ABSTRACT
Lysobacter species are a group of environmental bacteria that are emerging as a new source of antibiotics. One characteristic of Lysobacter is intrinsic resistance to multiple antibiotics, which had not been studied. To understand the resistance mechanism, we tested the effect of blocking two-component regulatory systems (TCSs) on the antibiotic resistance of Lysobacter enzymogenes, a prolific producer of antibiotics. Upon treatment with LED209, an inhibitor of the widespread TCS QseC/QseB, L. enzymogenes produced a large amount of an unknown metabolite that was barely detectable in the untreated culture. Subsequent structural elucidation by nuclear magnetic resonance (NMR) unexpectedly revealed that the metabolite was indole. Indole production was also markedly induced by adrenaline, a known modulator of QseC/QseB. Next, we identified two TCS genes, L. enzymogenes qseC (Le-qseC) and Le-qseB, in L. enzymogenes and found that mutations of Le-qseC and Le-qseB also led to a dramatic increase in indole production. We then chemically synthesized a fluorescent indole probe that could label the cells. While the Le-qseB (cytoplasmic response regulator) mutant was clearly labeled by the probe, the Le-qseC (membrane sensor) mutant was not labeled. It was reported previously that indole can enhance antibiotic resistance in bacteria. Therefore, we tested if the dramatic increase in the level of indole production in L. enzymogenes upon blocking of Le-qseC and Le-qseB would lead to enhanced antibiotic resistance. Surprisingly, we found that indole caused the intrinsically multiantibiotic-resistant bacterium L. enzymogenes to become susceptible. Point mutations at conserved amino acids in Le-QseC also led to antibiotic susceptibility. Because indole is known as an interspecies signal, these findings may have implications.
IMPORTANCE The environmental bacterium Lysobacter is a new source of antibiotic compounds and exhibits intrinsic antibiotic resistance. Here, we found that the inactivation of a two-component regulatory system (TCS) by an inhibitor or by gene deletion led to a remarkable increase in the level of production of a metabolite in L. enzymogenes, and this metabolite was identified to be indole. We chemically synthesized a fluorescent indole probe and found that it could label the wild type and a mutant of the TCS cytoplasmic response regulator but not a mutant of the TCS membrane sensor. Indole treatment caused the intrinsically multidrug-resistant bacterium L. enzymogenes to be susceptible to antibiotics. Mutations of the TCS sensor also led to antibiotic susceptibility. Because indole is known as an interspecies signal between gut microbiota and mammalian hosts, the observation that indole could render intrinsically resistant L. enzymogenes susceptible to common antibiotics may have implications.
KEYWORDS: indole, intrinsic antibiotic resistance, Lysobacter, natural products, two-component regulatory systems
INTRODUCTION
The search for new antibiotics is a pressing and continual need, as drug-resistant pathogens constantly emerge. Natural products are a main source of antibiotics, and Gram-positive bacteria of the Actinomycetes and filamentous fungi have been the primary sources of bioactive natural products for decades. In recent years, the Gram-negative bacterium Lysobacter is emerging as a new source of antibiotic natural products with new structural features and interesting modes of action (1, 2). For example, lysocins from Lysobacter sp. strain RH2180-5 are cyclic peptide antibiotics that exhibit a novel mode of action by binding to menaquinone in the cell membranes (3, 4). Lysobactin (katanosin B) from Lysobacter sp. strain ATCC 53042 is a potent antibacterial cyclic depsipeptide that binds to the reducing end of lipid-linked cell wall precursors and thus blocks peptidoglycan biosynthesis of the cell wall (5–7). WAP-8294A from Lysobacter sp. strain WAP-8294 and Lysobacter enzymogenes OH11 is a family of cyclic lipodepsipeptides with potent activity against methicillin-resistant Staphylococcus aureus (8–10). Tripropeptins from Lysobacter sp. strain BMK333-48F3 are another group of cyclic lipodepsipeptides that inhibit the lipid cycle of bacterial cell wall biosynthesis by forming a complex with Ca2+ and undecaprenyl pyrophosphate (11, 12). Cephabacins from Lysobacter lactamgenus contain a cephem core attached to an oligopeptide side chain (13–15). Heat-stable antifungal factor (HSAF) and analogs from L. enzymogenes strains C3 and OH11 are a group of polycyclic tetramate macrolactams with potent antifungal activity and a distinct mode of action (16–24). Phenazines from Lysobacter antibioticus OH13 are broad-spectrum antibiotics, including the well-known antibiotic myxin (25).
As ubiquitous environmental Gram-negative bacteria, Lysobacter species have two distinct characteristics. One is their unusual richness in peptide metabolites, and the other is their intrinsic resistance to multiple antibiotics. For the former characteristic, we and others have started to look into molecular mechanisms for the production and regulation of bioactive natural products. Several Lysobacter genomes have been sequenced, and most of these genomes contain multiple gene clusters (12 to 16 clusters) for natural product biosynthesis (2, 26). Unlike gene clusters of Gram-positive Streptomyces species that are also rich in bioactive natural products, most of the Lysobacter gene clusters are predicted to synthesize peptides. Lysobacter species are considered “peptide production specialists” (2). In addition, recent studies have shown that the production of antibiotic compounds is regulated by endogenous and/or xenogenous small molecules (27–31). For example, L. enzymogenes diffusible signaling factor 1 (LeDSF1) to LeDSF5, a group of fatty acids, were identified in L. enzymogenes strain OH11 and found to act as DSFs to regulate the biosynthesis of HSAF (27). The signaling is mediated by a two-component regulatory system (TCS), L. enzymogenes RpfC (Le-RpfC)/Le-RpfG, which is responsible for sensing the DSF and for conveying the signal for subsequent gene expression and HSAF production. Independent of DSF signaling, 4-hydroxbenzoic acid was recently shown to serve as a diffusible factor (DF) to regulate HSAF biosynthesis through a LysR family transcription factor (LysRLe) (28–31). Interestingly, PilS/PilR, another TCS that is known to regulate T4P (type IV pilus) production in many bacteria, was unexpectedly found to play a role in HSAF regulation in L. enzymogenes, and signaling was conveyed by the second messenger cyclic di-GMP (c-di-GMP) (32).
In contrast to the biosynthesis and regulation of natural products, essentially nothing is known about the molecular mechanism(s) underlying intrinsic multidrug resistance in Lysobacter. An understanding of the mechanism(s) is important for the better utilization of this largely underexplored source of bioactive natural products. In this work, we serendipitously found that a simple molecule, indole, is able to reverse the antibiotic resistance of Lysobacter through a QseC (quorum sensing Escherichia coli regulator C)/QseB-like TCS. QseC/QseB was originally found in E. coli (33, 34). QseC is the membrane-bound sensor protein of the TCS with histidine kinase activity, and QseB is the cytoplasmic response regulator that regulates the expression of downstream target genes. QseC/QseB homologs are present in many important human and plant pathogens and are important for the virulence of pathogens (35–37). In enterohemorrhagic E. coli, QseC/QseB regulates multiple virulence factors and is known to respond to human epinephrine/norepinephrine and bacterial AI-3 (autoinducer 3), a group of mysterious signals whose identity is yet to be discovered (35–40). QseC is considered a target for new antibiotic development, and a potent QseC inhibitor, LED209, was identified (35, 41, 42). Through QseC/QseB signaling, AI-3 signals regulate the expression of enterocyte effacement, Shiga toxin, and flagellum and motility genes in enterohemorrhagic E. coli (39). In this study, we describe the identification of indole as a small-molecule signal for Le-QseC/Le-QseB in L. enzymogenes OH11 and present evidence for the involvement of this signaling system in intrinsic multiantibiotic resistance.
RESULTS
Treatment of L. enzymogenes OH11 with the QseC inhibitor LED209 induces a dramatic increase in indole production.
