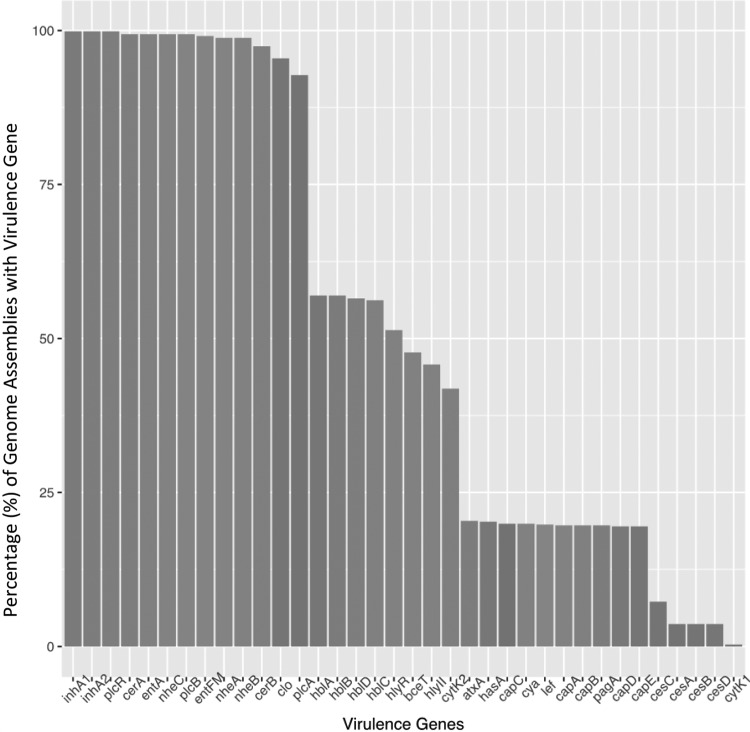
BTyper was used to perform virulence gene detection, MLST, panC clade typing, and rpoB allelic typing on 662 B. cereus group genome assemblies (157 assemblies labeled as B. anthracis, 353 assemblies as B. cereus s.s., 2 assemblies as B. cytotoxicus, 19 assemblies as B. mycoides, 2 assemblies as B. pseudomycoides, 94 assemblies as B. thuringiensis, 3 assemblies as B. toyonensis, 21 assemblies as B. weihenstephanensis, and 11 assemblies as B. wiedmannii). Within the 662 assemblies, 13 virulence genes were detected in more than 90% of all genomes when the default minimum amino acid sequence identity and coverage thresholds of 50 and 70% were used, respectively (Fig. 2). The least commonly detected gene was cytK1 (Fig. 2), which was detected in both available B. cytotoxicus genomes and no other WGS assemblies.
FIG 2.
Percentage (%) of B. cereus group assemblies in which a particular virulence gene was detected. Minimum identity and coverage thresholds of 50 and 70%, respectively, were used for virulence gene detection.
For in silico MLST, 544 assemblies were assigned to one of 213 B. cereus sequence types (STs), the most common of which was ST1 (n = 123 isolates). This was unsurprising, considering that ST1 is associated with B. anthracis (55), and B. anthracis makes up a considerable portion (23.7%) of the B. cereus group genome assemblies currently in NCBI's database. In silico rpoB allelic typing grouped the 662 isolates into one of 43 different, best-matching rpoB allelic types (ATs), with 185 isolates matching AT463 most closely. AT463 has been previously associated with clade III isolates (30), the phylogenetic clade that encompasses B. anthracis.
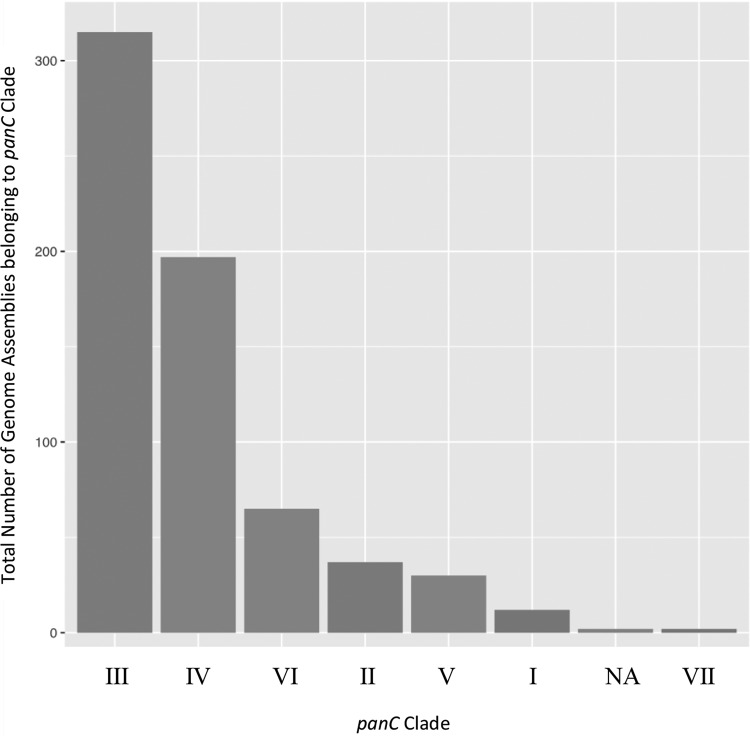
For panC-based phylogenetic clade typing, a panC locus was detected in 658 out of 662 genomes (Fig. 3). The most commonly assigned clade was clade III, a polyphyletic clade which contains B. anthracis, as well as some strains currently misclassified in the NCBI database as B. cereus s.s. and B. thuringiensis (30, 39, 40). Together, clade IV, which consists of some B. cereus s.s. and B. thuringiensis strains (30, 39, 40), as well as the type strains of these two species, and clade III accounted for more than 75% of all B. cereus group WGS assemblies in the NCBI database (Fig. 3). Clade VII, which contains the B. cytotoxicus (2) type strain, was the most poorly represented clade; the two available B. cytotoxicus assemblies were placed here.
FIG 3.
Closest-matching phylogenetic clade using the panC loci from 662 B. cereus group genome assemblies. A panC locus could not be assigned in 4 genome assemblies, which is denoted by “NA.”
Application of BTyper to identify B. anthracis-associated genes in non-anthracis Bacillus isolates reveals virulence gene heterogeneity within genome assemblies from anthrax toxin-encoding isolates.
When Fisher's exact test was used to determine if any virulence genes were significantly associated with a phylogenetic clade, virulence genes typically associated with B. anthracis were found to be significantly associated with members of clade III after a Bonferroni correction was applied (P < 0.05; Table 2). The B. anthracis toxin genes cya (edema factor-encoding), lef (lethal factor-encoding), and pagA (protective antigen-encoding), as well as their regulator gene atxA (56), were found only in clade III isolates (P < 0.05; Table 2). In addition, B. anthracis polyglutamate capsule synthesis genes capABCDE (57) were more commonly associated with clade III assemblies (P < 0.05; Table 2) and found primarily in genomes classified in the NCBI database as B. anthracis. Meanwhile, genes associated with diarrheal disease (8) were found to be significantly associated with clades II, IV, V, and VI (P < 0.05; Table 2); these included the diarrheal toxin genes hblCDAB, which were found to be significantly associated with clades II, IV, V, and VI (P < 0.05; Table 2), while being less common in members of clade III (P < 0.05; Table 2), driven by the large number of B. anthracis assemblies in this clade that did not possess these genes.
TABLE 2.
Virulence genes significantly associated with 5 B. cereus group phylogenetic clades after a Bonferroni correctiona
| Clade | Genes |
|---|---|
| II | hblCDAB |
| III | atxA,b capABCDE, cya,b hasA, hlyII, hlyR, lef,b pagAb |
| IV | bceT, cytK2, hblCDAB |
| V | bceT, hblCDABc |
| VI | bceT, cesC, hblCDABc |
Significant at a P value of <0.05. For exact corrected P values, see Table S7.
Indicates a virulence gene that was detected only in its respective clade (includes clades I and VII).
Indicates a virulence gene that was detected in all members of its respective clade.
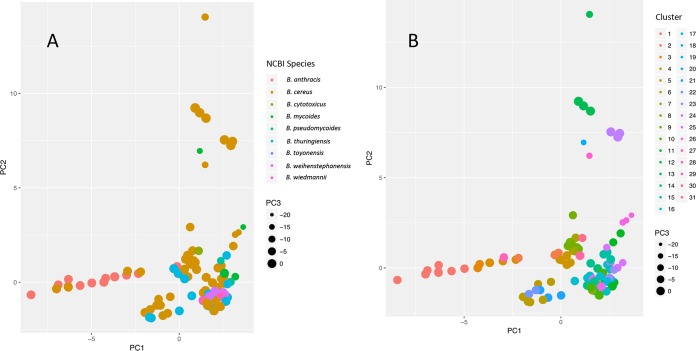
Principal-component analysis (PCA) based on the presence/absence of virulence genes using BMiner revealed several assemblies labeled as B. cereus and B. thuringiensis that clustered with B. anthracis assemblies (Fig. 4A). When k-medoids clustering was performed with an optimum k of 31, isolates classified in the NCBI database as B. anthracis were placed into clusters 1 through 8 (Fig. 4B). Additionally, clusters 17, 21, 22, and 29 did not contain any assemblies labeled in NCBI as B. anthracis, but they contained at least one assembly in which one or more of the B. anthracis-associated virulence genes identified using Fisher's exact test were detected (Fig. 5).
FIG 4.
