Abstract
Background
Despite evidence that genetic factors contribute to gestational length and preterm birth, robust associations with genetic variants have not been identified. We hypothesized that analyzing larger data sets with gestational length information by genomewide association would reveal trait-influencing variants.
Methods
We performed a genomewide association study in a discovery data set of 43,568 women of European ancestry from 23andMe, Inc., for gestational length as a continuous trait and for term or preterm (<37 weeks) birth as a dichotomous outcome. We used three Nordic data sets (8,643 women) for replication of 14 genomic loci achieving either genomewide (P < 5×10-8) or suggestive association (P < 1×10-6).
Results
In the discovery stage, for gestational length, four loci (EBF1, EEFSEC, AGTR2 and WNT4) achieved genomewide significance, all of which were replicated in the Nordic data sets. Functional analysis of the WNT4 locus indicated the likely causative variant alters the binding of ESR1. ADCY5 and RAP2C, which had suggestive significance in the discovery stage, were significantly replicated and achieved genomewide significance in joint analysis. Common variants in EBF1, EEFSEC and AGTR2 were also associated with preterm birth with genomewide significance. Analysis of mother-infant dyads indicated that these findings likely resulted from maternal genome actions.
Conclusions
Our study is the first to identify maternal genetic variants robustly associated with gestational length and preterm birth. Roles of these loci in uterine development, maternal nutrition, and vascular control support their mechanistic involvement and create opportunities to investigate new risk factors for prevention of preterm birth.
Keywords: Gestational length, preterm birth, genomewide association, single nucleotide polymorphism
Introduction
Preterm birth (defined as birth before 37 completed weeks of gestation) affects 9.6% of pregnancies in the United States 1 and over 15 million pregnancies worldwide each year. It is the leading global cause of mortality in children under five years of age. 2,3 The majority of preterm births arise by the spontaneous, idiopathic onset of uterine contractions or rupture of fetal membranes. 4 Despite the considerable morbidity and mortality arising from preterm birth, few interventions have proven effective in limiting its occurrence. The limited progress in preterm birth prevention may arise from the lack of understanding of the pathways regulating the timing of birth including the normal length of gestation. 5,6 A substantial body of evidence has accumulated demonstrating a contribution of genetic factors in gestational length and preterm birth risk. 7 For example, twin and family studies suggest that 30-40% of the variation in birth timing, or risk for preterm birth, arises from genetic factors, largely but not exclusively residing in the maternal genome. 8-12
Preterm birth, and gestational length in general, is a complicated phenotype with contributions from two genomes – maternal and fetal – that may have separate or interacting contributions. Furthermore, different genotypes may predispose to preterm birth at different gestational ages. Finally, defining preterm birth as a dichotomous trait based upon a somewhat arbitrary cutoff of 37 weeks, rather than time of birth for a specified level of fetal maturity or as a continuous trait, limits data interpretation. Therefore, defining the genetic variants associated with gestational length (a quantitative trait) as well as preterm birth (a dichotomous trait), will both yield important new insights. Further, analyzing gestational length as a continuous trait increases the power to detect associations that is limited when traits are dichotomized. 13 Control of timing of birth is multifactorial, and common polymorphisms involved in gestational length or preterm birth risk are likely to individually be of small effect size. Nonetheless, the insights they provide into essential genes and pathways may open novel avenues for intervention. 14 However, for genomewide association studies to reveal robustly associated variants, large sample sizes are required, 15 and particularly so for preterm birth given the complexity of the phenotype. To date, individual genomewide association studies of spontaneous preterm birth have included on the order of 1,000 case mothers or infants with control groups of similar size, but no replicated genomewide significant loci have yet emerged. 16-18
To overcome previous sample size limitations, we leverage data on gestational length and preterm birth in a large sample of women of European ancestry (approximately 44,000) collected as part of genotyping and phenotyping efforts by 23andMe, Inc., a genetics company. We then selected the top loci (P < 1×10-6) and performed replication analyses for gestational length and preterm birth in three data sets of Nordic women (8,643). Further, we provide evidence indicating the observed effect was due to an action in the maternal genome and provide functional data implicating the causative SNP underlying the WNT4 locus.
Methods
We performed a two-stage genomewide association study to discover and replicate genetic loci associated with gestational length and preterm birth. In the discovery stage, we performed genomewide association analyses on 43,568 European-ancestry females identified among 23andMe’s research participants. In the replication stage, the top significant loci from the discovery stage analyses were tested in three birth data sets collected from Nordic countries (Finland, Denmark, and Norway).
Discovery stage
Women in the discovery data set were participants in 23andMe’s research program. All women provided informed consent and answered surveys online following a human subjects protocol, reviewed and approved by Ethical & Independent Review Services, a private institutional review board (http://www.eandireview.com). Unrelated women of European ancestry who self-reported gestational length of their first live singleton birth were included in the analysis. Categories of preterm birth addressed on the survey were 1) spontaneous preterm labor, 2) planned or required delivery for medical reasons, 3) cervical problems, 4) other, or 5) none of the above. Women with a medical indication for their preterm delivery were excluded from the study; those that did not specify a medical indication on the survey were retained to optimize sample size. Preterm birth status was determined based on dichotomization of self-reported gestational length (preterm < 37 weeks; term ≥37 weeks). For those in the preterm group, 96.8% of women responded to the question regarding mode of delivery. For the term birth group, we ascertained information on aggregate outcomes of all pregnancies, and could not unambiguously determine spontaneous or medically indicated birth at more than 37 weeks.