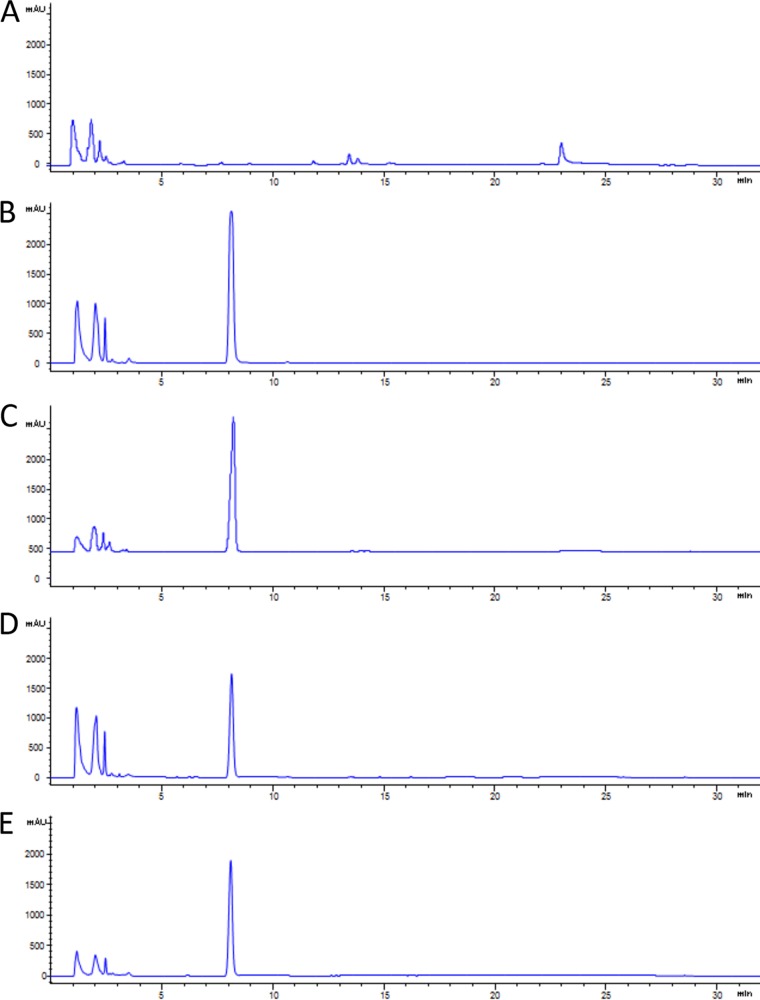
In bacteria, the TCS is the basic system for sensing and responding to environmental stimuli and signaling factors (43). Since antibiotic treatment could be regarded as a type of environmental stress to bacteria, we were curious about the potential consequences of blocking TCSs in L. enzymogenes. Because of the widespread occurrence of QseC/QseB and its importance in pathogenic bacteria, we chose to test LED209, a small synthetic compound that exhibited potent inhibitory activity against the binding of signals to QseC (35). After LED209 treatment, we examined the metabolite profile of L. enzymogenes OH11. The level of a new metabolite with a 7.8-min retention time upon high-performance liquid chromatography (HPLC), which was barely detectable in untreated OH11 cells, was dramatically increased in OH11 cells treated with LED209 (10 pM) (Fig. 1). We purified the compound from a scale-up culture and elucidated its chemical structure using nuclear magnetic resonance (NMR) (see Fig. S1 in the supplemental material). 1H-NMR (400 MHz, CDCl3) gave the following signals: δ 8.28 (s, 1H), 7.67 (d, J = 7.9 Hz, 1H), 7.42 (d, J = 8.1 Hz, 1H), 7.22 (m, 2H), 7.14 (t, J = 7.5 Hz, 1H), and 6.58 (s, 1H) ppm. 13C-NMR (100 MHz, CDCl3) gave the following signals: δ 135.81, 127.86, 124.13, 121.94, 120.71, 119.76, 111.02, and 102.57 ppm. The NMR data indicate that this compound is indole, which was confirmed by comparison with the spectra and HPLC data for standard indole.
FIG 1.
Indole production in Lysobacter enzymogenes OH11. (A) Wild-type strain OH11; (B) strain OH11 treated with LED209 (10 pM); (C) strain OH11 treated with epinephrine (5 mM); (D) Le-qseC deletion mutant; (E) Le-qseB deletion mutant.
The dramatic induction of indole by LED209 is intriguing, because indole and some of its derivatives are signal molecules involved in biofilm formation, virulence, motility, and antibiotic tolerance (44). The concentration of indole can reach ∼100 μM in the human gastrointestinal (GI) tract and 250 to 1,100 μM in human feces (45, 46). Cultured commensal E. coli cells can produce as much as 600 μM indole (47). For L. enzymogenes OH11, we estimated that upon LED209 treatment, the concentration of indole increased over 1,300-fold (from approximately 0.3 μM to about 410 μM). We were able to obtain ∼30 mg indole from 1 liter of culture (corresponding to approximately 256 μM), confirming the drastic induction of indole production by the QseC inhibitor LED209. Finally, one hallmark of the QseC/QseB-like TCSs is that they respond to adrenergic signals such as epinephrine (adrenaline). Indeed, treatment with epinephrine also led to a remarkable induction of indole production in L. enzymogenes OH11 (Fig. 1).
Deletion of Le0754-Le0752 in L. enzymogenes OH11 also leads to a large increase in indole production.
The remarkable induction of indole by the QseC inhibitor LED209 suggests that indole signaling may involve a QseC/QseB-like TSC in L. enzymogenes. Among the genes coding for putative TCSs in L. enzymogenes (32), we found that Le0754-Le0752 showed the highest similarity to genes for QseC/QseB of E. coli (strain K-12 and enterohemorrhagic strain O157:H7). Le0754 shares 31.1% identity and 43.3% similarity to QseC of E. coli, and Le0752 has 47.5% identity and 62.0% similarity to QseB of E. coli (see Fig. S2 in the supplemental material). We thus use the names Le-qseC and Le-qseB for Le0754 and Le0752, respectively, in this study. To test if this TSC plays a role in indole signaling, we examined the expression of the Le-qseC gene and the Le-qseB gene upon exogenous indole treatment or LED209 treatment. The results showed that indole slightly upregulated Le-qseB and Le-qseC, while LED209 clearly downregulated these two genes (Fig. S3). These results indicate that the induction and subsequent accumulation of indole by LED209 do not appear to have a significant effect on the gene expression of Le-qseB and Le-qseC.
To test whether indole signaling is mediated by Le-QseC/Le-QseB, we deleted the Le-qseC gene and the Le-qseB gene in OH11. Similarly to LED209 treatment, the deletion of this TCS also led to a remarkable increase in the level of indole production (estimated at 267 μM for the Le-qseC mutant and 323 μM for the Le-qseB mutant) (Fig. 1). These results clearly demonstrate that the loss of function of the Le-QseC/Le-QseB TCS leads to indole induction in OH11.
Fluorescently labeled indole binds to wild-type L. enzymogenes and the Le-QseB mutant but not to the Le-QseC mutant.
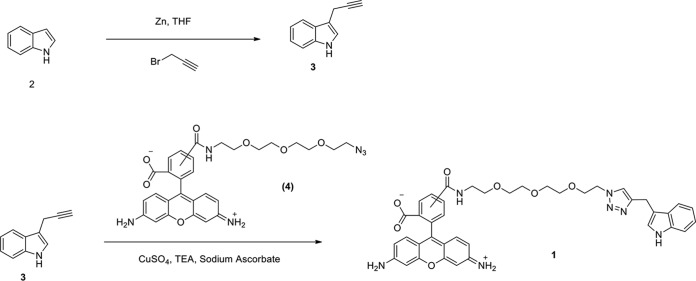
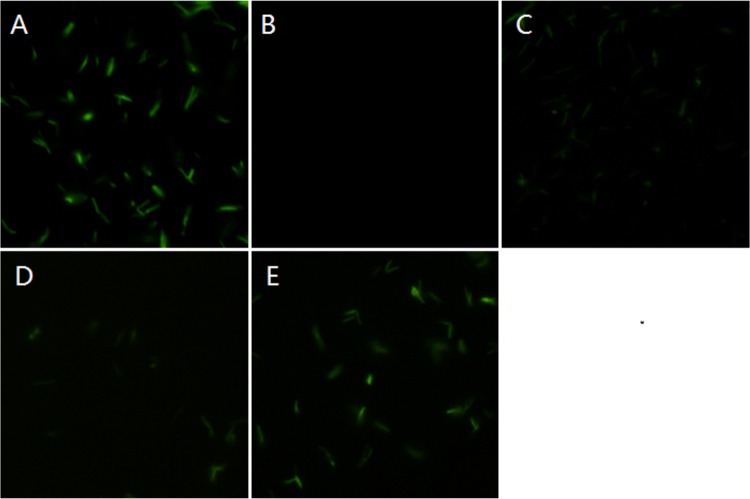
To obtain evidence for the involvement of Le-QseC/Le-QseB in indole signaling, we chemically synthesized a fluorescent indole probe (compound 1) in which indole was linked to the fluorophore Azide-fluor 488 (from Sigma-Aldrich; compound 4) using click chemistry (Fig. 2). When this probe was incubated with L. enzymogenes OH11 cells, wild-type cells and Le-qseB deletion mutant cells were clearly labeled with green fluorescence, while Le-qseC deletion mutant cells were not labeled (Fig. 3). As controls, L. enzymogenes cells gave no fluorescence in the absence of the probe or when the cells were incubated with the fluorophore alone (compound 4). These results suggest that indole binds to Le-QseC, the membrane sensor of the TCS, but not to Le-QseB, the cytoplasmic response regulator of the TCS.