Principal-component analysis (PCA) of 662 B. cereus group genome assemblies based on presence/absence of virulence genes. Virulence gene typing was carried out using BTyper, while PCA was performed using BMiner. Principal components 1 (PC1) and 2 (PC2) are plotted on the x and y axes, respectively, while principal component 3 (PC3) corresponds to point size. Plots are colored by isolate species, as found in NCBI (A), and assigned cluster using k-medoids (B). To view interactive versions of these plots containing isolate names and metadata, all BTyper final results files and metadata can be downloaded from https://github.com/lmc297/BTyper/tree/master/sample_data and viewed in BMiner.
FIG 5.
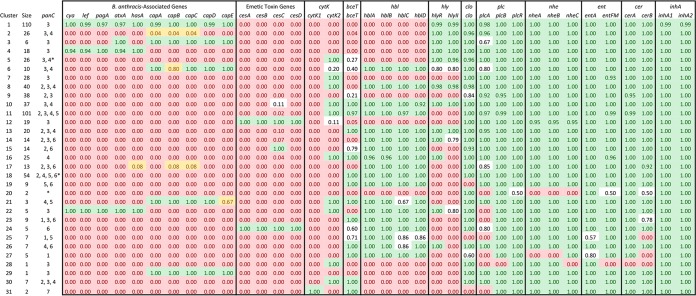
k-medoids clusters based on presence/absence of virulence genes detected using BTyper. Size corresponds to the number of assemblies assigned to a given cluster, while panC corresponds to panC clades found in the cluster, with an asterisk denoting one or more assemblies that could not be placed into a panC clade. Numbers within cells correspond to the proportion of assemblies in a given cluster in which the corresponding virulence gene was detected. Green shading corresponds to a virulence gene detected in more than 90% of all assemblies in a cluster, while red shading corresponds to a virulence gene detected in fewer than 10% of all assemblies in a cluster. Yellow shading corresponds to B. anthracis-associated genes detected in fewer than 90% but greater than 0% of assemblies in a cluster.
Cluster 1 (Fig. 4B), which contained the majority of isolates labeled as B. anthracis, contained 110 isolates, 107 of which were classified in the NCBI database as B. anthracis, and all of which belonged to panC clade III (Fig. 5). Assemblies derived from human and veterinary clinical isolates associated with anthrax disease populated a large proportion of the cluster, including assemblies associated with isolates from the 2001 anthrax bioterrorism attacks (58), European heroin users and an associated outbreak (59, 60), and a 2011 outbreak in Swedish cattle (61). Three assemblies labeled as B. cereus clustered among them (Fig. 4B). Two of these assemblies were labeled as B. cereus strain 03BB102, an isolate that was thought to cause fatal pneumonia in a welder in San Antonio, TX (Table 3), while the third was labeled as B. cereus biovar anthracis strain CI, which caused fatal anthrax in a chimpanzee in the rainforest of Taï National Park, Côte d'Ivoire (Table 3) (51). Consistent with these findings, placement into cluster 1 was driven largely by an assembly's possession of all, or nearly all, anthrax-associated genes identified using Fisher's exact test (Fig. 6); the anthrax toxin genes cya, lef, and pagA, toxin regulator gene atxA, hyaluronic acid capsule gene hasA, and B. anthracis polyglutamate capsule genes capABCDE were detected in nearly all (>97%) cluster 1 assemblies (Fig. 5).
TABLE 3.
Non-anthracis Bacillus assemblies in which anthrax toxin genes cya, lef, and/or pagA were detected using BTyper
| Clustera | NCBI species classification | panC cladeb | GenBank accession no.c | Strain | Isolate source (reference) | Gene(s) detected? |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| cya | lef | pagA | atxA | hasA | capABCDE | ||||||
| 1 | B. cereus | III | GCA_000022505.1, GCA_000832405.1 | 03BB102 | Human with fatal pneumonia, San Antonio, TX, USA (104) | + | + | + | − | + | + |
| 1 | B. cereus | III | GCA_000143605.1 | Biovar anthracis strain CI | Chimpanzee with fatal anthrax, Taï National Park, Côte d'Ivoire (51) | + | + | + | + | + | + |
| 22 | B. cereus | III | GCA_000167215.1, GCA_000832805.1 | G9241 | Human with pneumonia, nausea, and vomiting, LA, USA (65) | + | + | + | + | + | − |
| 22 | B. cereus | III | GCA_000688755.1 | BcFL2013 | Human with anthrax-like skin lesion, FL, USA (66) | + | + | + | + | + | − |
| 22 | B. cereus | III | GCA_000789315.1 | 03BB87 | Human with fatal pneumonia, Lubbock, TX, USA (67) | + | + | + | + | + | − |
| 22 | B. cereus | III | GCA_002007005.1 | LA2007 | Human with fatal pneumonia and septic shock, Galliano, LA, USA (68) | + | + | + | + | + | − |
Clusters were assigned using a k-medoids approach (k = 31).
panC clades were assigned using BTyper.
Multiple accession numbers are given for strains associated with multiple assemblies.
FIG 6.
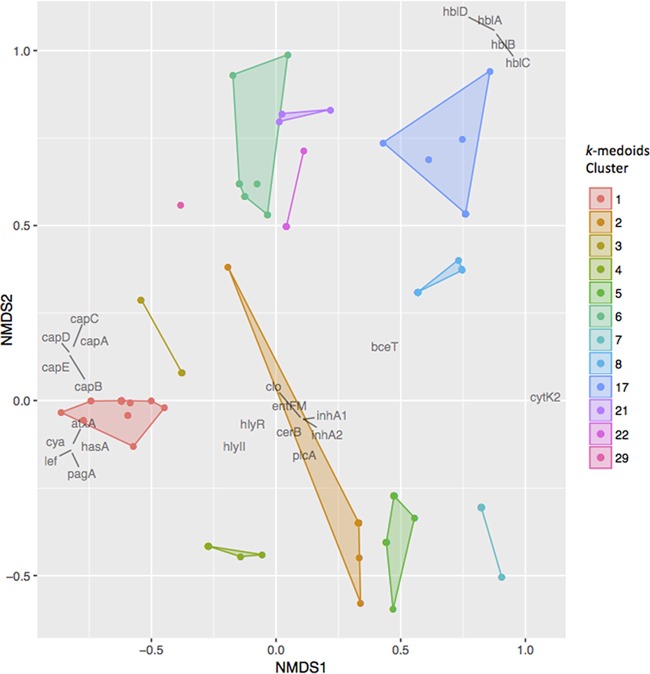
Nonmetric multidimensional scaling (NMDS) plot of Bacillus cereus group clusters that (i) possessed at least one assembly that was classified as Bacillus anthracis in NCBI, and/or (ii) possessed at least one assembly in which at least one B. anthracis-associated virulence gene (cya, lef, pagA, atxA, hasA, and/or capABCDE) was detected using BTyper. NMDS was performed in BMiner using virulence gene presence/absence data and a Jaccard dissimilarity metric. Isolates are represented by points, and convex hulls and shading correspond to the assigned k-medoids cluster. Virulence genes are plotted in dark gray.
Despite the fact that all assemblies classified in NCBI as B. anthracis were assigned to clusters 1 through 8, the only other clusters in addition to cluster 1 in which anthrax toxin genes were detected were clusters 4 and 22. Like cluster 1, all isolates in clusters 4 and 22 belonged to panC clade III, and nearly all possessed the anthrax toxin genes cya, lef, and pagA, regulator gene atxA, and hyaluronic acid capsule gene hasA (Fig. 5). However, the B. anthracis polyglutamate capsule genes capABCDE were not detected in any of the cluster 4 or cluster 22 assemblies at the default identity and coverage thresholds (Fig. 5). While cluster 4 (n = 18; Fig. 4B) contained only isolates classified in the NCBI database as B. anthracis, it contained assemblies from several strains with attenuated virulence, including several vaccine strains (62–64). Cluster 22 (n = 5; Fig. 4B), however, contained 5 anthrax-associated assemblies, all of which were classified in the NCBI database as B. cereus (Table 3). All assemblies in cluster 22 originated from human clinical isolates in which the isolate was classified as B. cereus, but the patient presented anthrax-like symptoms; two assemblies were of B. cereus strain G9241, a strain of Bacillus isolated from the sputum and blood of a patient with pneumonia, nausea, and vomiting (65). The isolate, which had been classified as B. cereus via biochemical tests and 16S rRNA gene sequencing, was found to possess the anthrax toxin gene pagA but not the polyglutamate capsule genes capABCDE (65), which is consistent with its classification using BTyper (Table 3). BTyper's classification of the three other assemblies in this cluster also aligned with their previously published descriptions and included the following: (i) a B. cereus assembly associated with an isolate from a patient in Florida possessing an anthrax-like skin lesion (66), which was found to possess anthrax toxin genes cya, lef, and pagA and the hyaluronic acid capsule gene hasA and belong to ST78 (66), (ii) a B. cereus isolate from a patient with a fatal case of pneumonia in Lubbock, TX, that was also found to possess B. anthracis virulence genes (67), and (iii) an assembly associated with a B. cereus isolate that was found to possess anthrax toxin genes and hasA and was isolated from a patient in Galliano, LA, who had a fatal case of pneumonia and septic shock (Table 3) (68).
While no anthrax toxin genes were detected outside clusters 1, 4, and 22, other B. anthracis-associated genes identified using Fisher's exact test were detected in several other clusters and assemblies. Cluster 3 (n = 6; Fig. 4B) contained 6 B. anthracis assemblies belonging to panC clade III in which the B. anthracis toxin regulator gene atxA and polyglutamate capsule genes capABCDE were detected (Fig. 5). Other assemblies in this cluster included B. anthracis strain Smith 1013, described as “Pasteur-like” in that it possessed plasmid pXO2 (the plasmid associated with cap genes) but not plasmid pXO1 (the plasmid associated with B. anthracis toxin genes) (69, 70), as well as B. anthracis strain Pasteur itself (Table 4).
TABLE 4.