DNA extraction and genotyping were performed on saliva samples by the National Genetics Institute. To minimize the effects of population stratification, we restricted analyses to women with >97% European ancestry, as determined through an analysis of local ancestry. 19 Participant genotype data were imputed against the 1000 Genomes Phase1 reference haplotypes. 20
Single-marker genetic associations with gestational length and preterm birth were tested by linear regression or logistic regression, respectively, using imputed allelic dosage data assuming additive allelic effects. Maternal age and the top five principal components to account for residual population structure were included as covariates.
We clustered SNPs into association regions (or loci). Specifically, we defined association regions by first identifying SNPs with P < 1×10-4, then grouping these into a region if they were adjacent to each other (<250kb). The SNP with smallest P value within each region was chosen as the index SNP. Regions that achieved suggestive significance (P < 1×10-6) were tested in the replication stage.
Replication stage
We used the data of 8,643 mothers from three independent Nordic birth studies (Table S1) of singleton pregnancies with spontaneous onset of labor. Briefly, the Finnish study (FIN) consisted of nearly 900 mothers and their infants recruited from the Helsinki University Hospital between 2004 and 2014 with gestational length confirmation by early ultrasound at 10-13 weeks of gestation. The Mother Child Cohort of Norway (MoBa) is a nationwide pregnancy cohort study administered by the Norwegian Institute of Public Health. 21 The genotype data were derived from a genomewide association study of preterm birth, with gestational length determination by second trimester ultrasound in more than 95% of participants. 22 For the current study, 1,834 mothers and 1,143 infants that passed QC were included in the analysis. The Danish National Birth Cohort (DNBC) data is a cohort including mothers and their children from more than 100,000 pregnancies recruited between 1996 and 2002. 23 Gestational length in this cohort was assigned by combining all available information from multiple sources: self-reported date of last menstrual period, self-reported delivery date, and gestational length at birth registered in the Medical Birth Register and the National Patient Register. The genotype data were derived from two genomewide association studies of preterm birth 24 and obesity, 25,26 respectively. In the current study, data from 5,921 mothers and 2,130 infants that passed QC were analyzed.
Genotyping of the Nordic studies was conducted using various SNP arrays as previously described. 27 Similar genotype QC procedures were used across the three studies. Subjects of non-European-ancestry were identified and excluded using principal components analysis (PCA). Genomewide imputation for the replication data sets was conducted using the reference haplotypes extracted from the Phase I 1000 Genomes Project. 20
Single-marker genetic association tests were conducted in each replication data set, using regression methods and imputation dosage similar to the discovery stage. Genotypic association tests (d.f. = 2) were also performed to examine possible dominance effect. The replication P values (inflation adjusted) combining results from the three Nordic data sets were calculated using the fixed-effects inverse-variance method. Significant replication P values and the same direction of effect at the index or other significant SNPs (P < 1×10-6, discovery stage) in the region or their close proxies (r 2 > 0.8) were regarded as statistical evidence of replication of a putative locus. The significance level of each region was corrected by the effective number of independent SNPs tested in the region and the total number of regions that underwent replication attempts (Table S5 and Supplementary Text). A region was considered successfully replicated and genomewide significant after replication if the most significant replication P value was below the significance level and had a combined discovery and replication P value less than 5×10-8.
We also performed association tests in 4,090 infant samples and joint maternal/fetal genetic association analysis in 3,184 (FIN: 769; MoBa: 1019 and DNBC: 1396) mother/infant pairs from the Nordic data sets to evaluate whether the observed significant associations were likely to be of maternal or fetal origin.
Functional annotation and other statistical analyses
We checked whether the SNPs associated with gestational length or preterm overlap with previously reported genomewide association SNPs in the GWAS catalog 28 and used the GTEx 29 database to search for associations with tissue-specific gene expression. We examined whether multiple independent variants at a given locus influenced birth timing by an approximate conditional and joint multiple-SNP (COJO) analysis. 30 We estimated the fraction of phenotype variance in the replication data sets explained by all common SNPs 31 by GCTA 32 or sets of SNPs associated at different significance thresholds in the discovery cohort using a genetic score approach. 33 We also performed gene-centric associations and gene-set enrichment analyses. Detailed description of these analyses and associated results are described in the Supplementary Text.
Functional follow-up
We performed experimental functional follow-up of the WNT4 locus, one of the most significant loci with plausible functional relevance in pregnancy. First we examined the expression level of WNT4 in human endometrial stromal cells, before and after decidualization using mRNA-seq technology. We predicted specific transcription factors binding using a Bioinformatic approach and studied the presence of H3K4me3 marks and open chromatin domains overlapping the hypothetical causal SNP by ChIP-seq and ATAC-seq, respectively. We performed electrophoretic mobility shift assays (EMSA) to determine whether the variant differentially affected specific transcription factor binding. Detailed description of these analyses can be found in the Supplementary Text (Functional analyses of the WNT4 locus).
Results
Study data sets
The discovery data set included 43,568 women identified through 23andMe (Table S1). Most of the women (86.8%, N=37,803) delivered at term (37 to 42 weeks); 7.6% (N=3,331) delivered preterm (<37 weeks) and 5.6% (N=2,434) delivered post-term (>42 weeks) (Figure S1 and Table S2). Maternal age was strongly associated with gestational length (P = 2.3×10-41) with older mothers having shorter gestational length (Table S3).