FIG 2.
Chemical synthesis of a fluorescent indole probe (compound 1). Indole (compound 2) was linked to the fluorophore Azide-fluor 488 (compound 4) by using click chemistry via an alkyne intermediate (compound 3).
FIG 3.
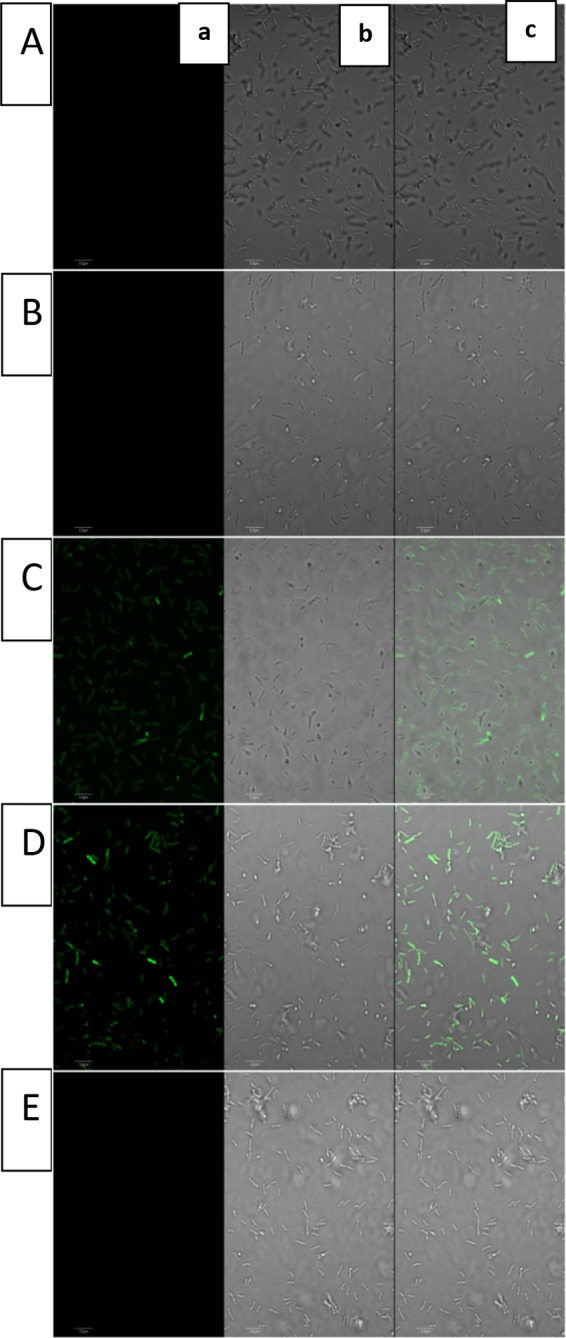
Fluorescence imaging of Lysobacter enzymogenes OH11, the Le-qseB deletion mutant, and the Le-qseC deletion mutant. (A) Strain OH11; (B) strain OH11 treated with compound 4 (the fluorophore alone); (C) strain OH11 treated with compound 1 (the fluorophore with the indole probe); (D) Le-qseB deletion mutant treated with compound 1; (E) Le-qseC deletion mutant treated with compound 1. Each of the picture panels shows a picture taken at a 488-nm excitation wavelength and a 509-nm emission wavelength (a), a picture taken under transmitted light (b), and a merged image of panels a and b (c).
To obtain more evidence, we generated four other Le-qseC mutants, each of which contained a single amino acid change at four conserved residues within the N-terminal ligand-binding domain (but not the C-terminal kinase domain [see Fig. S2 in the supplemental material]). While the wild type clearly showed fluorescence, the Le-qseC D85V, Le-qseC F126A, and Le-qseC F154A single point mutants gave barely detectable fluorescence signals when incubated with the indole probe (Fig. 4). On the other hand, the Le-qseC W164E mutant exhibited fluorescence similar to that of the wild type. Due to the lack of structural information on Le-QseC, it is difficult to predict the potential contributions of these conserved amino acid residues to the function of Le-QseC. Nonetheless, these data support that the binding of indole to L. enzymogenes cells is Le-QseC dependent.
FIG 4.
Fluorescence imaging of Lysobacter enzymogenes OH11 and Le-qseC single-amino-acid mutants. OH11 and the mutants were treated with compound 1 (indole fluorophore probe), and images of the cells were taken at a 488-nm excitation wavelength and a 509-nm emission wavelength. (A) Wild-type strain OH11; (B) Le-qseC D85V mutant; (C) Le-qseC F126A mutant; (D) Le-qseC F154A mutant; (E) Le-qseC W164E mutant.
Indole reverses antibiotic resistance in L. enzymogenes OH11.
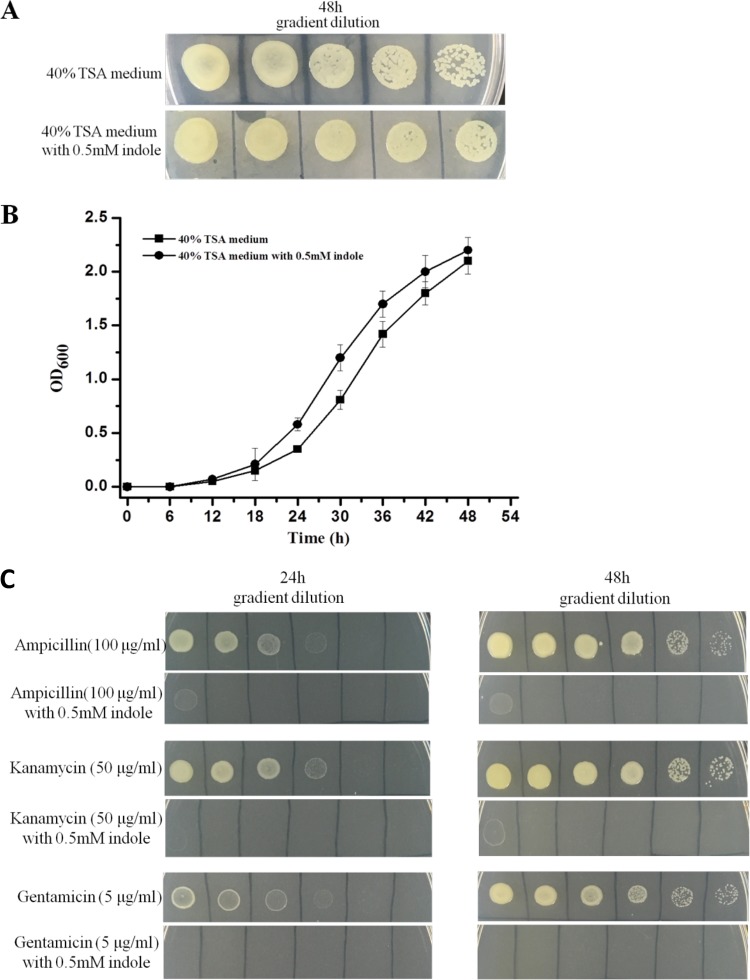
To evaluate potential physiological consequences of Le-QseC/Le-QseB-mediated indole signaling, we examined antibiotic resistance in L. enzymogenes OH11, because this is one of the distinct characteristics of Lysobacter species. First, we checked whether exogenous indole would have a toxic effect on OH11 growth. In the absence of an antibiotic, indole (0.5 mM) exhibited no obvious effect on OH11 growth in both solid culture and liquid culture (Fig. 5). In contrast, indole almost completely stopped the growth of OH11 in the presence of several common antibiotics, although OH11 is intrinsically resistant to these antibiotics (Fig. 5). These results show that indole treatment causes a multiantibiotic-resistant Lysobacter strain to become susceptible.
FIG 5.