Non-anthracis Bacillus assemblies in which B. anthracis-associated genes were detected, excluding anthrax toxin genes cya, lef, and pagA and regulator atxA
| Cluster | NCBI species classification | panC clade | GenBank accession no.c | Strain/isolate IDa | Isolate source (reference) | Gene detected? |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| hasA | capA | capB | capC | capD | capE | ||||||
| 2 | B. cereus | III | GCA_001286905.1 | JRS1 | Rhazya stricta rhizosphere, Jeddah, Saudi Arabia (105) | − | + | + | + | − | − |
| 6 | B. cereus | III | GCA_000003955.1 | AH1273 | Human blood, Iceland (52) | − | + | + | + | + | + |
| 6 | B. cereus | III | GCA_000161395.1 | AH1272 | Amniotic fluid, Iceland (52) | − | + | − | + | + | + |
| 6 | B. cereus | III | GCA_000181655.1, GCA_000832865.1 | 03BB108 | Dust containing pneumonia-causing B. cereus strain 03BB012 (106) | − | + | + | + | + | + |
| 6 | B. cereus | IV | GCA_000398945.1 | Schrouff | Food (107) | − | + | + | + | + | + |
| 6 | B. cereus | IV | GCA_000399185.1 | K-5975c | Food (107) | − | + | + | + | + | + |
| 6 | B. cereus | IV | GCA_000399305.1 | HuB4-4 | Soil, Belgium (107) | − | + | − | + | + | + |
| 6 | B. thuringiensis | III | GCA_000161595.1 | Serovar Monterrey strain BGSC 4AJ1 | Mexico (108) | − | + | + | + | + | + |
| 6 | B. thuringiensis | IV | GCA_001640965.1 | BGSC 4C1 | Bombyx mori, Czechoslovakia (109) | − | + | + | + | + | + |
| 17 | B. cereus | VI | GCA_002014585.1 | FSL H8-0485 | Soil, USA (110) | + | − | − | − | − | − |
| 17 | B. thuringiensis | III | GCA_000948155.1 | Et10/1 | Geothermal spring, Lirima thermal springs, Chile (111) | − | − | + | + | − | − |
| 21 | B. cereus | IV | GCA_000161315.1 | F65185 | Open fracture, NY, USA (112) | − | + | + | + | + | + |
| 21 | B. cereus | V | GCA_000290835.1 | VD115 | Soil, Guadeloupe (107) | − | + | + | + | + | + |
| 21 | B. thuringiensis | IV | GCA_001677055.1b | BGSC 4BT1 | Red soil, China (113) | − | + | + | + | + | − |
| 29 | B. cereus | III | GCA_001913295.1 | MOD1_Bc119 | Whole black pepper, USA (114) | − | + | + | + | + | + |
ID, identification.
capE was detected at a lower amino acid identity (47.7%, compared to the default threshold of 50%).
Multiple accession numbers are given for strains associated with multiple assemblies.
The polyglutamate capsule genes capABCDE were also detected in assemblies assigned to clusters 6, 21, and 29 (Table 4). Cluster 6 (n = 10; Fig. 4B) contained 10 assemblies: 1 assembly classified in NCBI as B. anthracis, 7 assemblies classified as B. cereus, and 2 assemblies classified as B. thuringiensis. Members of this cluster belonged to panC clades III and IV, and consistent with the detection of cap genes in this cluster, one of the B. thuringiensis assemblies in this group had been shown to produce a polyglutamate capsule (71). Cluster 21 (n = 3; Fig. 4B) contained 2 assemblies labeled as B. cereus and 1 assembly labeled as B. thuringiensis. One of the B. cereus assemblies came from B. cereus strain F65185, which was confirmed to belong to ST168 and was isolated from a patient in New York with an open fracture wound (Table 4). Members of this group belonged to either panC clade IV or V. Cluster 29 (n = 1; Fig. 4B) consisted of a single B. cereus assembly belonging to panC clade III and associated with a strain isolated from whole black pepper in the United States in 2015 (Table 4).
Additionally, cap genes were detected in a single isolate in clusters 2 and 17 (n = 26 and 13, respectively; Fig. 4B). However, B. anthracis-associated genes were not detected in any other assemblies in this cluster, despite being composed primarily of assemblies classified as B. anthracis (21, 4, and 1 assemblies labeled in NCBI as B. anthracis, B. cereus, and B. thuringiensis, respectively). Consistent with a lack of virulence genes, this cluster contained the genome of the avirulent strain B. anthracis Ames, which is commonly used in laboratory settings and does not possess B. anthracis plasmid pXO1 or pXO2 (72). All non-anthracis Bacillus assemblies in this group were isolated from either food or environmental sources, and all belonged to either panC clade III or IV.
Application of BTyper to identify assemblies associated with emetic B. cereus group isolates.
Assemblies possessing emetic toxin genes cesABCD were grouped into two clusters using k-medoids. Cluster 12 (n = 19; Fig. 4B) consisted of 19 assemblies classified as B. cereus in NCBI. All belonged to panC clade III, cesABCD were detected in all assemblies, and hblCDAB were not detected in any assemblies (Fig. 5). Included in this cluster was strain AH187, an isolate from the United Kingdom that was responsible for a 1972 emetic outbreak (Table 5). This isolate tested positive for emetic toxin (cereulide) formation and nonhemolytic enterotoxin (NHE) and negative for HBL hemolytic enterotoxin and cytotoxin K, and it belonged to MLST ST26 (Table 5) (73); these findings were confirmed using BTyper. Other notable strains in this cluster included (i) emetic strain B. cereus H3081.97, a B. cereus strain of sequence type 144 (ST144) which is closely related to strain AH187, and (ii) emetic strain B. cereus NC7401 (74).
TABLE 5.
B. cereus group assemblies in which emetic toxin genes cesABCD were detected
| Cluster | NCBI species classification | panC clade | GenBank accession no. | Strain | Isolate source (reference) |
|---|---|---|---|---|---|
| 12 | B. cereus | III | GCA_000021225.1 | AH187 | Vomit of a person who ate cooked rice; isolate was associated with an emetic outbreak in 1972 (73) |
| 12 | B. cereus | III | GCA_000161075.1 | BDRD-ST26 | BDRD stock strain (52)a |
| 12 | B. cereus | III | GCA_000171035.2 | H3081.97 | Food; emetic toxin-producing isolate from 1997 outbreak linked to rice, TX, USA (115) |
| 12 | B. cereus | III | GCA_000283675.1 | NC7401 | Emetic isolate (74) |
| 12 | B. cereus | III | GCA_000290935.2 | IS075 | Wild mammal (vole) (116) |
| 12 | B. cereus | III | GCA_000290995.1 | AND1407 | Black currant (53) |
| 12 | B. cereus | III | GCA_000291235.1 | MSX-A12 | Not available (107) |
| 12 | B. cereus | III | GCA_000399205.1 | IS845/00 | Bank vole, Poland (107, 117) |
| 12 | B. cereus | III | GCA_000399225.1 | IS195 | Bank vole, Poland (107, 117) |
| 12 | B. cereus | III | GCA_000743195.1 | F1-15 | Foodborne source (118) |
| 12 | B. cereus | III | GCA_001566375.1 | MB.15 | Food, Munich, Germany (119) |
| 12 | B. cereus | III | GCA_001566385.1 | MB.18 | Food, Munich, Germany (119) |
| 12 | B. cereus | III | GCA_001566435.1 | MB.16 | Food, Munich, Germany (119) |
| 12 | B. cereus | III | GCA_001566445.1 | MB.17 | Food, Munich, Germany (119) |
| 12 | B. cereus | III | GCA_001566455.1 | MB.21 | Food, Munich, Germany (119) |
| 12 | B. cereus | III | GCA_001566465.1 | MB.8 | Food, Munich, Germany (119) |
| 12 | B. cereus | III | GCA_001566515.1 | MB.8-1 | Food, Munich, Germany (119) |
| 12 | B. cereus | III | GCA_001566525.1 | MB.20 | Food, Munich, Germany (119) |
| 12 | B. cereus | III | GCA_001566535.1 | MB.22 | Food, Munich, Germany (119) |
| 24 | B. cereus | VI | GCA_000291155.1 | MC67 | Sandy loam, Møn, Denmark (75, 107, 120) |
| 24 | B. cereus | VI | GCA_000291315.1 | CER074 | Raw milk (53) |
| 24 | B. cereus | VI | GCA_000291335.1 | CER057 | Parsley (53) |
| 24 | B. cereus | VI | GCA_000293605.1 | BtB2-4 | Forest soil (53) |
| 24 | B. cereus | VI | GCA_000399245.1 | MC118 | Sandy loam, Møn, Denmark (75, 107, 120) |
BDRD, Biological Defense Research Directorate.
The other cluster in which all cesABCD genes were detected in all assemblies was cluster 24 (n = 5; Fig. 4B). This cluster contained 5 assemblies classified as B. cereus, all of which belonged to panC clade VI (Table 5). Unlike cluster 12, hblCDAB genes were detected in all assemblies in this cluster (Fig. 5). The assemblies in this cluster originated from food and environmental isolates (Table 5). Despite their assemblies being classified in the NCBI database as B. cereus, all 5 strains in this cluster were classified as emetic B. weihenstephanensis in their respective manuscripts, and all were capable of growth at 8°C (53, 75).
DISCUSSION
Accessible whole-genome sequence analysis tools can facilitate improved taxonomic classification and characterization of B. cereus group isolate virulence potential.