Three Nordic birth studies 27 were used in combination for replication. In total, phenotype and genotype data were available from 8,643 mothers and 4,090 infants (Table S1). These data sets were case/control studies, in which samples from preterm births were enriched and samples with post-term or close to the preterm-term boundary (37-38 weeks) were excluded (Figure S2). In these studies infant gender and maternal height were associated with gestational length (Table S4).
Discovery stage findings in mothers
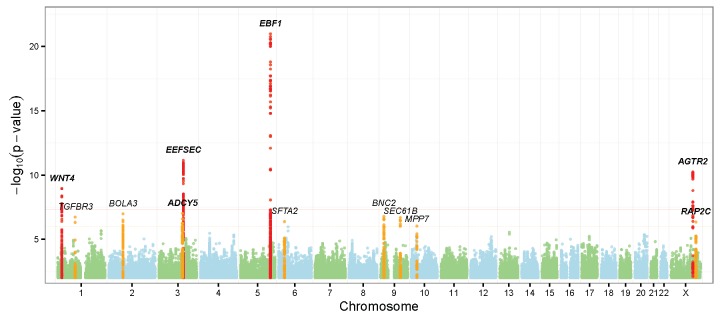
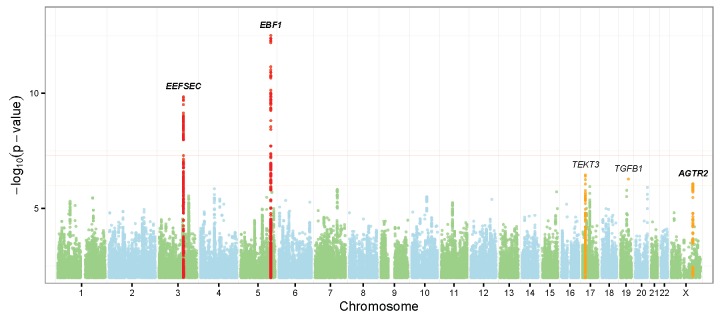
Single-marker association tests were performed across 15,635,593 SNPs that passed the 23andMe QC (Supplementary Methods). We focused our analysis on 9,042,878 markers with MAF>0.01. Test results were adjusted for genomic inflation factors (Figure S3). For gestational length, 12 loci were identified with P < 1×10-6 (suggestive significance). Of these, four had an association P < 5×10-8 (Figure 1A, Table 1 and Table S5). For preterm birth, 5 loci were identified with P < 1×10-6, two of which achieved genomewide significance (Figure 1B, Table 1 and Table S5). The top three loci associated with gestational length (EBF1, EEFSEC and AGTR2) shared association loci for preterm birth risk. Altogether, 14 independent loci were taken forward for replication. To confirm the robustness of the association signals, we conducted similar association tests in a subset of discovery subjects who explicitly checked “spontaneous delivery” in the questionnaire (excluding those who did not specify a choice of spontaneous or medically indicated delivery) and the results were similar to those obtained from the full discovery data sets (Table S6).
Figure 1. Manhattan plots of discovery stage genomewide-associated results.
Top: gestational length as quantitative trait; bottom: preterm birth as dichotomous trait. Regions reached genome wide significance (P < 5×10-8) and suggestive significance (P < 1×10-6) were highlighted in red and orange respectively. The six replicated loci were highlighted in bold.
Table 1. Discovery and replication of loci associated with gestational length or preterm birth.
For each locus, the most significant SNP in discovery stage (index SNP) and the most significant SNP in replication stage are shown.
| No | Char | Genes@ | SNP Information# | Discovery Phase | Replication Phase | Joint analysis | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs | pos | A/B | Freq | Eff$ | P-value% | Rank& | r 2 | Freq | Eff$ | P-value% | Directions* | P–value% | |||
| Gestational age | |||||||||||||||
| 1 | 5 | EBF1 | rs2963463 | 157895049 | C/T | 0.272 | -1.29 | 1.0E-21 | 1 | 0.264 | -1.11 | 0.0017 | +-+ | 7.7E-24 | |
| rs2946171 | 157921940 | T/G | 0.219 | -1.24 | 1.1E-17 | 24 | 0.71 | 0.206 | -1.46 | 0.00014 | +-+ | 8.1E-21 | |||
| 2 | 3 | EEFSEC | rs2955117 | 127881613 | G/A | 0.286 | 0.911 | 7.2E-12 | 1 | 0.279 | 1.33 | 0.00016 | +++ | 9.5E-15 | |
| rs200745338 | 127869457 | D/I | 0.237 | 0.986 | 1.5E-11 | 8 | 0.72 | 0.232 | 1.91 | 7.6E-07 | +++ | 7.5E-16 | |||
| 3 | X | AGTR2 | rs201226733 | 115164770 | I/D | 0.422 | -0.820 | 5.7E-11 | 1 | 0.420 | -1.67 | 9.2E-08 | +++ | 7.2E-16 | |
| rs5950491 | 115129714 | C/A | 0.423 | -0.826 | 6.8E-11 | 5 | 0.92 | 0.425 | -1.75 | 4.7E-08 | +++ | 6.6E-16 | |||