Effect of indole on antibiotic resistance of L. enzymogenes OH11. (A and B) Indole exhibits little effect on the growth of L. enzymogenes OH11 in solid (A) and liquid (B) media. OD600, optical density at 600 nm. (C) In contrast, indole nearly completely stopped the growth of L. enzymogenes OH11 in the presence of several antibiotics, although Lysobacter species are intrinsically resistant to these antibiotics.
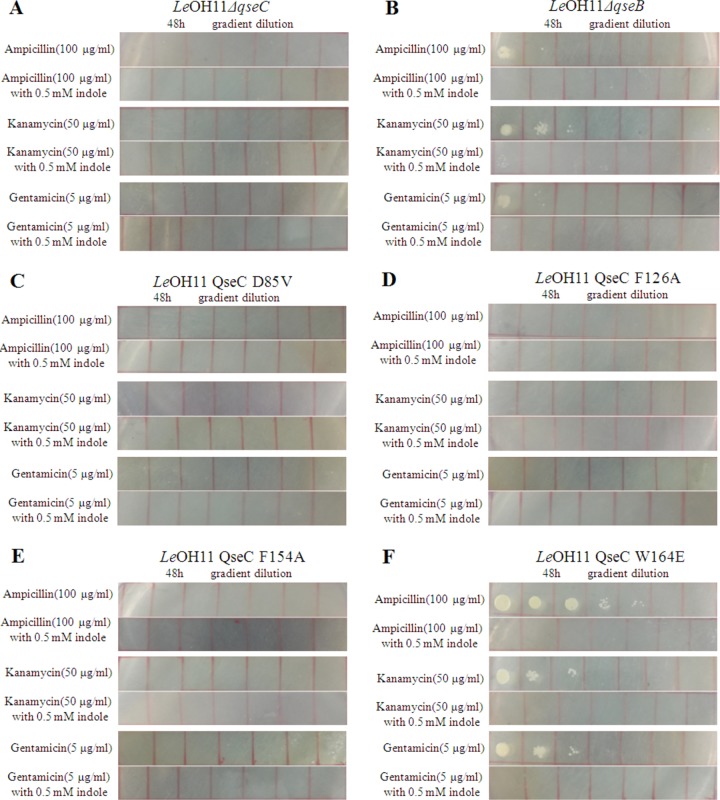
To further look into the reversed antibiotic resistance, we examined the TCS Le-qseC–Le-qseB mutants, including gene deletion mutants and single point mutants. While the wild type grew normally in medium containing ampicillin (100 μg/ml), kanamycin (50 μg/ml), or gentamicin (5 μg/ml), the Le-qseC deletion mutant was unable to grow under the same conditions, regardless of the presence or absence of indole (Fig. 6). This confirms the involvement of the Le-QseC sensor of the TCS in antibiotic susceptibility. In contrast, the Le-qseB deletion mutant was able to grow, albeit more slowly than the wild type, in medium containing ampicillin, kanamycin, or gentamicin, in the absence of indole (Fig. 6). However, in the presence of indole, the Le-qseB deletion mutant became totally susceptible to the antibiotics. This is consistent with data from the fluorescent indole binding experiments, which showed that the Le-QseB response regulator of the TCS does not directly interact with indole. As long as the Le-QseC sensor is functional, indole signaling could be transmitted to the cells, perhaps via an alternative response regulator of the 54 putative TCSs found in OH11 (32). This could explain why the Le-qseB deletion mutant grew more slowly than the wild type in the absence of indole, probably due to a less optimal cytoplasmic response regulator for the membrane sensor Le-QseC.
FIG 6.
Effect of mutations in Le-qseC–Le-qseB on the antibiotic resistance of L. enzymogenes OH11 in the absence or presence of indole. (A) Le-qseC deletion mutant; (B) Le-qseB deletion mutant; (C) Le-qseC D85V mutant; (D) Le-qseC F126A mutant; (E) Le-qseC F154A mutant; (F) Le-qseC W164E mutant.
To obtain more evidence, we also tested the Le-QseC single-amino-acid mutants (Fig. 6). As expected, no growth was observed for the Le-qseC D85V mutant, the Le-qseC F126A mutant, or the Le-qseC F154A mutant in medium containing ampicillin, kanamycin, or gentamicin, with or without indole. In contrast, the Le-qseC W164E mutant grew nearly as well as the wild type in medium containing the antibiotics but without indole. However, when indole was added to the medium, the Le-qseC W164E mutant could not grow in medium containing any of the antibiotics (Fig. 6). These results are in agreement with those from fluorescent indole binding experiments with the Le-qseC single-amino-acid mutants.
Indole stimulates HSAF production.
Because L. enzymogenes is known to produce several antibiotic metabolites, we examined indole's effect on the biosynthesis of HSAF, the major antifungal metabolite in this strain. The exogenous addition of indole markedly increased the production of HSAF and its analogs in L. enzymogenes (see Fig. S6 in the supplemental material). These results suggest that HSAF biosynthesis is affected by indole signaling.
DISCUSSION
Indole signaling in bacteria has been investigated by several groups. Early studies reported that SdiA, a quorum sensing regulator of E. coli, mediated indole signaling, in addition to sensing acyl homoserine lactones (44, 48, 49). Indole was shown to induce multidrug exporter gene expression, and its signaling was transmitted via BaeSR/CpxAR, another TCS in E. coli (50). A more recent study found that while indole had effects on gene expression and biofilm formation, these effects were not dependent on SdiA in E. coli and Salmonella enterica serovar Typhimurium (51). Furthermore, indole was shown to be freely diffusible across bacterial membranes (52). Thus, it was not entirely clear whether indole signaling would require a sensor/receptor protein and, if so, whether the protein would be a TCS-type sensor.
In this study, we found that LED209, an inhibitor of the TCS sensor QseC, and epinephrine, a known modulator of QseC, dramatically induced indole production in L. enzymogenes. We further discovered that the deletion of the Le-qseC gene or the Le-qseB gene in L. enzymogenes also remarkably stimulated indole production. Using a chemically synthesized fluorescent indole probe, we obtained evidence to support that indole could bind to the sensor protein Le-QseC in L. enzymogenes. The QseC/QseB signaling system is very important for the virulence and motility of enterohemorrhagic E. coli, which uses epinephrine/norepinephrine and AI-3 as signal molecules (35–40). However, the nature of this so-called AI-3 remains unclear, other than that AI-3 was suggested to be a group of bacterial aromatic compounds. The QseC inhibitor LED209 reduces the in vivo virulence of enterohemorrhagic E. coli and Salmonella enterica serovar Typhimurium. Targeted blocking of QseC activity has been proposed as a strategy against Gram-negative pathogens (42).
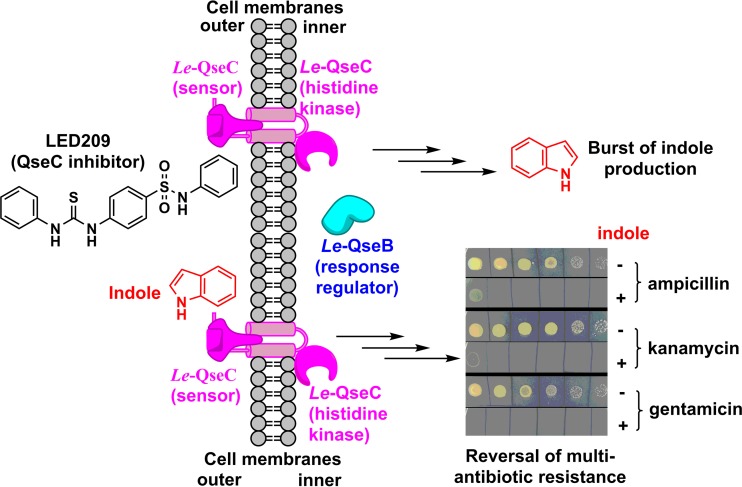
In contrast to pathogenic E. coli and Salmonella enterica, L. enzymogenes is a nonpathogenic environmental bacterium. Its QseC/QseB-like system is not expected to be associated with virulence, and our results show that this TSC is involved in antibiotic resistance (Fig. 7). In E. coli, indole was shown to serve as a signal to turn on drug efflux pumps and oxidative stress-protective mechanisms, which conferred population-based antibiotic resistance (53). Indole signaling also induced E. coli persistence through activating stress responses in the bacterial community (54). As an interspecies signal, indole caused Salmonella to become less susceptible to antibiotics (55). In the presence of antibiotics, E. coli cells produced a higher level of indole that enhanced cell survival during antibiotic stress (56). It appears that indole generally enhances antibiotic resistance in these pathogenic bacteria. Therefore, in light of the dramatic increase in indole production in L. enzymogenes upon blocking of Le-QseC or mutation of Le-qseC–Le-qseB, our first instinct was to expect enhanced antibiotic resistance. However, to our surprise, we found that indole caused the intrinsically multidrug resistant bacterium L. enzymogenes to become susceptible to several common antibiotics. Similarly, the Le-qseC–Le-qseB mutants also became antibiotic susceptible. In the absence of an antibiotic, indole itself did not show toxicity to the growth of L. enzymogenes.