As whole-genome sequencing becomes more widely used in the realms of public health and food safety, the ability to classify potential pathogenic microorganisms quickly and effectively becomes increasingly important. A number of bioinformatics tools already exist for this purpose, including SRST2, which can be used to perform MLST and detect antimicrobial resistance genes using Illumina reads (54); SeqSero, which performs in silico serotyping using Illumina reads or nucleotide assemblies from Salmonella enterica isolates (76); PlasmidFinder, which can be used to detect plasmids in isolates using Illumina reads or nucleotide assemblies (77); and VirulenceFinder, which can be used to detect virulence genes in Listeria monocytogenes, Staphylococcus aureus, Escherichia coli, and Enterococcus (78). Recently, methods such as in silico MLST and virulence gene detection have been combined into single computational pipelines that can be used to characterize numerous bacterial species (79). Here, we have created a bioinformatics tool specific to the Bacillus cereus group that combines virulence gene detection using a curated database of B. cereus virulence factors with in silico manifestations of established molecular and virulence typing methods to phylogenetically classify and rapidly assess the virulence potential of any B. cereus group isolate. Additionally, we have provided a companion application, BMiner, that allows users to interact with data from hundreds of genomes at once, which we anticipate will become increasingly valuable as more B. cereus group genomes are sequenced.
The in silico typing methods employed by BTyper and other bioinformatics tools are valuable from a public health and food safety perspective, due to their (i) speed, as BTyper and similar tools can be used to perform gene detection and typing tasks in seconds using assembled genomes (76, 77); (ii) scalability, with the ability to provide users with information about a single isolate or hundreds from the command line (54, 76); and (iii) ability to output concise and easily interpretable summaries of large amounts of data (54), making it easy for a user to understand their results, share data with colleagues, and make informed decisions about an isolate in question (i.e., is it pathogenic or not). Additionally, the use of virulence gene-based typing as employed by BTyper offers the advantage that isolates can be classified according to their virulence potential, which means that one does not have to make any prior assumptions about the taxonomic classification of an isolate in question. This marks a valuable step forward in distinguishing pathogenic B. cereus group isolates from their nonpathogenic counterparts; however, marked improvements could be made to BTyper and similar tools through the integration of phenotypic data. By associating genotypic characteristics of B. cereus group isolates with phenotypic data, such as host illness and symptoms and growth temperature, BTyper and other tools used to genotype foodborne pathogens may become more valuable from a risk assessment perspective.
Analysis of publicly available B. cereus group assemblies using BTyper and BMiner identifies virulence gene-based clusters that capture phylogenetic heterogeneity in isolates with similar phenotypes.
Using the output of BTyper and BMiner, virulence gene profiles of 662 B. cereus group genomes were assigned to one of 31 clusters by employing a k-medoids approach, without making unnecessary prior assumptions about an assembly's taxonomic classification in the public domain. This allowed for the identification of several well-defined clusters with clinical or taxonomic relevance, including (i) fully virulent B. anthracis and B. anthracis-like B. cereus (cluster 1), (ii) capABCDE-negative anthrax-causing B. cereus strains (cluster 22), (iii) B. anthracis with attenuated virulence (clusters 3 and 4), (iv) 2 emetic clusters (clusters 12 and 24), and (v) B. cytotoxicus (cluster 31). The clustering of the emetic assemblies into 2 separate clusters reflected the observed heterogeneity among emetic strains of B. cereus and B. weihenstephanensis: Hoton et al. (53) described two distinct clusters formed by emetic toxin-producing B. cereus group strains, with psychrotolerant B. weihenstephanensis strains belonging to a distinct emetic cluster (referred to in its respective manuscript as cluster II) (53, 80). Assemblies from these strains were placed into a single cluster (k-medoids cluster 24) consisting of B. weihenstephanensis assemblies belonging to panC clade VI, while members of Hoton et al.'s emetic cluster I were placed into a second cluster (k-medoids cluster 12) containing assemblies belonging to panC clade III. For B. cytotoxicus, the two available assemblies, both of which were the only panC clade VII representatives, were placed into a single cluster composed of only themselves (k-medoids cluster 31), driven largely by their possession of cytK1, as described by Guinebretière et al. (40). For B. anthracis, strains possessing both anthrax virulence plasmids (pXO1 and pXO2) were assigned to cluster 1, distinguishing them from attenuated strains in which one or neither plasmid was detected, as well as B. cereus strains that caused anthrax-like disease (cluster 22). Despite lacking the polyglutamate capsule genes capABCDE, B. cereus strains in cluster 22 were able to cause anthrax-like symptoms using a second capsule encoded by B. cereus exopolysaccharide genes bpsXABCDEFGH (bpsX-H) on a different plasmid, pBC218 (81). The bpsX-H operon in its entirety was detected in 4 of the 5 anthrax-causing, capABCDE-negative B. cereus assemblies in cluster 22 (all but strain BcFL2013) and in no other cluster. It is likely that results like this from additional studies will be able to further resolve clade assignments and disease phenotypes with BTyper; recently, Bazinet identified numerous genes associated with phenotypic traits, such as anthrax and food poisoning (82). Here, we found associations between B. cereus group virulence genes and the panC clade, and virulence gene heterogeneity within disease phenotypes was identified. As more B. cereus group WGS and associated metadata become available, the potential for identifying new virulence alleles or phylogenetic markers that can further identify alleles or genes that are not only associated with a particular disease, but with specific symptoms or a clinical outcome using BTyper, becomes promising. For example, future work will be needed to better define specific genetic markers that can classify B. cereus group strains and clusters that are likely to cause diarrheal illnesses. Future epidemiological studies that assess the associations between different clusters and disease outcomes and symptoms will also provide an opportunity to further define and refine the types of disease outcomes and public health risks associated with different B. cereus group strains.
MATERIALS AND METHODS
Database construction.
To construct a virulence gene database specific to B. cereus group isolates, amino acid sequences from a total of 36 virulence genes (see Table S1 in the supplemental material) were collected from the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/). For an MLST database, the 7-gene MLST database for Bacillus cereus was downloaded from PubMLST (https://pubmlst.org/bcereus/). For panC typing, chromosomes of 45 B. cereus group strains were downloaded from the NCBI database (Table S2). panC genes were extracted from each strain using nucleotide BLAST (BLASTn) (83) and the panC genes of various B. cereus group type strains, and the online tool available at https://tools.symprevius.org/Bcereus/english.php was used to ensure that at least one representative from each of the seven panC clades was present in the collection (40) (Table S2). For rpoB allelic typing, the rpoB allelic type database created and curated by Cornell University's Food Safety Lab and Milk Quality Improvement Program (CUFSL/MQIP; Ithaca, NY) was used. While 16S rRNA gene typing is not performed by default (see “Construction of BTyper tool,” below), 16S rRNA gene typing can be performed using reference 16S rRNA gene sequences from nine different B. cereus group type strain genomes. To obtain these sequences, the 16S rRNA gene sequence from a cultured B. cereus type strain was downloaded from the Ribosomal Database Project (RDP) (84) and used in conjunction with BLASTn (83) to extract 16S rRNA gene genes from each of nine different B. cereus group species type strain genomes (Table S3). All database files can be downloaded from https://github.com/lmc297/BTyper.
Construction of BTyper tool.
BTyper was created with the following dependencies: Python version 2.7 (https://www.python.org/), Biopython version 1.6.8 (85), BLAST version 2.4.0 (83), SPAdes version 3.9.0 (86), and SRA toolkit version 2.8.0 (87, 88). The whole-genome sequences of 22 previously characterized B. cereus group isolates (30) were downloaded from the NCBI and used as a training set to optimize parameters (referred to here as the “training set”; Table S4). For virulence gene detection using translated nucleotide BLAST (tBLASTn) (83), default minimum coverage and minimum identity thresholds of 70 and 50%, were chosen, respectively, as they correlated highly with previously published PCR results (30), and the allele with the highest corresponding bit score was reported. For MLST, rpoB allelic typing, and panC clade typing, the highest-scoring allele in the respective database was selected using its associated BLAST bit score, with no minimum threshold applied (Fig. 1). Virulence gene detection, MLST, rpoB allelic typing, and panC clade typing methods were chosen to be performed by default, as these methods are valuable for their discriminatory power (30). 16S rRNA gene typing, although not performed by default due to its inability to discriminate between phylogenetic clades and species (34, 89, 90), was added as an option as well, as many users may be interested in this locus. For this method, the highest-scoring 16S rRNA gene of the nine type strain 16S rRNA genes was selected using its BLAST bit score, with no minimum threshold applied.
PCR detection of virulence genes.
To assess the accuracy of BTyper's in silico virulence gene detection, each of the 24 isolates in the validation set was screened for eight virulence genes (hblA, hblC, hblD, nheA, nheB, nheC, cytK, and entFM) using PCR. Bacterial DNA used as the template in PCRs was extracted by inoculating single colonies into 100 μl of sterile water; lysates were then heated at 95°C for 10 min in a thermocycler. For PCRs, 1 μl of dirty lysate was added to a master mix containing sterile water, 2× GoTaq Green master mix (Promega, Madison, WI), and primers at a concentration of 0.4 μM each (Table S5). The PCRs included an initial denaturation time of 3 min at 94°C, followed by 30 cycles of amplification; each cycle consisted of denaturation at 94°C for 30 s, annealing (see Table S5 for annealing temperatures) for 30 s, and elongation for 1 min at 72°C, with a final extension at 72°C for 7 min. PCR products were electrophoresed in 1% agarose gels, followed by ethidium bromide staining to confirm specific amplification. For isolates that did not yield a PCR amplicon for a given gene, the PCR was repeated at least once in order to confirm the negative PCR result.
MLST.