| 4 | 1 | WNT4 | rs56318008 | 22470407 | C/T | 0.139 | 1.05 | 1.2E-09 | 1 | 0.153 | 2.27 | 1.8E-07 | +++ | 3.4E-14 | |
| rs12037376 | 22462111 | G/A | 0.145 | 1.00 | 4.5E-09 | 4 | 0.91 | 0.157 | 2.41 | 2.1E-08 | +++ | 5.6E-14 | |||
| 5 | 3 | ADCY5 | rs4383453 | 123068359 | G/A | 0.200 | -0.808 | 9.6E-08 | 1 | 0.197 | -0.587 | 0.15 | -++ | 3.7E-08 | |
| rs9861425 | 123072883 | A/C | 0.453 | -0.598 | 6.1E-07 | 5 | 0.34 | 0.470 | -1.38 | 9.5E-06 | +++ | 4.2E-10 | |||
| 6 | 2 | BOLA3 | rs4853012 | 74361290 | G/A | 0.141 | -0.920 | 1.1E-07 | 1 | 0.145 | -0.355 | 0.42 | -++ | 1.6E-07 | |
| rs17009553 | 74220035 | G/A | 0.0567 | -1.28 | 1.1E-06 | 6 | 0.13 | 0.0565 | -2.11 | 0.0020 | -++ | 1.6E-08 | |||
| 7 | 9 | BNC2 | rs717267 | 16408826 | G/A | 0.399 | -0.637 | 1.7E-07 | 1 | 0.423 | -0.12 | 0.70 | +-+ | 5.3E-07 | |
| rs9298764 | 16431230 | A/G | 0.456 | -0.55 | 5.2E-06 | 19 | 0.81 | 0.474 | -0.443 | 0.16 | +-+ | 2.0E-06 | |||
| 8 | 1 | TGFBR3 | rs4658267 | 92240753 | C/A | 0.319 | 0.679 | 1.9E-07 | 1 | 0.319 | 0.0562 | 0.87 | -++ | 8.7E-07 | |
| rs4658265 | 92240685 | C/T | 0.312 | 0.662 | 4.8E-07 | 2 | 0.91 | 0.311 | 0.125 | 0.71 | -++ | 1.4E-06 | |||
| 9 | 9 | SEC61B | rs182704 | 102068912 | T/C | 0.344 | 0.658 | 2.1E-07 | 1 | 0.397 | 0.145 | 0.67 | --+ | 5.4E-07 | |
| 10 | 6 | SFTA2 | rs2532929 | 30897774 | A/G | 0.397 | -0.622 | 4.2E-07 | 1 | 0.402 | -0.0486 | 0.88 | +-+ | 1.8E-06 | |
| rs2532926 | 30898441 | A/G | 0.363 | -0.556 | 9.0E-06 | 3 | 0.87 | 0.374 | -0.0653 | 0.84 | +-+ | 2.5E-05 | |||
| 11 | X | RAP2C | rs200879388 | 131300571 | I/D | 0.351 | -0.662 | 4.5E-07 | 1 | 0.364 | -1.1 | 0.00092 | +++ | 3.4E-09 | |
| 12 | 10 | MPP7 | rs2253165 | 28337017 | A/G | 0.440 | -0.594 | 9.3E-07 | 1 | 0.429 | 0.454 | 0.15 | +-- | 4.5E-05 | |
| rs2245244 | 28316456 | T/C | 0.438 | -0.561 | 3.7E-06 | 2 | 0.86 | 0.428 | 0.507 | 0.11 | +-- | 0.00019 | |||
| Preterm birth | |||||||||||||||
| 1 | 5 | EBF1 | rs2963463 | 157895049 | C/T | 0.272 | 1.23 | 3.2E-13 | 1 | 0.265 | 1.13 | 0.0015 | +-+ | 4.5E-15 | |
| rs2946169 | 157918959 | C/T | 0.217 | 1.22 | 1.1E-10 | 19 | 0.68 | 0.207 | 1.16 | 0.00055 | +-+ | 2.2E-13 | |||
| 2 | 3 | EEFSEC | rs201450565 | 128058610 | D/I | 0.233 | 0.810 | 1.4E-10 | 1 | 0.135 | 0.824 | 0.0017 | +++ | 1.9E-12 | |
| rs200745338 | 127869457 | D/I | 0.237 | 0.829 | 9.0E-09 | 95 | 0.24 | 0.232 | 0.797 | 3.5E-07 | +++ | 3.3E-14 | |||
| 3 | 17 | TEKT3 | rs7217780 | 15191024 | T/C | 0.336 | 1.15 | 3.5E-07 | 1 | 0.341 | 1.09 | 0.025 | +++ | 4.9E-08 | |
| rs179521 | 15173221 | C/A | 0.359 | 1.13 | 3.8E-06 | 11 | 0.81 | 0.357 | 1.10 | 0.012 | +++ | 1.6E-07 | |||
| 4 | 19 | TGFB1 | rs11466328 | 41851042 | G/A | 0.0288 | 0.567 | 5.3E-07 | 1 | 0.0311 | 0.711 | 0.038 | +++ | 5.5E-07 | |
| 5 | X | AGTR2 | rs201386833 | 115164281 | D/I | 0.410 | 1.15 | 8.5E-07 | 1 | 0.41 | 1.18 | 2.3E-06 | +++ | 1.0E-11 | |
| rs5950506 | 115175748 | G/A | 0.420 | 1.14 | 1.2E-06 | 10 | 0.92 | 0.418 | 1.18 | 1.E-06 | +++ | 1.1E-11 | |||
For each suggestive locus (P < 1×10-6, discovery stage), the SNP showing the strongest association in the replication stage is shown below the index SNP (the most significant SNP in discovery stage). Only SNPs with P < 1×10-6 (discovery stage) and their close proxies (r 2 > 0.8) were tested for replication. Replicated regions are highlighted in bold.