FIG 7.
Indole-mediated reversal of multiantibiotic resistance in L. enzymogenes OH11. The two-component regulatory system Le-QseC/Le-QseB is involved in this process.
Indole has been shown to induce the production of efflux pumps in both E. coli and Salmonella (53, 57). The change in efflux pump activity could be a main reason for the observed change in antibiotic resistance in these bacteria. However, indole's effect is to increase antibiotic resistance in the above-described pathogenic bacteria. For the environmental bacterium L. enzymogenes, we found the opposite effect, that indole caused this intrinsically resistant bacterium become susceptible (Fig. 7). This may suggest that the direction of antibiotic transportation by the indole-induced efflux pumps in L. enzymogenes is probably the opposite of that in the pathogenic bacteria described above. The increase in the level of indole may lead to enhanced antibiotic transport from outside to inside the cells so that an inhibitory concentration of the antibiotic could be reached. Whether this is a feature specific only to the environmental Gram-negative bacterium Lysobacter or a property present in broader microorganisms is worth further investigation, particularly for members of the gut microbiota, because they are known to produce a large amount of indole and to be involved in diseases in humans (45–47). In addition, QseC homologs are present in many important human and plant pathogens, and the QseC/QseB signaling pathway is important for the virulence of these pathogens. Thus, the finding that indole could cause an antibiotic-resistant bacterium to be antibiotic susceptible has important implications in terms of understanding the pathology as well as developing potentially novel approaches toward fighting against multidrug-resistant superbugs, particularly Gram-negative pathogens, against which there are very few weapons available.
Finally, Le-QseC/Le-QseB is the third TCS among the 54 putative TCSs whose signal molecules have been identified (27, 32). As Lysobacter species produce a large number of bioactive natural products and their regulatory mechanism remains largely unexplored (1, 2), it will be interesting to test if Le-QseC/Le-QseB-mediated indole signaling could also play a role in regulating antibiotic production by Lysobacter.
MATERIALS AND METHODS
Bacterial strains, plasmids, and general methods.
Escherichia coli strains DH5α and S17-1 were used as the hosts for general DNA manipulation and for conjugation, respectively. Lysobacter enzymogenes OH11 (CGMCC 1978) and the derived mutants were grown in 40%-strength tryptic soy broth (TSB). Plasmid miniprep and DNA fragment extraction were performed according to the instructions of the kits from Qiagen. All other manipulations were carried out according to methods described previously (21). PCR primers were synthesized by Eurofins MWG Operon (distributed by Fisher Scientific). Restriction enzymes and other molecular biology reagents were purchased from NEB. LED209 was purchased from Santa Cruz Biotechnology, Azide-fluor 488 was obtained from Sigma-Aldrich, and all other general chemicals were purchased from Sigma-Aldrich or Fisher Scientific.
Isolation and structural determination of a new signal molecule in L. enzymogenes OH11.
L. enzymogenes OH11 was grown in 40% TSB (50 ml, containing 5 pM LED209) at 28°C at 200 rpm for 3 days. The culture was extracted with the same volume of ethyl acetate. The organic phase was concentrated under a vacuum, and the residues were dissolved with 0.5 ml methanol (MeOH) to afford the crude extract. An aliquot (20 μl) of the crude extract was injected into an HPLC instrument to analyze the metabolite prolife of the culture containing LED209, in comparison with that of the control culture. The HPLC instrument (1120 system; Agilent, Santa Clara, CA) was equipped with a reverse-phase (RP) C18 column (4.6 by 150 mm, 3.5 μm), using the solvent system described in Table S1 in the supplemental material. This led to the identification of a new peak that was dramatically increased upon LED209 treatment. To identify this new compound, the culture was scaled up to 1 liter (40% TSB, 5 pM LED209) and grown at 28°C at 200 rpm for 3 days. The culture was filtered, and the filtrate was extracted with 1 liter of ethyl acetate. The organic solution was collected and concentrated by using a Rotavapor instrument. The crude extract was dissolved in methanol (2 ml) and loaded onto a column containing 10 g of silica gel 60 (Merck, Darmstadt, Germany). The column was eluted with CHCl3 and MeOH (from 100% CHCl3 to 100% MeOH) to afford 5 fractions, fraction 1 (100% CHCl3), fraction 2 (CHCl3-MeOH at a 100:1 dilution), fraction 3 (95% CHCl3–5% MeOH), fraction 4 (90% CHCl3–10% MeOH), and fraction 5 (100% MeOH). Thin-layer chromatography (TLC) (precoated silica gel 60 F254 plates; Merck, Darmstadt, Germany) was used to monitor the new compound in the fractions. The fraction (fraction 3) containing the compound was concentrated, and the residues were redissolved in methanol (2 ml). The solution was applied to a preparative HPLC column (RP-C18 [10.0 by 250 mm, 5 μm]), which was developed with 50% acetonitrile. The peak containing this new metabolite was collected to obtain ∼30 mg of pure compound. To determine the chemical structure, the compound was subjected to spectral analysis by NMR (Bruker Advance 400 spectrometer, 400/100 MHz; Bruker, Fällanden, Switzerland) and mass spectrometry (MS) (liquid chromatography quadrupole [LCQ] mass spectrometer; Thermo, West Palm Beach, FL, USA).
Quantitative PCR (qPCR).
The wild type and mutants of L. enzymogenes were grown on 100 ml 1/10-strength TSB medium for 24 h. An aliquot of 3 ml of cells was transferred to sterile centrifuge tubes and centrifuged for 3 min at 15,300 × g. The TRIzol solution was used to isolate the total RNA from the cells according to the manufacturer's instructions. A PrimerScript RT reagent kit with a genomic DNA (gDNA) Eraser kit (TaKaRa Biocompany) was used to remove DNA and for reverse transcription of the mRNA. An iQ SYBR green supermix kit (Bio-Rad) was used to quantitatively determine the expression levels of target genes, with 16S RNA as the reference (27).
Chemical synthesis of the fluorescent indole probe (compound 1).
Zinc dust (1.3 g; 20 mmol) was added to a solution containing indole 2 (585 mg; 5 mmol) and propargyl bromide (2.4 ml; 20 mmol) in 6 ml of tetrahydrofuran (THF) at ambient temperature. After stirring for 12 h, the reaction mixture was diluted with ethyl acetate (EtOAc), filtered through a pad of Celite, and concentrated in vacuo. Quick flash chromatography (10:1 hexane-EtOAc) yielded 542 mg of the target compound 3-(prop-2-yn-1-yl)-1H-indole (compound 3) (70%), and the compound was characterized as follows: Rf = 0.87 (3:1 hexane-EtOAc); 1H NMR (400 MHz, CDCl3), δ 8.03 (br s, 1H), 7.68 (d, 2H, J = 7.8 Hz), 7.39 (d, 1H, J = 8.1 Hz), 7.25 (t, 1H, J = 7.6 Hz), 7.20 (s, 1H), 7.18 (t, 1H, J = 7.6 Hz), 3.76 to 3.72 (m, 2H), and 2.19 to 2.16 (m, 1H).
A stirring solution of THF-water (1:2; 0.3 ml) at 25°C was charged with Azide-fluor 488 (compound 4) (0.574 mg; 0.001 mmol), compound 3 (0.155 mg; 0.001 mmol), copper sulfate (0.1 mg; 0.005 mmol), triethylamine (TEA) (0.05 mg; 0.005 mmol), and sodium ascorbate (0.2 mg; 0.001 mmol). The resulting mixture was stirred at room temperature for 24 h. Reaction progress was monitored by electrospray ionization (ESI)-mass spectrometry. Sephadex LH-20 chromatography (75% methanol) yielded 0.526 mg of the target compound (72%). The mass was calculated to be 729.79 for C40H40N7O7 and found m/z 730.67 [M + H]+ by ESI-MS. More spectral characterization of the synthetic compounds is included in Fig. S4 and S5 in the supplemental material.
Detection of fluorescence in various strains of L. enzymogenes using the synthetic indole probe.