Multilocus sequence typing (MLST) was performed for all 24 isolates in the validation set using a 7-housekeeping-gene scheme available through the PubMLST website (https://pubmlst.org/bcereus/). The PCRs consisted of 1 μl of dirty lysate as the DNA template added to a master mix containing sterile water, 2× GoTaq Green master mix (Promega), and primers at a final concentration of 0.4 μM each. The PCR cycles included an initial denaturation (3 min at 94°C), followed by 20 cycles of denaturation (94°C for 30 s), annealing for 30 s with a touchdown scheme (annealing temperatures that decrease by 0.5°C per cycle, starting with 55°C and reaching 45°C at the last cycle), and elongation at 72°C for 45 s. The 20 cycles of touchdown PCR were followed by an additional 20 cycles using an annealing temperature of 45°C. A final extension at 72°C for 5 min was included at the end of the 40 cycles. After amplification, the PCR products were sequenced at the Biotechnology Resource Center (BRC; Cornell University, Ithaca, NY), and ATs and sequence types (STs; based on all 7 genes) were assigned using the PubMLST website. All isolates were submitted to the B. cereus PubMLST database (30).
rpoB allelic typing.
A 632-nucleotide (nt) internal sequence of rpoB, encoding the β-subunit of the RNA polymerase, was used for assigning rpoB allelic types (ATs), as described previously (11). The sequences of all rpoB ATs are available in the Food Microbe Tracker database (91).
Validation of BTyper using additional B. cereus group whole-genome sequences.
The genomes of 24 additional B. cereus group isolates were sequenced and assembled according to Miller et al. (referred to here as the “validation set”; Table S6) (6). BTyper was used to perform virulence gene detection, MLST, rpoB allelic typing, and panC clade typing on each draft genome using the chosen default settings (see “Construction of BTyper tool,” above). The same analyses were performed using the Illumina paired-end reads associated with each isolate, again using BTyper's default settings. To assess the accuracy of the panC clades assigned by BTyper, clade assignments provided by BTyper were compared to the isolates' whole-genome sequence clades provided by Kovac et al. (30) and Miller et al. (R. A. Miller, J. Jian, S. M. Beno, L. M. Carroll, M. Wiedmann, and J. Kovac, unpublished data) for the training and validation sets, respectively. A current version of the command line tool, as well as the curated virulence gene and rpoB allelic type databases, can be found at https://github.com/lmc297/BTyper. A link to a Web-based version of BTyper will also be made available at https://github.com/lmc297/BTyper at a later time.
Construction of BMiner companion application.
BMiner, a companion application for parsing, viewing, and analyzing multiple BTyper files in aggregate, was created with the following dependencies: R version 3.3.2 (92) and R packages shiny version 1.01 (93), ggplot2 version 2.2.1 (94), readr version 1.1.0 (95), stringr version 1.2.0 (96), vegan version 2.4-2 (97), plyr version 1.8.4 (98), dplyr version 0.5.0 (99), cluster version 2.0.6 (100), ggrepel version 0.6.5 (101), and magrittr version 1.5 (102). BMiner is freely available at https://github.com/lmc297/BMiner.
Application of BTyper and BMiner to whole-genome sequencing data.
The latest assembly versions for all (n = 651) B. cereus group genome assemblies available in GenBank were downloaded on 6 April 2017. Genome assemblies were assigned to one of nine taxa according to their GenBank classification: B. anthracis (n = 157), B. cereus s.s. (n = 343), B. cytotoxicus (n = 2), B. mycoides (n = 19), B. pseudomycoides (n = 2), B. thuringiensis (n = 93), B. toyonensis (n = 3), B. weihenstephanensis (n = 21), and B. wiedmannii (n = 11). BTyper was used to perform virulence gene detection, MLST, rpoB allelic typing, and panC clade typing on all 651 isolates, as well as an additional 11 isolates that were part of the validation set but did not have assemblies in the NCBI database at the time (total number of B. cereus group genomes, 662). All available metadata associated with each assembly's BioSample were downloaded from the NCBI (103). Data mining using BTyper results from all 662 B. cereus group assemblies was conducted using BMiner. The final results files for all 662 B. cereus group genome assemblies, as well as the associated metadata, can be found at https://github.com/lmc297/BTyper.
Post hoc statistical analyses.
Post hoc statistical analyses were conducted in R version 3.3.2 (92). Fisher's exact test was used to test for associations between virulence genes and panC-based phylogenetic clades using the fisher.test function in R's stats package (Table S7). Phylogenetic clades I and VII were excluded from this analysis, due to both being underrepresented among B. cereus group genomes in the NCBI database (12 and 2 isolates, respectively), while rare and common virulence genes present in fewer than 20 and more than n − 20 assemblies (where n corresponds to the total number of assemblies being tested), respectively, were also excluded. A Bonferroni correction was used to correct for multiple comparisons. To find members of the B. cereus group that clustered with B. anthracis isolates based on their virulence gene presence-absence profiles, as well as to assess within-species virulence heterogeneity, k-medoids clustering was performed using the clara function in R's cluster package (100) and a Euclidean distance metric. To find an optimum value for k, k-medoids clustering was performed for each value of k for 2 ≤ k ≤ (n − 1), where n is 662, the total number of assembled genomes. A k value of 31 was selected, as it corresponded to the largest average silhouette width.
Supplementary Material
ACKNOWLEDGMENTS
This material is based on work supported by the National Science Foundation Graduate Research Fellowship Program under grant no. DGE-1144153. Partial funding for this project was provided by the New York State Dairy Promotion Advisory Board through the New York State Department of Agriculture and Markets.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01096-17.
REFERENCES
- 1.Logan NA, Carman JA, Melling J, Berkeley RC. 1985. Identification of Bacillus anthracis by API tests. J Med Microbiol 20:1–19. doi: 10.1099/00222615-20-1-75. [DOI] [PubMed] [Google Scholar]
- 2.Guinebretière MH, Auger S, Galleron N, Contzen M, De Sarrau B, De Buyser ML, Lamberet G, Fagerlund A, Granum PE, Lereclus D, De Vos P, Nguyen-The C, Sorokin A. 2013. Bacillus cytotoxicus sp. nov. is a novel thermotolerant species of the Bacillus cereus group occasionally associated with food poisoning. Int J Syst Evol Microbiol 63:31–40. doi: 10.1099/ijs.0.030627-0. [DOI] [PubMed] [Google Scholar]
- 3.Lechner S, Mayr R, Francis KP, Pruss BM, Kaplan T, Wiessner-Gunkel E, Stewart GS, Scherer S. 1998. Bacillus weihenstephanensis sp. nov. is a new psychrotolerant species of the Bacillus cereus group. Int J Syst Bacteriol 48:1373–1382. doi: 10.1099/00207713-48-4-1373. [DOI] [PubMed] [Google Scholar]
- 4.Nakamura LK. 1998. Bacillus pseudomycoides sp. nov. Int J Syst Bacteriol 48:1031–1035. [DOI] [PubMed] [Google Scholar]
- 5.Jiménez G, Urdiain M, Cifuentes A, Lopez-Lopez A, Blanch AR, Tamames J, Kampfer P, Kolsto AB, Ramon D, Martinez JF, Codoner FM, Rossello-Mora R. 2013. Description of Bacillus toyonensis sp. nov., a novel species of the Bacillus cereus group, and pairwise genome comparisons of the species of the group by means of ANI calculations. Syst Appl Microbiol 36:383–391. doi: 10.1016/j.syapm.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 6.Miller RA, Beno SM, Kent DJ, Carroll LM, Martin NH, Boor KJ, Kovac J. 2016. Bacillus wiedmannii sp. nov., a psychrotolerant and cytotoxic Bacillus cereus group species isolated from dairy foods and dairy environments. Int J Syst Evol Microbiol 66:4744–4753. doi: 10.1099/ijsem.0.001421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. Anthrax. Centers for Disease Control and Prevention, Atlanta, GA. https://www.cdc.gov/anthrax/.