For each region, the gene closest to the index SNP was shown.
SNP positions were based on GRCh37/hg19. Alleles were given based on positive strand of reference genome. Allele B is used as the reference allele for frequency and effect.
For gestational length, effect is unstandardized regression coefficient, which shows the estimated changes in gestational days per allele (B). For preterm birth, effect is the estimated odds ratio of the reference allele (B).
Discovery stage P-values were adjusted by inflation factors. The replication stage P-values were calculated from the inflation adjusted effect sizes and standard error of the three Nordic studies using fixed-effect meta-analysis. Joint-analysis P-values were calculated from 23andMe and combined Nodic studies using the inverse variance method.
Directions represent whether the effects observed in the three Nordic studies (FIN/MoBa/DNBC) are same (+) or different (-) from the effects estimated from the 23andMe discovery cohort.
For each locus, the rank (based on the P-value in discovery stage) of the most significant SNP in replication stage (show in italic) together with the r 2 with the index SNP was provided. The r 2 was estimated from haplotype data of the Phase 1 1000 Genomes EUR samples.
Replication of suggestive genomewide-associated loci
For each of the 14 loci from the discovery stage, we examined the replication association signals (P value and direction of effect) at the index SNP and other SNPs with P < 1×10-6 in the discovery stage, and their close proxies (r 2 > 0.8). Six loci (Table 1 and Table S7, S8) replicated given a significance threshold adjusted for the effective number of independent SNPs at a locus as well as the number of loci tested (Table S5) and the direction of the effect. The 6 loci include EBF1, EEFSEC, and AGTR2, which were associated with both gestational length and preterm birth; WNT4, ADCY5 and RAP2C, which were associated with gestational length but not with preterm birth at the significance level for genomewide discovery (P < 1×10-6). In addition, associations of the BOLA3 locus with gestational length, and the TEKT3 and the TGFB1 loci with preterm birth showed marginal significance (P < 0.05). At the EBF1, EEFSEC, AGTR2, WNT4 and RAP2C loci, the most significant SNPs in the replication stage were either same as or in substantial LD (r 2 > 0.6) with the most significant SNPs in discovery stage. However, at the ADCY5 locus, the LD between the most significant SNPs in replication stage and the discovery stage is less substantial (r 2 < 0.4). SNPs at the EEFSEC locus showed nominally significant dominant effects (P.dom < 0.05) (Table S7, S8).
Annotation of SNPs at significant loci
A number of SNPs with potentially functional impact (i.e. nonsense, missense and splicing SNPs) are encompassed by the loci we identified as potentially important (Table S5). However, none of those potentially functionally important variants are in close LD (r 2 >0.8) with SNPs significantly associated with gestational length or preterm birth in the discovery stage. Within these loci, there are SNPs reportedly associated with complex traits (GWAS Catalog 28 ) (Table S5). Among these, three previously identified SNPs (rs10934853, rs2999052 and rs2687729) in the EEFSEC locus were significantly associated with gestational length and preterm birth. The alleles that were associated with longer gestational length (or reduced risk of preterm birth) have also been associated with increased risk of prostate cancer (rs10934853-A) 34 , reduced risk of hypospadias (rs2999052-C) 35 and later age of menarche (rs2687729-G). 36,37 Five significant SNPs in the WNT4 locus were previously associated with endometriosis, 38 ovarian cancer 39 and bone mineral density. 40 The alleles that increased gestational length in our analysis have also been identified as high-risk alleles for endometriosis, ovarian cancer or low bone mineral density (Table S9). Our eQTL analyses showed that some significant SNPs at the associated loci can significantly influence expression level of nearby genes (cis-expression QTLs) based on GTEx data 29 (Table S10 and S11).
SNPs at the ADCY5 locus have been reported to be associated with birth weight 41 and blood glucose traits. 42 More recently, a large meta-analysis has revealed SNPs at the ADCY5, WNT4 and EBF1 loci that are associated with birth weight. 43 The SNPs at the ADCY5 and WNT4 loci appear to influence birth weight through the fetal genome and none of them were in close LD with the SNPs showing significant association with gestational length; while the SNP (rs7729301) at the EBF1 locus seems to influence birth weight through the maternal effect, and the allele (G) associated with reduced birth weight was also associated with shorter gestational length (Table S5).
Maternal or fetal genetic effect
Association analyses of the top regions in the infant samples from our Nordic data sets (Table S12 and S13) yielded weaker associations. The results showed the same direction of effect but smaller effect sizes for the top significantly replicated SNPs (Table S14), supporting the inference that the loci identified in this study are “maternal” loci. The effect sizes estimated from infant samples were highly correlated (ρ = 0.95) and approximately half of the effect sizes estimated from maternal samples (Figure S4), supporting that the effect observed in infants is due to sharing of one maternal allele by descent. In addition, joint association analysis in mother/infant pairs with both maternal and fetal genotypes as predictors demonstrated significant associations exclusively with maternal genotypes but not with fetal genotypes (Table S15), which again indicated the maternal origin of the observed genetic associations.
We also evaluated our findings for detection of allelic heterogeneity, dominance effects, percentage of the variance explained, and gene set enrichment/pathway analyses. These results are presented in the Supplementary Text and include Figures S5-9 and Tables S16-19.