To test the binding of the fluorescent indole probe, various strains of L. enzymogenes, including wild-type strain OH11, the Le-qseC and Le-qseB deletion mutants, and the Le-qseC single-amino-acid point mutants, were grown in 3 ml LB at 30°C at 250 rpm overnight. The cells were harvested by centrifugation and resuspended in a 1-ml solution of the probe (50 μM) in phosphate-buffered saline (PBS) (pH 7.4) at 4°C. The cells were then incubated at 30°C for 2 h, pelleted, and washed 2 times with PBS (pH 7.4) at 4°C. Finally, the cells were fixed by using 1/10 formaldehyde in PBS for 1 h at 25°C. The fluorescence of the cells was detected and photographed on an Olympus IX 81 inverted confocal microscope (Olympus, Tokyo, Japan). A 488-nm argon laser was utilized for continuous spectral detection. When indole binds to Le-QseC on the cell membranes of L. enzymogenes, the cells will be labeled with green fluorescence due to the fluorescent reporter of the probe.
Generation of in-frame Le-qseB and Le-qseC gene deletion mutants.
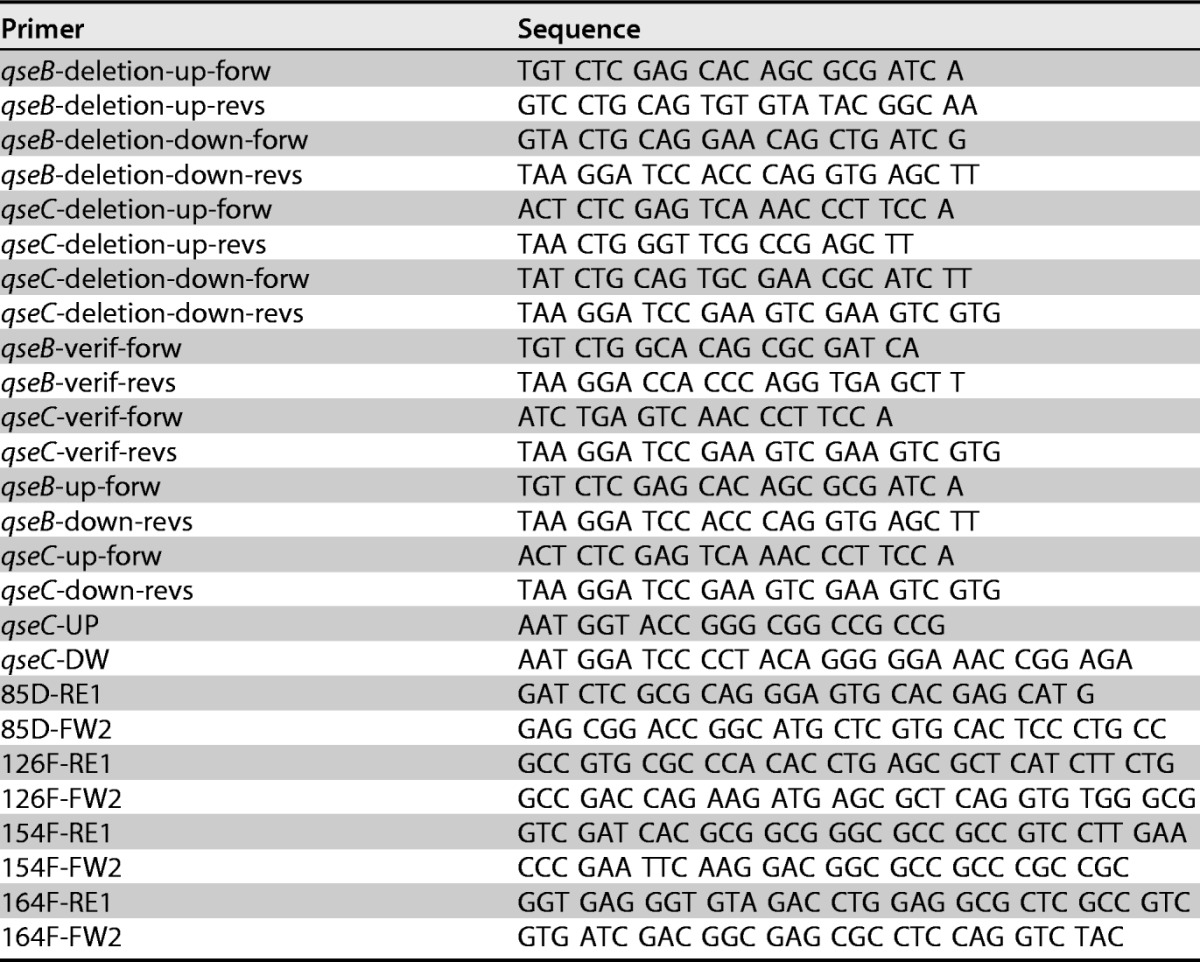
To construct vectors for the in-frame deletion of Le-qseB and Le-qseC in L. enzymogenes OH11, two DNA fragments were amplified from the upstream and downstream regions of each of these two genes by using the primer pairs shown in Table 1. Genomic DNA from wild-type L. enzymogenes OH11 was used as the PCR template. Each of the upstream fragments was digested with XhoI/PstI, and each of the downstream fragments was digested with PstI/BamHI. The upstream and downstream fragments of Le-qseB were cloned into the conjugation vector pJQ200SK to produce pJQ200SK-qseBupdown, and those of Le-qseC were cloned into pJQ200SK to generate pJQ200-qseCupdown. The resulting vectors were transferred into L. enzymogenes according to procedures described previously (17), and gentamicin-resistant colonies were selected for PCR verification by using primers listed in Table 1. The confirmed colonies resulting from single-crossover homologous recombination were subjected to a double crossover, which led to the loss of the vector through a second homologous recombination. The Le-qseB double-crossover mutants were screened by using primers qseB-up-forw and qseB-down-revs, and the qseC double-crossover mutants were screened by using primers qseC-up-forw and qseC-down-revs (Table 1). The qseB double-crossover mutants generated the expected 620-bp fragment, and the qseC double-crossover mutants generated the expected 833-bp fragment. Genomic DNA of wild-type L. enzymogenes and water were used as controls.
TABLE 1.
Primers used in this study
Generation of Le-qseC single-amino-acid mutants.
To construct vectors for Le-qseC single-amino-acid mutants in L. enzymogenes OH11, two DNA fragments that overlap and contain a single codon change at each of the four conserved residues in Le-qseC, as shown in Fig. S2 in the supplemental material, were amplified by PCR using primer pairs (qseC-UP and qseC-DW) listed in Table 1. Genomic DNA from OH11 was used as the template, and the two fragments were pieced together through a second PCR. Each of the resultant fragments was digested with KpnI/BamHI and then cloned into the conjugation vector pET18Gm to produce pET18Gm-qseCupdown. The constructs were confirmed by sequencing and then transferred into L. enzymogenes OH11 according to procedures described previously (17), and gentamicin-resistant colonies were selected for PCR verification of single-crossover homologous recombination using primers listed in Table 1. The confirmed colonies resulting from single-crossover homologous recombination were subjected to a double crossover, which led to the loss of the vector through a second homologous recombination. The double-crossover mutants were screened by using test primers (qsec-UP and qsec-DW). The PCR products were digested with the selected restriction enzymes (ApaLI for the Le-QseC D85V mutant, AfeI for the Le-QseC F126A mutant, EheI for the Le-QseC F154A mutant, and BpmI for the Le-QseC W164E mutant) to verify the mutants.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by the NSFC (31329005), the NIH (R01AI097260), the Nebraska Research Initiative, a UNL Research Council Faculty Seed Grant, and the Taishan Scholars Program of Shandong Province.
We thank Joe Zhou and Terri Fangman for technical assistance.
L.D., H.Y., Y.W., and Y.S. conceived and designed experiments. H.Y., Y.W., Y.Y., and H.C. carried out experiments. L.D., Y.H., and Y.W. analyzed data. L.D. wrote the manuscript. Y.H., Y.W., L.D., and Y.S. reviewed and revised the manuscript.
We declare that we have no conflicts of interest with the contents of this article.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00995-17.