- 8.Stenfors Arnesen LP, Fagerlund A, Granum PE. 2008. From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiol Rev 32:579–606. doi: 10.1111/j.1574-6976.2008.00112.x. [DOI] [PubMed] [Google Scholar]
- 9.Lücking G, Stoeckel M, Atamer Z, Hinrichs J, Ehling-Schulz M. 2013. Characterization of aerobic spore-forming bacteria associated with industrial dairy processing environments and product spoilage. Int J Food Microbiol 166:270–279. doi: 10.1016/j.ijfoodmicro.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 10.Doll EV, Scherer S, Wenning M. 2017. Spoilage of microfiltered and pasteurized extended shelf life milk is mainly induced by psychrotolerant spore-forming bacteria that often originate from recontamination. Front Microbiol 8:135. doi: 10.3389/fmicb.2017.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ivy RA, Ranieri ML, Martin NH, den Bakker HC, Xavier BM, Wiedmann M, Boor KJ. 2012. Identification and characterization of psychrotolerant sporeformers associated with fluid milk production and processing. Appl Environ Microbiol 78:1853–1864. doi: 10.1128/AEM.06536-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong HA, Duc le H, Cutting SM. 2005. The use of bacterial spore formers as probiotics. FEMS Microbiol Rev 29:813–835. doi: 10.1016/j.femsre.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 13.Jiménez G, Blanch AR, Tamames J, Rossello-Mora R. 2013. Complete genome sequence of Bacillus toyonensis BCT-7112T, the active ingredient of the feed additive preparation toyocerin. Genome Announc 1(6):e01080-13. doi: 10.1128/genomeA.01080-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu K, Holzel CS, Cui Y, Mayer R, Wang Y, Dietrich R, Didier A, Bassitta R, Martlbauer E, Ding S. 2016. Probiotic Bacillus cereus strains, a potential risk for public health in China. Front Microbiol 7:718. doi: 10.3389/fmicb.2016.00718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jouzani GS, Valijanian E, Sharafi R. 2017. Bacillus thuringiensis: a successful insecticide with new environmental features and tidings. Appl Microbiol Biotechnol 101:2691–2711. doi: 10.1007/s00253-017-8175-y. [DOI] [PubMed] [Google Scholar]
- 16.Aceves-Diez AE, Estrada-Castaneda KJ, Castaneda-Sandoval LM. 2015. Use of Bacillus thuringiensis supernatant from a fermentation process to improve bioremediation of chlorpyrifos in contaminated soils. J Environ Manage 157:213–219. doi: 10.1016/j.jenvman.2015.04.026. [DOI] [PubMed] [Google Scholar]
- 17.Dash HR, Mangwani N, Das S. 2014. Characterization and potential application in mercury bioremediation of highly mercury-resistant marine bacterium Bacillus thuringiensis PW-05. Environ Sci Pollut Res Int 21:2642–2653. doi: 10.1007/s11356-013-2206-8. [DOI] [PubMed] [Google Scholar]
- 18.Armada E, Azcon R, Lopez-Castillo OM, Calvo-Polanco M, Ruiz-Lozano JM. 2015. Autochthonous arbuscular mycorrhizal fungi and Bacillus thuringiensis from a degraded Mediterranean area can be used to improve physiological traits and performance of a plant of agronomic interest under drought conditions. Plant Physiol Biochem 90:64–74. doi: 10.1016/j.plaphy.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 19.Wang G, Feng G, Snyder AB, Manns DC, Churey JJ, Worobo RW. 2014. Bactericidal thurincin H causes unique morphological changes in Bacillus cereus F4552 without affecting membrane permeability. FEMS Microbiol Lett 357:69–76. doi: 10.1111/1574-6968.12486. [DOI] [PubMed] [Google Scholar]
- 20.Lee H, Churey JJ, Worobo RW. 2009. Biosynthesis and transcriptional analysis of thurincin H, a tandem repeated bacteriocin genetic locus, produced by Bacillus thuringiensis SF361. FEMS Microbiol Lett 299:205–213. doi: 10.1111/j.1574-6968.2009.01749.x. [DOI] [PubMed] [Google Scholar]
- 21.Ohba M, Mizuki E, Uemori A. 2009. Parasporin, a new anticancer protein group from Bacillus thuringiensis. Anticancer Res 29:427–433. [PubMed] [Google Scholar]
- 22.Ammons DR, Short JD, Bailey J, Hinojosa G, Tavarez L, Salazar M, Rampersad JN. 2016. Anti-cancer parasporin toxins are associated with different environments: discovery of two novel parasporin 5-like genes. Curr Microbiol 72:184–189. doi: 10.1007/s00284-015-0934-3. [DOI] [PubMed] [Google Scholar]
- 23.Duc LH, Hong HA, Barbosa TM, Henriques AO, Cutting SM. 2004. Characterization of Bacillus probiotics available for human use. Appl Environ Microbiol 70:2161–2171. doi: 10.1128/AEM.70.4.2161-2171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huys G, Botteldoorn N, Delvigne F, De Vuyst L, Heyndrickx M, Pot B, Dubois JJ, Daube G. 2013. Microbial characterization of probiotics–advisory report of the working group “8651 Probiotics” of the Belgian Superior Health Council (SHC). Mol Nutr Food Res 57:1479–1504. doi: 10.1002/mnfr.201300065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenquist H, Smidt L, Andersen SR, Jensen GB, Wilcks A. 2005. Occurrence and significance of Bacillus cereus and Bacillus thuringiensis in ready-to-eat food. FEMS Microbiol Lett 250:129–136. doi: 10.1016/j.femsle.2005.06.054. [DOI] [PubMed] [Google Scholar]
- 26.Panel on Biological Hazards. 2016. Risks for public health related to the presence of Bacillus cereus and other Bacillus spp. including Bacillus thuringiensis in foodstuffs. EFSA J 14:e4524. doi: 10.2903/j.efsa.2016.4524. [DOI] [Google Scholar]
- 27.Tallent SM, Kotewicz KM, Strain EA, Bennett RW. 2012. Efficient isolation and identification of Bacillus cereus group. J AOAC Int 95:446–451. doi: 10.5740/jaoacint.11-251. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y, Lai Q, Goker M, Meier-Kolthoff JP, Wang M, Sun Y, Wang L, Shao Z. 2015. Genomic insights into the taxonomic status of the Bacillus cereus group. Sci Rep 5:14082. doi: 10.1038/srep14082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fox GE, Wisotzkey JD, Jurtshuk P Jr. 1992. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol 42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- 30.Kovac J, Miller RA, Carroll LM, Kent DJ, Jian J, Beno SM, Wiedmann M. 2016. Production of hemolysin BL by Bacillus cereus group isolates of dairy origin is associated with whole-genome phylogenetic clade. BMC Genomics 17:581. doi: 10.1186/s12864-016-2883-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swiecicka I, Van der Auwera GA, Mahillon J. 2006. Hemolytic and nonhemolytic enterotoxin genes are broadly distributed among Bacillus thuringiensis isolated from wild mammals. Microb Ecol 52:544–551. doi: 10.1007/s00248-006-9122-0. [DOI] [PubMed] [Google Scholar]
- 32.Pruss BM, Dietrich R, Nibler B, Martlbauer E, Scherer S. 1999. The hemolytic enterotoxin HBL is broadly distributed among species of the Bacillus cereus group. Appl Environ Microbiol 65:5436–5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ehling-Schulz M, Messelhausser U. 2013. Bacillus “next generation” diagnostics: moving from detection toward subtyping and risk-related strain profiling. Front Microbiol 4:32. doi: 10.3389/fmicb.2013.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caamaño-Antelo S, Fernandez-No IC, Bohme K, Ezzat-Alnakip M, Quintela-Baluja M, Barros-Velazquez J, Calo-Mata P. 2015. Genetic discrimination of foodborne pathogenic and spoilage Bacillus spp. based on three housekeeping genes. Food Microbiol 46:288–298. doi: 10.1016/j.fm.2014.08.013. [DOI] [PubMed] [Google Scholar]
- 35.Ko KS, Kim JW, Kim JM, Kim W, Chung SI, Kim IJ, Kook YH. 2004. Population structure of the Bacillus cereus group as determined by sequence analysis of six housekeeping genes and the plcR gene. Infect Immun 72:5253–5261. doi: 10.1128/IAI.72.9.5253-5261.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ko KS, Kim JM, Kim JW, Jung BY, Kim W, Kim IJ, Kook YH. 2003. Identification of Bacillus anthracis by rpoB sequence analysis and multiplex PCR. J Clin Microbiol 41:2908–2914. doi: 10.1128/JCM.41.7.2908-2914.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinez BA, Stratton J, Bianchini A. 2017. Isolation and genetic identification of spore-forming bacteria associated with concentrated-milk processing in Nebraska. J Dairy Sci 100:919–932. doi: 10.3168/jds.2016-11660. [DOI] [PubMed] [Google Scholar]
- 38.Miller RA, Kent DJ, Watterson MJ, Boor KJ, Martin NH, Wiedmann M. 2015. Spore populations among bulk tank raw milk and dairy powders are significantly different. J Dairy Sci 98:8492–8504. doi: 10.3168/jds.2015-9943. [DOI] [PubMed] [Google Scholar]
- 39.Guinebretière MH, Thompson FL, Sorokin A, Normand P, Dawyndt P, Ehling-Schulz M, Svensson B, Sanchis V, Nguyen-The C, Heyndrickx M, De Vos P. 2008. Ecological diversification in the Bacillus cereus group. Environ Microbiol 10:851–865. doi: 10.1111/j.1462-2920.2007.01495.x. [DOI] [PubMed] [Google Scholar]