Functional evidence implicating the WNT4 locus
The genetic loci we identified fall in noncoding regions of the genome, suggesting that they will affect gene regulation rather than protein function. To dissect the consequences of these variants, knowledge of cell-type context in which they are active is essential. The WNT4 locus provides an especially attractive region, as unlike the other loci we identified, it implicates a particular tissue context related to its role in pregnancy, the endometrium. 44 The variants we identified also associate with risk for endometriosis, 45 and WNT4 function is critical for decidualization of the endometrium and subsequently implantation and establishment of pregnancy. 46 Therefore, we sought to analyze the expression of the WNT4 gene in human endometrial stromal cells, before and after decidualization (Supplementary Methods). Using RNA sequencing, we confirmed a substantial induction of WNT4 mRNA with decidualization – average of 0.0 transcripts per million (TPM) prior to decidualization in vitro to 29.5 TPM after decidualization in samples run in duplicate from two different endometrial stromal cell lines.
We next sought to identify particular regulatory mechanisms controlling the expression of WNT4 that might be altered by variants associated with gestational length. To this end, we used the CisBP web server 47 to predict the specific transcription factors whose binding might be altered by any of the six gestational length-associated variants that localize to the WNT4 locus. These analyses indicate that rs3820282 (r 2=0.94 with the index SNP rs56318008), which is located in the first intron of WNT4 is capable of altering the binding of the estrogen receptor (ESR1). Specifically, the underlying quantitative data from protein binding microarray (PBM) assays 48 indicate that the minor allele (T) of rs3820282 “creates” a near-perfect half-site for ESR1 (Figure 2) – the PBM-derived E-score for the major allele (C) is 0.09 (no binding), whereas the minor allele (T) is 0.46 (strong binding). Importantly, ESR1 and ESR2 are the only two human nuclear receptors that bind GGTCA half-sites with an "IR3" (Inverted Repeat 3) pattern, 49 eliminating the ~50 other nuclear receptors from consideration. Further, ChIP-seq peaks for ESR1 are present in four different experiments performed in MCF7 cells 50 , indicating that ESR1 is capable of binding to this locus in a cellular context. We confirmed the presence of H3K4me3 marks and an open chromatin domain by ATAC-seq overlapping rs3820282 (Figure 2b) in an immortalized endometrial stromal cell line (Supplementary Methods), demonstrating that the chromatin over this locus is likely accessible and active in these cells. Importantly, we observed enhanced binding of ESR1 to the T allele of rs3820282 in electrophoretic mobility shift assays, as predicted by the in silico analysis (Figure 2c). Collectively, these data suggest that the likely mechanism underlying the gestational length association in the WNT4 locus is modulation (via rs3820282) of the binding of ESR1. rs3820282 is also strongly associated with epithelial ovarian cancer 39 (Table S9), suggesting that this same mechanism might be acting in multiple diseases.
Figure 2. ESR1 binding at the WNT4 locus.
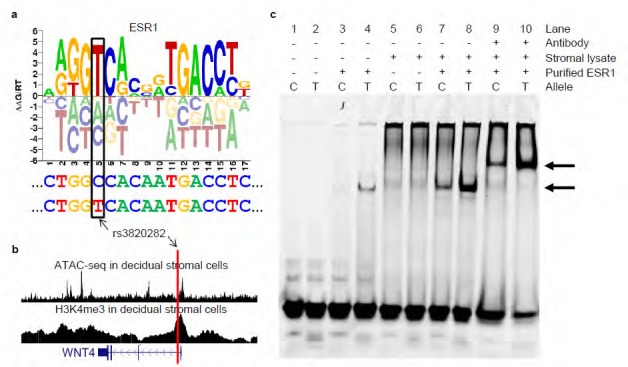
a. The rs3820282 T allele creates a stronger ESR1 binding site. The ESR1 binding motif ‘sequence logo’ (taken from the CisBP web server) illustrates the DNA binding preferences of ESR1. Tall nucleotides above the X-axis indicate DNA bases preferred by ESR1. Bases below the X-axis are disfavored. The sequence located in the WNT4 promoter is shown below, with the T allele for rs3820282 shown at the bottom. Note that the T allele changes the sequence from C (most disfavored) to T (most preferred). b. rs3820282 overlaps ATAC-seq and H3K4me3 signals in decidual stromal cells at the WNT4 locus. The red vertical line indicates the position of rs3820282. The location of the WNT4 gene is depicted at the bottom. Tall blocks indicate exons, medium height blocks indicate UTRs, and thin lines indicate introns. Arrows within introns indicate the direction of transcription. c. Experimental validation of allele-dependent binding of ESR1 to rs3820282 by electrophoretic mobility shift assay (EMSA). Fluorescently-labeled rs3820282 probe with either the C or T allele was incubated with nuclear extracts of decidual stromal endometrial cells in the presence or absence of purified ESR1 and/or antibody against ESR1. Lane pairs indicate C and T alleles. Preferential binding of ESR1 to the T allele is observed through an increased signal intensity of the shifted band. Upper and lower arrows indicate the locations of supershifted and shifted bands, respectively. Left to right - Lanes 1+2: negative control lanes containing only oligos; Lanes 3+4: increased binding of purified ESR1 to the T allele. Lanes 5+6: limited binding in the presence of nuclear extract only, due to low expression of ESR1 in these cells; Lanes 7+8: substantial allelic binding is detected with the addition of purified ESR1 to the nuclear extract; Lanes 9+10: Supershift using an ESR1 antibody
Discussion
Our genomewide association study is the first to identify human genetic polymorphisms that are significantly and reproducibly associated with gestational length and preterm birth, the single greatest contributor to mortality in children younger than five years and a common source of morbidity throughout life for those who survive. Our approach demonstrates the utility of using data collected as part of direct-to-consumer genotyping and phenotyping to rapidly assemble large data sets capable of revealing contributing loci in particularly complex phenotypes such as preterm birth where both maternal and fetal genomes, and many genes, are likely to contribute to the outcome. By combining the power of a large 43,568-person discovery data set and stringent replication by the well phenotyped Nordic data sets, we identified and replicated six maternal genomic loci robustly associated with gestational length and three of them also associated with preterm birth with genomewide significance (P<5E-8) in the joint analysis.