REFERENCES
- 1.Xie Y, Wright S, Shen Y, Du L. 2012. Bioactive natural products from Lysobacter. Nat Prod Rep 19:1–14. doi: 10.1039/c2np20064c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Panthee S, Hamamoto H, Paudel A, Sekimizu K. 2016. Lysobacter species: a potential source of novel antibiotics. Arch Microbiol 198:839–845. doi: 10.1007/s00203-016-1278-5. [DOI] [PubMed] [Google Scholar]
- 3.Hamamoto H, Urai M, Ishii K, Yasukawa J, Paudel A, Murai M, Kaji T, Kuranaga T, Hamase K, Katsu T. 2015. Lysocin E is a new antibiotic that targets menaquinone in the bacterial membrane. Nat Chem Biol 11:127–133. doi: 10.1038/nchembio.1710. [DOI] [PubMed] [Google Scholar]
- 4.Murai M, Kaji T, Kuranaga T, Hamamoto H, Sekimizu K, Inoue M. 2015. Total synthesis and biological evaluation of the antibiotic lysocin E and its enantiomeric, epimeric, and N-demethylated analogues. Angew Chem Int Ed Engl 54:1556–1560. doi: 10.1002/anie.201410270. [DOI] [PubMed] [Google Scholar]
- 5.O'Sullivan J, McCullough JE, Tymiak AA, Kirsch DR, Trejo WH, Principe PA. 1988. Lysobactin, a novel antibacterial agent produced by Lysobacter sp. I. Taxonomy, isolation and partial characterization. J Antibiot 41:1740–1744. doi: 10.7164/antibiotics.41.1740. [DOI] [PubMed] [Google Scholar]
- 6.Li W, Rokni-Zadeh H, De Vleeschouwer M, Ghequire MG, Sinnaeve D, Xie G-L, Rozenski J, Madder A, Martins JC, De Mot R. 2013. The antimicrobial compound xantholysin defines a new group of Pseudomonas cyclic lipopeptides. PLoS One 8:e62946. doi: 10.1371/journal.pone.0062946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee W, Schaefer K, Qiao Y, Srisuknimit V, Steinmetz H, Müller R, Kahne RD, Walker S. 2015. The mechanism of action of lysobactin. J Am Chem Soc 138:100–103. doi: 10.1021/jacs.5b11807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kato A, Nakaya S, Ohashi Y, Hirata H, Fujii K, Harada K. 1997. WAP-8294A2, a novel anti-MRSA antibiotic produced by Lysobacter sp. J Am Chem Soc 119:6680–6681. doi: 10.1021/ja970895o. [DOI] [Google Scholar]
- 9.Zhang W, Li Y, Qian G, Wang Y, Chen H, Li Y-Z, Liu F, Shen Y, Du L. 2011. Identification and characterization of the anti-methicillin-resistant Staphylococcus aureus WAP-8294A2 biosynthetic gene cluster from Lysobacter enzymogenes OH11. Antimicrob Agents Chemother 55:5581–5589. doi: 10.1128/AAC.05370-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen H, Olson AS, Su W, Dussault PH, Du L. 2015. Fatty acyl incorporation in the biosynthesis of WAP-8294A, a group of potent anti-MRSA cyclic lipodepsipeptides. RSC Adv 5:105753–105759. doi: 10.1039/C5RA20784C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hashizume H, Igarashi M, Hattori S, Hori M, Hamada M, Takeuchi T. 2001. Tripropeptins, novel antimicrobial agents produced by Lysobacter sp. I. Taxonomy, isolation and biological activities. J Antibiot 54:1054–1059. doi: 10.7164/antibiotics.54.1054. [DOI] [PubMed] [Google Scholar]
- 12.Hashizume H, Takahashi Y, Harada S, Nomoto A. 2015. Natural lipopeptide antibiotic tripropeptin C revitalizes and synergistically potentiates the activity of beta-lactams against methicillin-resistant Staphylococcus aureus. J Antibiot 68:373–378. doi: 10.1038/ja.2014.169. [DOI] [PubMed] [Google Scholar]
- 13.Sohn Y-S, Nam D-H, Ryu DD. 2001. Biosynthetic pathway of cephabacins in Lysobacter lactamgenus: molecular and biochemical characterization of the upstream region of the gene clusters for engineering of novel antibiotics. Metab Eng 3:380–392. doi: 10.1006/mben.2001.0200. [DOI] [PubMed] [Google Scholar]
- 14.Demirev AV, Lee C-H, Jaishy BP, Nam D-H, Ryu DD. 2006. Substrate specificity of nonribosomal peptide synthetase modules responsible for the biosynthesis of the oligopeptide moiety of cephabacin in Lysobacter lactamgenus. FEMS Microbiol Lett 255:121–128. doi: 10.1111/j.1574-6968.2005.00067.x. [DOI] [PubMed] [Google Scholar]
- 15.Lee JS, Vladimirova MG, Demirev AV, Kim BG, Lim SK, Nam DH. 2008. Expression and characterization of polyketide synthase module involved in the late step of cephabacin biosynthesis from Lysobacter lactamgenus. J Microbiol Biotechnol 18:427–433. [PubMed] [Google Scholar]
- 16.Li S, Du L, Yuen G, Harris SD. 2006. Distinct ceramide synthases regulate polarized growth in the filamentous fungus Aspergillus nidulans. Mol Biol Cell 17:1218–1227. doi: 10.1091/mbc.E05-06-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu F, Zaleta-Rivera K, Zhu X, Huffman J, Millet JC, Harris SD, Yuen G, Li X-C, Du L. 2007. Structure and biosynthesis of heat-stable antifungal factor (HSAF), a broad-spectrum antimycotic with a novel mode of action. Antimicrob Agents Chemother 51:64–72. doi: 10.1128/AAC.00931-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lou L, Qian G, Xie Y, Hang J, Chen H, Zaleta-Rivera K, Li Y, Shen Y, Dussault PH, Liu F. 2010. Biosynthesis of HSAF, a tetramic acid-containing macrolactam from Lysobacter enzymogenes. J Am Chem Soc 133:643–645. doi: 10.1021/ja105732c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, Huffman J, Li Y, Du L, Shen Y. 2012. 3-Hydroxylation of the polycyclic tetramate macrolactam in the biosynthesis of antifungal HSAF from Lysobacter enzymogenes C3. MedChemComm 3:982–986. doi: 10.1039/c2md20026k. [DOI] [Google Scholar]
- 20.Lou L, Chen H, Cerny RL, Li Y, Shen Y, Du L. 2012. Unusual activities of the thioesterase domain for the biosynthesis of the polycyclic tetramate macrolactam HSAF in Lysobacter enzymogenes C3. Biochemistry 51:4–6. doi: 10.1021/bi2015025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Chen H, Ding Y, Xie Y, Wang H, Cerny RL, Shen Y, Du L. 2014. Iterative assembly of two separate polyketide chains by the same single-module bacterial polyketide synthase in the biosynthesis of HSAF. Angew Chem Int Ed Engl 53:7524–7530. doi: 10.1002/anie.201403500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu L, Wu P, Wright SJ, Du L, Wei X. 2015. Bioactive polycyclic tetramate macrolactams from Lysobacter enzymogenes and their absolute configurations by theoretical ECD calculations. J Nat Prod 78:1841–1847. doi: 10.1021/acs.jnatprod.5b00099. [DOI] [PubMed] [Google Scholar]
- 23.Ding Y, Li Z, Li Y, Lu C, Wang H, Shen Y, Du L. 2016. HSAF-induced antifungal effects in Candida albicans through ROS-mediated apoptosis. RSC Adv 6:30895–30904. doi: 10.1039/C5RA26092B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding Y, Li Y, Li Z, Zhang J, Lu C, Wang H, Shen Y, Du L. 2016. Alteramide B is a microtubule antagonist of inhibiting Candida albicans. Biochim Biophys Acta 1860:2097–2106. doi: 10.1016/j.bbagen.2016.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao Y, Qian G, Ye Y, Wright S, Chen H, Shen Y, Liu F, Du L. 2016. Heterocyclic aromatic N-oxidation in the biosynthesis of phenazine antibiotics from Lysobacter antibioticus. Org Lett 18:2495–2498. doi: 10.1021/acs.orglett.6b01089. [DOI] [PubMed] [Google Scholar]