- 40.Guinebretière MH, Velge P, Couvert O, Carlin F, Debuyser ML, Nguyen-The C. 2010. Ability of Bacillus cereus group strains to cause food poisoning varies according to phylogenetic affiliation (groups I to VII) rather than species affiliation. J Clin Microbiol 48:3388–3391. doi: 10.1128/JCM.00921-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Warda AK, Siezen RJ, Boekhorst J, Wells-Bennik MH, de Jong A, Kuipers OP, Nierop Groot MN, Abee T. 2016. Linking Bacillus cereus genotypes and carbohydrate utilization capacity. PLoS One 11:e0156796. doi: 10.1371/journal.pone.0156796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmid D, Rademacher C, Kanitz EE, Frenzel E, Simons E, Allerberger F, Ehling-Schulz M. 2016. Elucidation of enterotoxigenic Bacillus cereus outbreaks in Austria by complementary epidemiological and microbiological investigations, 2013. Int J Food Microbiol 232:80–86. doi: 10.1016/j.ijfoodmicro.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 43.Sorokin A, Candelon B, Guilloux K, Galleron N, Wackerow-Kouzova N, Ehrlich SD, Bourguet D, Sanchis V. 2006. Multiple-locus sequence typing analysis of Bacillus cereus and Bacillus thuringiensis reveals separate clustering and a distinct population structure of psychrotrophic strains. Appl Environ Microbiol 72:1569–1578. doi: 10.1128/AEM.72.2.1569-1578.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang Y, Yu XF, Zhan L, Chen JC, Zhang YY, Zhang JY, Chen HH, Zhang Z, Zhang YJ, Lu YY, Mei LL. 2017. Multilocus sequence type profiles of Bacillus cereus isolates from infant formula in China. Food Microbiol 62:46–50. doi: 10.1016/j.fm.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 45.Yang Y, Gu H, Yu X, Zhan L, Chen J, Luo Y, Zhang Y, Zhang Y, Lu Y, Jiang J, Mei L. 2017. Genotypic heterogeneity of emetic toxin producing Bacillus cereus isolates from China. FEMS Microbiol Lett 364:fnw237. doi: 10.1093/femsle/fnw237. [DOI] [PubMed] [Google Scholar]
- 46.Drewnowska JM, Swiecicka I. 2013. Eco-genetic structure of Bacillus cereus sensu lato populations from different environments in northeastern Poland. PLoS One 8:e80175. doi: 10.1371/journal.pone.0080175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tourasse NJ, Helgason E, Klevan A, Sylvestre P, Moya M, Haustant M, Okstad OA, Fouet A, Mock M, Kolsto AB. 2011. Extended and global phylogenetic view of the Bacillus cereus group population by combination of MLST, AFLP, and MLEE genotyping data. Food Microbiol 28:236–244. doi: 10.1016/j.fm.2010.06.014. [DOI] [PubMed] [Google Scholar]
- 48.Hoffmaster AR, Novak RT, Marston CK, Gee JE, Helsel L, Pruckler JM, Wilkins PP. 2008. Genetic diversity of clinical isolates of Bacillus cereus using multilocus sequence typing. BMC Microbiol 8:191. doi: 10.1186/1471-2180-8-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cardazzo B, Negrisolo E, Carraro L, Alberghini L, Patarnello T, Giaccone V. 2008. Multiple-locus sequence typing and analysis of toxin genes in Bacillus cereus food-borne isolates. Appl Environ Microbiol 74:850–860. doi: 10.1128/AEM.01495-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Böhm ME, Huptas C, Krey VM, Scherer S. 2015. Massive horizontal gene transfer, strictly vertical inheritance and ancient duplications differentially shape the evolution of Bacillus cereus enterotoxin operons hbl, cytK and nhe. BMC Evol Biol 15:246. doi: 10.1186/s12862-015-0529-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klee SR, Brzuszkiewicz EB, Nattermann H, Bruggemann H, Dupke S, Wollherr A, Franz T, Pauli G, Appel B, Liebl W, Couacy-Hymann E, Boesch C, Meyer FD, Leendertz FH, Ellerbrok H, Gottschalk G, Grunow R, Liesegang H. 2010. The genome of a Bacillus isolate causing anthrax in chimpanzees combines chromosomal properties of B. cereus with B. anthracis virulence plasmids. PLoS One 5:e10986. doi: 10.1371/journal.pone.0010986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zwick ME, Joseph SJ, Didelot X, Chen PE, Bishop-Lilly KA, Stewart AC, Willner K, Nolan N, Lentz S, Thomason MK, Sozhamannan S, Mateczun AJ, Du L, Read TD. 2012. Genomic characterization of the Bacillus cereus sensu lato species: backdrop to the evolution of Bacillus anthracis. Genome Res 22:1512–1524. doi: 10.1101/gr.134437.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hoton FM, Fornelos N, N′Guessan E, Hu X, Swiecicka I, Dierick K, Jaaskelainen E, Salkinoja-Salonen M, Mahillon J. 2009. Family portrait of Bacillus cereus and Bacillus weihenstephanensis cereulide-producing strains. Environ Microbiol Rep 1:177–183. doi: 10.1111/j.1758-2229.2009.00028.x. [DOI] [PubMed] [Google Scholar]
- 54.Inouye M, Dashnow H, Raven LA, Schultz MB, Pope BJ, Tomita T, Zobel J, Holt KE. 2014. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med 6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Helgason E, Tourasse NJ, Meisal R, Caugant DA, Kolsto AB. 2004. Multilocus sequence typing scheme for bacteria of the Bacillus cereus group. Appl Environ Microbiol 70:191–201. doi: 10.1128/AEM.70.1.191-201.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dai Z, Sirard JC, Mock M, Koehler TM. 1995. The atxA gene product activates transcription of the anthrax toxin genes and is essential for virulence. Mol Microbiol 16:1171–1181. doi: 10.1111/j.1365-2958.1995.tb02340.x. [DOI] [PubMed] [Google Scholar]
- 57.Candela T, Mock M, Fouet A. 2005. CapE, a 47-amino-acid peptide, is necessary for Bacillus anthracis polyglutamate capsule synthesis. J Bacteriol 187:7765–7772. doi: 10.1128/JB.187.22.7765-7772.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.NCBI. 2003. BioProject PRJNA299. Bacillus anthracis strain A2012, Endospore-forming bacteria that causes anthrax. https://www.ncbi.nlm.nih.gov/bioproject?LinkName=biosample_bioproject&from_uid=2435829.
- 59.Rückert C, Licht K, Kalinowski J, Espirito Santo C, Antwerpen M, Hanczaruk M, Reischl U, Holzmann T, Gessner A, Tiemann C, Grass G. 2012. Draft genome sequence of Bacillus anthracis UR-1, isolated from a German heroin user. J Bacteriol 194:5997–5998. doi: 10.1128/JB.01410-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Price EP, Seymour ML, Sarovich DS, Latham J, Wolken SR, Mason J, Vincent G, Drees KP, Beckstrom-Sternberg SM, Phillippy AM, Koren S, Okinaka RT, Chung WK, Schupp JM, Wagner DM, Vipond R, Foster JT, Bergman NH, Burans J, Pearson T, Brooks T, Keim P. 2012. Molecular epidemiologic investigation of an anthrax outbreak among heroin users, Europe. Emerg Infect Dis 18:1307–1313. doi: 10.3201/eid1808.111343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ågren J, Finn M, Bengtsson B, Segerman B. 2014. Microevolution during an anthrax outbreak leading to clonal heterogeneity and penicillin resistance. PLoS One 9:e89112. doi: 10.1371/journal.pone.0089112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lekota KE, Mafofo J, Madoroba E, Rees J, van Heerden H, Muchadeyi FC. 2015. Draft genome sequences of two South African Bacillus anthracis strains. Genome Announc 3:e01313-15. doi: 10.1128/genomeA.01313-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.NCBI. 2016. BioSample SAMN06270273. Vaccine-associated Bacillus anthracis. https://www.ncbi.nlm.nih.gov/biosample/SAMN06270273/.
- 64.Okinaka RT, Challacombe J, Drees K, Birdsell DN, Janke N, Naumann A, Seymour M, Hornstra H, Schupp J, Sahl J, Foster JT, Pearson T, Turnbull P, Keim P. 2014. Genome sequence of Bacillus anthracis STI, a Sterne-like Georgian/Soviet vaccine strain. Genome Announc 2(5):e00853-14. doi: 10.1128/genomeA.00853-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hoffmaster AR, Ravel J, Rasko DA, Chapman GD, Chute MD, Marston CK, De BK, Sacchi CT, Fitzgerald C, Mayer LW, Maiden MC, Priest FG, Barker M, Jiang L, Cer RZ, Rilstone J, Peterson SN, Weyant RS, Galloway DR, Read TD, Popovic T, Fraser CM. 2004. Identification of anthrax toxin genes in a Bacillus cereus associated with an illness resembling inhalation anthrax. Proc Natl Acad Sci U S A 101:8449–8454. doi: 10.1073/pnas.0402414101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gee JE, Marston CK, Sammons SA, Burroughs MA, Hoffmaster AR. 2014. Draft genome sequence of Bacillus cereus strain BcFL2013, a clinical isolate similar to G9241. Genome Announc 2(3):e00469-14. doi: 10.1128/genomeA.00469-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Johnson SL, Minogue TD, Teshima H, Davenport KW, Shea AA, Miner HL, Wolcott MJ, Chain PS. 2015. Finished genome sequence of Bacillus cereus strain 03BB87, a clinical isolate with B. anthracis virulence genes. Genome Announc 3(1):e01446-14. doi: 10.1128/genomeA.01446-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pena-Gonzalez A, Marston CK, Rodriguez RL, Kolton CB, Garcia-Diaz J, Theppote A, Frace M, Konstantinidis KT, Hoffmaster AR. 2017. Draft genome sequence of Bacillus cereus LA2007, a human-pathogenic isolate harboring anthrax-like plasmids. Genome Announc 5(16):e00181-17. doi: 10.1128/genomeA.00181-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rasko DA, Altherr MR, Han CS, Ravel J. 2005. Genomics of the Bacillus cereus group of organisms. FEMS Microbiol Rev 29:303–329. doi: 10.1016/j.fmrre.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 70.von Terzi B, Turnbull PC, Bellan SE, Beyer W. 2014. Failure of Sterne- and Pasteur-like strains of Bacillus anthracis to replicate and survive in the urban bluebottle blow fly Calliphora vicina under laboratory conditions. PLoS One 9:e83860. doi: 10.1371/journal.pone.0083860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cachat E, Barker M, Read TD, Priest FG. 2008. A Bacillus thuringiensis strain producing a polyglutamate capsule resembling that of Bacillus anthracis. FEMS Microbiol Lett 285:220–226. doi: 10.1111/j.1574-6968.2008.01231.x. [DOI] [PubMed] [Google Scholar]
- 72.NCBI. 2010. BioProject 57909. Bacillus anthracis strain Ames. https://www.ncbi.nlm.nih.gov/bioproject/57909.
- 73.NCBI. 2008. BioProject 17715. Bacillus cereus AH187 associated with an emetic outbreak in 1972. https://www.ncbi.nlm.nih.gov/bioproject/17715.