The top four replicating genomewide significant SNPs for gestational length are in biologically plausible genes. EBF1 (early B-cell factor 1), also achieving genomewide significance for preterm birth, has been demonstrated to be essential for normal B cell development, 51 and recent genomewide association studies have implicated it in control of blood pressure, 52,53 carotid artery intima media thickness, 54 hypospadias, 35 and metabolic risk. 55 Whether EBF1 confers its effect on birth timing through pregnancy-specific mechanisms, or by contributing to more general cardiovascular or metabolic traits that influence gestation remains to be determined. In addition, the association between this locus and gestational length may explain the effect of this locus on birth weight reported by Horikoshi et al. 43
EEFSEC (eukaryotic elongation factor, selenocysteine tRNA-specific), also genomewide significant for both gestational length and preterm birth risk, participates in the incorporation of selenocysteine into selenoproteins. Selenoproteins, such as the glutathione peroxidases and thioredoxin reductases, serve critical cellular homeostatic functions in maintaining redox status and antioxidant defenses, as well as modulating inflammatory responses. 56 These physiologic functions have previously been linked to the parturition process and preterm birth. 5,57,58 Moreover, the SNPs we identified in EEFSEC are in high LD with SNPs that have previously been associated with age of onset of menarche, expression quantitative trait loci (eQTLs) for EEFSEC abundance, risk of prostate cancer 34 and hypospadias. 35 Intriguingly, the identification of the selenocysteine pathway suggests the potential benefit for further evaluating the role of maternal selenium micronutrient status on prematurity risk. While a recent Cochrane review of multiple micronutrient supplementation did not demonstrate a reduction in preterm birth risk, 59 the studies included for analysis did not all utilize selenium as part of their supplement. Indeed, a recent evaluation of maternal serum selenium concentration in early pregnancy demonstrated reduced selenium concentration in association with preterm birth, 60 and, while of multi-factorial etiology, the country with the highest global preterm birth risk, Malawi, 61 demonstrates a high frequency of selenium-deficiency. 62
AGTR2 (angiotensin II receptor, type 2), the coding gene nearest to a group of X chromosome SNPs achieving genomewide significance in the gestational length analysis, had suggestive association with preterm birth discovery stage, and genomewide significance for preterm birth in the joint analysis with robust association in the Nordic replication. AGTR2 has been suggested to play a role in modulating uteroplacental circulation, and harbors variants that may contribute to the risk of preeclampsia. 63,64 The involvement of the renin-angiotensin system in blood flow at the maternal-fetal interface and oxidative stress, interacting with the selenoprotein glutathione perioxidase, a target for EEFSEC, is a potential shared mechanism for these genes in spontaneous preterm birth. 65 It is unlikely that our association detects risk for preeclampsia rather than spontaneous preterm birth, because women with preeclampsia as a reason for their delivery were excluded in the Nordic studies, and were removed from the 23andMe discovery data set if medical indications for delivery were reported.
The final gene locus achieving genomewide significance in the discovery stage for gestational length was WNT4 (wingless-type MMTV integration site family member 4), with strong replication in the Nordic populations. WNT4 mutations have been found in women with Mullerian duct abnormalities, primary amenorrhea, and hyperandrogenism, 66 and common variants in WNT4, in high LD with our index SNPs, are associated with risk for endometriosis 38 , ovarian cancer 39 and bone mineral density. 40 Our analysis indicates that the minor allele (T) of the putative causative variant rs3820282 in the Nordic populations is associated with longer gestational length and is protective for preterm birth. rs3820282 is located in an active chromatin domain in the first intron of WNT4, and the T allele generates a strong ESR1 binding site, and as such likely alters estrogen-based regulation of WNT4 and/or adjacent genes. The role of estrogen signaling as the functional consequence of the polymorphism is further supported by the association of the same region with endometriosis and ovarian cancer, both hormone-responsive disorders. Further, the parallel of the spectrum of disorders associated with the WNT locus mirrors that of ARID1A, also critical for endometrial function early in pregnancy, with loss of function variants causing atypical endometriosis and ovarian cancer, and enhanced estrogen activity. 67,68 Lastly, the population frequencies for endometriosis (Asian>European>African ancestry) trend in the same direction as does the T allele for rs3820282 (EAS 0.49 > EUR 0.14 > AFR 0.01 based on 1000 Genomes). 69,70 WNT4 did not achieve genomewide significance or suggestive association in the preterm birth risk dichotomous trait analysis, suggesting its role may be largely exerted near term gestation.