- 26.de Bruijn I, Cheng X, de Jager V, Expósito RG, Watrous J, Patel N, Postma J, Dorrestein PC, Kobayashi D, Raaijmakers JM. 2015. Comparative genomics and metabolic profiling of the genus Lysobacter. BMC Genomics 16:991. doi: 10.1186/s12864-015-2191-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han Y, Wang Y, Tombosa S, Wright S, Huffman J, Yuen G, Qian G, Liu F, Shen Y, Du L. 2015. Identification of a small molecule signaling factor that regulates the biosynthesis of the antifungal polycyclic tetramate macrolactam HSAF in Lysobacter enzymogenes. Appl Microbiol Biotechnol 99:801–811. doi: 10.1007/s00253-014-6120-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qian G, Wang Y, Liu Y, Xu F, He Y-W, Du L, Venturi V, Fan J, Hu B, Liu F. 2013. Lysobacter enzymogenes uses two distinct cell-cell signaling systems for differential regulation of secondary-metabolite biosynthesis and colony morphology. Appl Environ Microbiol 79:6604–6616. doi: 10.1128/AEM.01841-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Zhao Y, Zhang J, Zhao Y, Shen Y, Su Z, Xu G, Du L, Huffman JM, Venturi V. 2014. Transcriptomic analysis reveals new regulatory roles of Clp signaling in secondary metabolite biosynthesis and surface motility in Lysobacter enzymogenes OH11. Appl Microbiol Biotechnol 98:9009–9020. doi: 10.1007/s00253-014-6072-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang R, Xu H, Du L, Chou S-H, Liu H, Liu Y, Liu F, Qian G. 2016. A TonB-dependent receptor regulates antifungal HSAF biosynthesis in Lysobacter. Sci Rep 6:26881. doi: 10.1038/srep26881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su Z, Chen H, Wang P, Tombosa S, Du L, Han Y, Shen Y, Qian G, Liu F. 2017. Hydroxybenzoic acid is a diffusible factor that connects metabolic shikimate pathway to the biosynthesis of a unique antifungal metabolite in Lysobacter enzymogenes. Mol Microbiol 104:163–178. doi: 10.1111/mmi.13619. [DOI] [PubMed] [Google Scholar]
- 32.Chen Y, Xia J, Su Z, Xu G, Gomelsky M, Qian G, Liu F. 2017. Lysobacter PilR, the regulator of type IV pilus synthesis, controls antifungal antibiotic production via a cyclic di-GMP pathway. Appl Environ Microbiol 83:e03397-16. doi: 10.1128/AEM.03397-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sperandio V, Torres AG, Kaper JB. 2002. Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol Microbiol 43:809–821. doi: 10.1046/j.1365-2958.2002.02803.x. [DOI] [PubMed] [Google Scholar]
- 34.Sperandio V, Torres AG, Jarvis B, Nataro JP, Kaper JB. 2003. Bacteria-host communication: the language of hormones. Proc Natl Acad Sci U S A 100:8951–8956. doi: 10.1073/pnas.1537100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rasko DA, Moreira CG, Li DR, Reading NC, Ritchie JM, Waldor MK, Williams N, Taussig R, Wei S, Roth M. 2008. Targeting QseC signaling and virulence for antibiotic development. Science 321:1078–1080. doi: 10.1126/science.1160354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharma VK, Casey T. 2014. Escherichia coli O157:H7 lacking the qseBC-encoded quorum-sensing system outcompetes the parental strain in colonization of cattle intestines. Appl Environ Microbiol 80:1882–1892. doi: 10.1128/AEM.03198-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma VK, Casey TA. 2014. Determining the relative contribution and hierarchy of hha and qseBC in the regulation of flagellar motility of Escherichia coli O157:H7. PLoS One 9:e85866. doi: 10.1371/journal.pone.0085866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clarke MB, Hughes DT, Zhu C, Boedeker EC, Sperandio V. 2006. The QseC sensor kinase: a bacterial adrenergic receptor. Proc Natl Acad Sci U S A 103:10420–10425. doi: 10.1073/pnas.0604343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hughes DT, Clarke MB, Yamamoto K, Rasko DA, Sperandio V. 2009. The QseC adrenergic signaling cascade in enterohemorrhagic E. coli (EHEC). PLoS Pathog 5:e1000553. doi: 10.1371/journal.ppat.1000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hughes DT, Sperandio V. 2008. Inter-kingdom signalling: communication between bacteria and their hosts. Nat Rev Microbiol 6:111–120. doi: 10.1038/nrmicro1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Curtis MM, Russell R, Moreira CG, Adebesin AM, Wang C, Williams NS, Taussig R, Stewart D, Zimmern P, Lu B. 2014. QseC inhibitors as an antivirulence approach for Gram-negative pathogens. mBio 5:e02165-14. doi: 10.1128/mBio.02165-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rooks MG, Veiga P, Reeves AZ, Lavoie S, Yasuda K, Asano Y, Yoshihara K, Michaud M, Wardwell-Scott L, Gallini CA. 2017. QseC inhibition as an antivirulence approach for colitis-associated bacteria. Proc Natl Acad Sci U S A 114:142–147. doi: 10.1073/pnas.1612836114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu Rev Biochem 69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 44.Lee J, Jayaraman A, Wood TK. 2007. Indole is an inter-species biofilm signal mediated by SdiA. BMC Microbiol 7:42. doi: 10.1186/1471-2180-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karlin D, Mastromarino A, Jones R, Stroehlein J, Lorentz O. 1985. Fecal skatole and indole and breath methane and hydrogen in patients with large bowel polyps or cancer. J Cancer Res Clin Oncol 109:135–141. doi: 10.1007/BF00391888. [DOI] [PubMed] [Google Scholar]
- 46.Zuccato E, Venturi M, Di Leo G, Colombo L, Bertolo C, Mussini E. 1993. Role of bile acids and metabolic activity of colonic bacteria in increased risk of colon cancer after cholecystectomy. Dig Dis Sci 38:514–519. doi: 10.1007/BF01316508. [DOI] [PubMed] [Google Scholar]
- 47.Domka J, Lee J, Wood TK. 2006. YliH (BssR) and YceP (BssS) regulate Escherichia coli K-12 biofilm formation by influencing cell signaling. Appl Environ Microbiol 72:2449–2459. doi: 10.1128/AEM.72.4.2449-2459.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee J, Maeda T, Hong SH, Wood TK. 2009. Reconfiguring the quorum-sensing regulator SdiA of Escherichia coli to control biofilm formation via indole and N-acylhomoserine lactones. Appl Environ Microbiol 75:1703–1716. doi: 10.1128/AEM.02081-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee J, Zhang X-S, Hegde M, Bentley WE, Jayaraman A, Wood TK. 2008. Indole cell signaling occurs primarily at low temperatures in Escherichia coli. ISME J 2:1007–1023. doi: 10.1038/ismej.2008.54. [DOI] [PubMed] [Google Scholar]
- 50.Hirakawa H, Inazumi Y, Masaki T, Hirata T, Yamaguchi A. 2005. Indole induces the expression of multidrug exporter genes in Escherichia coli. Mol Microbiol 55:1113–1126. doi: 10.1111/j.1365-2958.2004.04449.x. [DOI] [PubMed] [Google Scholar]
- 51.Sabag-Daigle A, Soares JA, Smith JN, Elmasry ME, Ahmer BM. 2012. The acyl homoserine lactone receptor, SdiA, of Escherichia coli and Salmonella enterica serovar Typhimurium does not respond to indole. Appl Environ Microbiol 78:5424–5431. doi: 10.1128/AEM.00046-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pinero-Fernandez S, Chimerel C, Keyser U, Summers D. 2011. Indole transport across Escherichia coli membranes. J Bacteriol 193:1793–1798. doi: 10.1128/JB.01477-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Allison KR, Brynildsen MP, Collins JJ. 2011. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 473:216–220. doi: 10.1038/nature10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vega NM, Allison KR, Khalil AS, Collins JJ. 2012. Signaling-mediated bacterial persister formation. Nat Chem Biol 8:431–433. doi: 10.1038/nchembio.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vega NM, Allison KR, Samuels AN, Klempner MS, Collins JJ. 2013. Salmonella Typhimurium intercepts Escherichia coli signaling to enhance antibiotic tolerance. Proc Natl Acad Sci U S A 110:14420–14425. doi: 10.1073/pnas.1308085110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Han TH, Lee J-H, Cho MH, Wood TK, Lee J. 2011. Environmental factors affecting indole production in Escherichia coli. Res Microbiol 162:108–116. doi: 10.1016/j.resmic.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nikaido E, Giraud E, Baucheron S, Yamasaki S, Wiedemann A, Okamoto K, Takagi T, Yamaguchi A, Cloeckaert A, Nishino K. 2012. Effects of indole on drug resistance and virulence of Salmonella enterica serovar Typhimurium revealed by genome-wide analyses. Gut Pathog 4:5. doi: 10.1186/1757-4749-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.