- 74.Takeno A, Okamoto A, Tori K, Oshima K, Hirakawa H, Toh H, Agata N, Yamada K, Ogasawara N, Hayashi T, Shimizu T, Kuhara S, Hattori M, Ohta M. 2012. Complete genome sequence of Bacillus cereus NC7401, which produces high levels of the emetic toxin cereulide. J Bacteriol 194:4767–4768. doi: 10.1128/JB.01015-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thorsen L, Hansen BM, Nielsen KF, Hendriksen NB, Phipps RK, Budde BB. 2006. Characterization of emetic Bacillus weihenstephanensis, a new cereulide-producing bacterium. Appl Environ Microbiol 72:5118–5121. doi: 10.1128/AEM.00170-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang S, Yin Y, Jones MB, Zhang Z, Deatherage Kaiser BL, Dinsmore BA, Fitzgerald C, Fields PI, Deng X. 2015. Salmonella serotype determination utilizing high-throughput genome sequencing data. J Clin Microbiol 53:1685–1692. doi: 10.1128/JCM.00323-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, Moller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Joensen KG, Scheutz F, Lund O, Hasman H, Kaas RS, Nielsen EM, Aarestrup FM. 2014. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol 52:1501–1510. doi: 10.1128/JCM.03617-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Thomsen MC, Ahrenfeldt J, Cisneros JL, Jurtz V, Larsen MV, Hasman H, Aarestrup FM, Lund O. 2016. A bacterial analysis platform: an integrated system for analyzing bacterial whole genome sequencing data for clinical diagnostics and surveillance. PLoS One 11:e0157718. doi: 10.1371/journal.pone.0157718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Castiaux V, N′Guessan E, Swiecicka I, Delbrassinne L, Dierick K, Mahillon J. 2014. Diversity of pulsed-field gel electrophoresis patterns of cereulide-producing isolates of Bacillus cereus and Bacillus weihenstephanensis. FEMS Microbiol Lett 353:124–131. doi: 10.1111/1574-6968.12423. [DOI] [PubMed] [Google Scholar]
- 81.Oh SY, Budzik JM, Garufi G, Schneewind O. 2011. Two capsular polysaccharides enable Bacillus cereus G9241 to cause anthrax-like disease. Mol Microbiol 80:455–470. doi: 10.1111/j.1365-2958.2011.07582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bazinet AL. 2017. Pan-genome and phylogeny of Bacillus cereus sensu lato. bioRXiv doi: 10.1101/119420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, Tiedje JM. 2014. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 42:D633–42. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cock PJ, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, de Hoon MJ. 2009. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kodama Y, Shumway M, Leinonen R, International Nucleotide Sequence Database Consortium . 2012. The Sequence Read Archive: explosive growth of sequencing data. Nucleic Acids Res 40:D54–D56. doi: 10.1093/nar/gkr854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Leinonen R, Sugawara H, Shumway M, International Nucleotide Sequence Database Consortium . 2011. The sequence read archive. Nucleic Acids Res 39:D19–D21. doi: 10.1093/nar/gkq1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rossi-Tamisier M, Benamar S, Raoult D, Fournier PE. 2015. Cautionary tale of using 16S rRNA gene sequence similarity values in identification of human-associated bacterial species. Int J Syst Evol Microbiol 65:1929–1934. doi: 10.1099/ijs.0.000161. [DOI] [PubMed] [Google Scholar]
- 90.Chen ML, Tsen HY. 2002. Discrimination of Bacillus cereus and Bacillus thuringiensis with 16S rRNA and gyrB gene based PCR primers and sequencing of their annealing sites. J Appl Microbiol 92:912–919. doi: 10.1046/j.1365-2672.2002.01606.x. [DOI] [PubMed] [Google Scholar]
- 91.Vangay P, Fugett EB, Sun Q, Wiedmann M. 2013. Food microbe tracker: a Web-based tool for storage and comparison of food-associated microbes. J Food Prot 76:283–294. doi: 10.4315/0362-028X.JFP-12-276. [DOI] [PubMed] [Google Scholar]
- 92.R Core Team 2016. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.r-project.org/. [Google Scholar]
- 93.Chang W, Cheng J, Allaire JJ, Xie Y, McPherson J. 2017. shiny: Web application framework for R. R package version 1.0.1. R Core Development Team, Vienna, Austria: https://cran.r-project.org/package=shiny. [Google Scholar]
- 94.Wickham H. 2009. Ggplot2: elegant graphics for data analysis. Springer, New York, NY. [Google Scholar]
- 95.Wickham H, Hester J, Francois R. 2017. readr: read rectangular text data. R Core Development Team, Vienna, Austria: https://cran.r-project.org/package=readr. [Google Scholar]
- 96.Wickham H. 2017. stringr: simple, consistent wrappers for common string operations. R Core Development Team, Vienna, Austria: https://cran.r-project.org/package=stringr. [Google Scholar]
- 97.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Henry M, Stevens H, Szoecs E, Wagner H. 2017. vegan: community ecology package. R Core Development Team, Vienna, Austria: https://cran.r-project.org/package=vegan. [Google Scholar]
- 98.Wickham H. 2011. The split-apply-combine strategy for data analysis. J Stat Softw 40:1–29. doi: 10.18637/jss.v040.i01. [DOI] [Google Scholar]
- 99.Wickham H, Francois R. 2016. dplyr: a grammar of data manipulation. R Core Development Team, Vienna, Austria: https://cran.r-project.org/package=dplyr. [Google Scholar]
- 100.Maechler M, Rousseeuw P, Struyf A, Hubert M, Hornik K, Studer M, Roudier P, Gonzalez J. 2017. cluster: “finding groups in data”: cluster analysis basics and extensions. R Core Development Team, Vienna, Austria: https://cran.r-project.org/web/packages/cluster/index.html. [Google Scholar]
- 101.Slowikowski K, Irisson J-O. 2016. ggrepel: repulsive text and label geoms for ‘ggplot2,’ v0.6.5. R Core Development Team, Vienna, Austria: https://cran.r-project.org/package=ggrepel. [Google Scholar]
- 102.Bache SM, Wickham H. 2014. magrittr: a forward-pipe operator for R. R Core Development Team, Vienna, Austria: https://cran.r-project.org/package=magrittr. [Google Scholar]
- 103.Barrett T, Clark K, Gevorgyan R, Gorelenkov V, Gribov E, Karsch-Mizrachi I, Kimelman M, Pruitt KD, Resenchuk S, Tatusova T, Yaschenko E, Ostell J. 2012. BioProject and BioSample databases at NCBI: facilitating capture and organization of metadata. Nucleic Acids Res 40:D57–D63. doi: 10.1093/nar/gkr1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.NCBI. 2009. BioProject 31307. Bacillus cereus 03BB102, clinical isolate. https://www.ncbi.nlm.nih.gov/bioproject/31307.
- 105.NCBI. 2015. BioProject 290051. Draft genome sequences of Bacillus species from the rhizosphere of the desert plant Rhazya stricta. https://www.ncbi.nlm.nih.gov/bioproject/290051. [DOI] [PMC free article] [PubMed]
- 106.NCBI. 2007. BioProject 19959. Bacillus cereus 03BB108 isolated from the same area as a pathogenic B. cereus strain. https://www.ncbi.nlm.nih.gov/bioproject/19959.
- 107.Van der Auwera GA, Feldgarden M, Kolter R, Mahillon J. 2013. Whole-genome sequences of 94 environmental isolates of Bacillus cereus sensu lato. Genome Announc 1(5):e00380-13. doi: 10.1128/genomeA.00380-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.NCBI. 2009. BioProject 29709. Bacillus thuringiensis serovar Monterrey BGSC 4AJ1 produces polyglutamate capsule. https://www.ncbi.nlm.nih.gov/bioproject/29709.
- 109.NCBI. 2016. BioSample SAMN04628222. Microbe sample from Bacillus thuringiensis serovar alesti strain BGSC 4C1. https://www.ncbi.nlm.nih.gov/biosample/SAMN04628222/.
- 110.NCBI. 2017. BioSample SAMN06242081. Pathogen: environmental/food/other sample for Bacillus cereus. https://www.ncbi.nlm.nih.gov/biosample/SAMN06242081.
- 111.NCBI. 2014. BioSample SAMN03025783. Microbe sample Bacillus thuringiensis Et10/1. https://www.ncbi.nlm.nih.gov/biosample/SAMN03025783.
- 112.NCBI. 2009. BioProject 29689. Bacillus cereus F65185 encapsulated wound isolate. https://www.ncbi.nlm.nih.gov/bioproject/29689.
- 113.NCBI. 2015. BioSample SAMN04000100. Microbe sample from Bacillus thuringiensis. https://www.ncbi.nlm.nih.gov/biosample/SAMN04000100.
- 114.NCBI. 2016. BioSample SAMN05608051. Pathogen: environmental/food/other sample from Bacillus cereus. https://www.ncbi.nlm.nih.gov/biosample/SAMN05608051.
- 115.University of Oslo. 2013. Full information on strain B. cereus H3081.97. University of Oslo, Oslo, Norway: http://mlstoslo.uio.no/cgi-bin/mlstdb/mlstdbnet5.pl?dbase=superdb&page=superstraininfo&file=bcereusgrp_isolates.xml&strain=%27H3081.97%27. [Google Scholar]
- 116.Ladeuze S, Lentz N, Delbrassinne L, Hu X, Mahillon J. 2011. Antifungal activity displayed by cereulide, the emetic toxin produced by Bacillus cereus. Appl Environ Microbiol 77:2555–2558. doi: 10.1128/AEM.02519-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Swiecicka I, De Vos P. 2003. Properties of Bacillus thuringiensis isolated from bank voles. J Appl Microbiol 94:60–64. doi: 10.1046/j.1365-2672.2003.01790.x. [DOI] [PubMed] [Google Scholar]
- 118.Zhong W, Shou Y, Yoshida TM, Marrone BL. 2007. Differentiation of Bacillus anthracis, B. cereus, and B thuringiensis by using pulsed-field gel electrophoresis. Appl Environ Microbiol 73:3446–3449. doi: 10.1128/AEM.02478-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Crovadore J, Calmin G, Tonacini J, Chablais R, Schnyder B, Messelhausser U, Lefort F. 2016. Whole-genome sequences of several strains of Bacillus cereus isolated from foodstuff or poisoning incidents. Genome Announc 4:e00435-16. doi: 10.1128/genomeA.00435-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hendriksen NB, Hansen BM, Johansen JE. 2006. Occurrence and pathogenic potential of Bacillus cereus group bacteria in a sandy loam. Antonie Van Leeuwenhoek 89:239–249. doi: 10.1007/s10482-005-9025-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.