ADCY5 (adenylyl cyclase type 5) and RAP2C (member of the RAS oncogene family) achieved near genomewide significance in the discovery stage and were successfully replicated (Table 1). SNPs at the ADCY5 locus have been reported to be associated with birth weight 41 and type 2 diabetes; 42 however, none of them were in close LD with the SNPs showing significant association with gestational length, suggesting shared mechanisms coordinating the duration of gestation with growth. The SNP rs2747022 in the RAP2C region (in gene FRMD7) was previously reported to be associated with spontaneous preterm delivery in Danish/Norwegian studies (the samples used in this previous study overlap with our replication samples). 22 Several additional loci (BOLA3, TEKT3 and TGFB1), while showing marginal evidence of replication, remain suggestive and await the addition of further studies for analysis.
The primary limitation of our study centers on the characteristics of our study data sets. The gestational length information of the 23andMe samples was self-reported, and 3.2% of women in the preterm group did not respond as to whether the labor and delivery was spontaneous or medically indicated. In the term group, we were not able to unambiguously determine spontaneous from medically-indicated births. Despite these limitations, we included these samples in order to dramatically increase the sample size of the discovery stage, recognizing that our replication data sets would be more precisely phenotyped for spontaneous preterm birth. A previous study suggested that approximately 90% of mother-reported gestational lengths agreed with their associated medical records. 71 In addition, other than maternal age and ancestry inferred by genotypes, other covariates were not available for the 23andMe samples.
Our study demonstrates the utility of combining large samples with self-reported phenotyping with more modestly sized but precisely phenotyped replication studies to reveal maternal loci associated with gestational length and preterm birth. With this foundation, future expansion of maternal and fetal genotyped samples associated with gestational length information is anticipated to further refine our understanding of human pregnancy, risk for adverse pregnancy outcomes, and targeting of new preventive strategies for preterm birth. As the National Institutes of Health expand “Precision Medicine” initiatives in the years ahead, we would argue that the optimal time to advance human health is before and during pregnancy. Our work suggests that integration of genomic information on women, and likely their offspring, with birth timing, may allow development of new options for preventative and therapeutic measures.
Supplementary Material
Acknowledgements
The authors thank the participants in the Finnish birth cohort (FIN), Mother Child Cohort of Norway (MoBa), and Danish National Birth Cohort (DNBC) and dbGAP for depositing and hosting the phenotype and genotype data. We would also like to thank the research participants and employees of 23andMe for making this work possible.
Funding support for establishing the Danish National Birth Cohort (DNBC) was provided by the Danish National Research Foundation, the Danish Pharmacists' Fund, the Egmont Foundation, the March of Dimes Birth Defects Foundation, the Augustinus Foundation, and the Health Fund of the Danish Health Insurance Societies. GWAS data for the DNBC obesity study (Genomics of Obesity in Young Adults) were funded by the Wellcome Trust (WT 084762MA). GWAS data for the preterm birth project DNBC samples were generated within the GENEVA consortium with funding provided through the NIH Genes, Environment, and Health Initiative (GEI, preterm birth: U01HG004423, dbGaP accession number phs000103.v1.p1). Assistance with genotype cleaning and general study coordination was provided by the GENEVA Coordinating Center (U01HG004446).
Professor Hugh Taylor, Department of Obstetrics, Gynecology and Reproductive Sciences, Yale Medical School, provided biopsy samples of endometrial stromal fibroblasts for RNAseq analysis. Immortalized human endometrial stromal fibroblasts used here for ChIPseq analysis were provided by the lab of Professor Gil More, also Yale OB/GYN. GPW lab is grateful to both of them for their support. Work in the Wagner lab was supported by a grant from the John Templeton Foundation (grant #54860). The opinions expressed in this article are not those of the John Templeton Foundation.
This work was supported by grants from the March of Dimes (22-FY15-003, 21-FY16-121) National Institutes of Health, Cincinnati Children’s Hospital Medical Center, Fifth Third Foundation, Bill and Melinda Gates Foundation (OPP1113966), Jane and Aatos Erkko Foundation, Norwegian Research Council (FUGE 183220/S10, FRIMEDKLI-05 ES236011), Jane and Dan Olsson Foundations and Swedish government grants to researchers in the public health service (ALFGBG-507701), and the European Community’s Seventh Framework Programme (FP7/2007-2013), grant agreement HEALTH-F4-2007-201413. BF is supported by an Oak Foundation fellowship. XL is supported by the Nordic Center of Excellence in Health- Related e-Sciences (NIASC). The Norwegian Mother and Child Cohort Study was supported by the Norwegian Ministry of Health and the Ministry of Education and Research, NIH/NIEHS (contract no N01-ES-75558), NIH/NINDS (grant no.1 UO1 NS 047537-01 and grant no.2 UO1 NS 047537-06A1), and Norwegian Research Council/FUGE (grant no. 151918/S10; FRIMEDBIO 249779) and the Swedish Research Council (2015-02559). Support from the Functional Genomics Core at Cincinnati Children’s Hospital Medical Center was made possible through NIH P30 AR070549.
Footnotes
Publisher's Disclaimer: This is an Author Final Manuscript, which is the version after external peer review and before publication in the Journal. The publisher’s version of record, which includes all New England Journal of Medicine editing and enhancements, is available at 10.1056/NEJMoa1612665..
Contributor Information
Ge Zhang, Division of Human Genetics, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, USA; Center for Prevention of Preterm Birth, Perinatal Institute, Cincinnati Children’s Hospital Medical Center and March of Dimes Prematurity Research Center Ohio Collaborative, Cincinnati, OH, USA.
Bjarke Feenstra, Department of Epidemiology Research, Statens Serum Institut, Copenhagen, Denmark.
Jonas Bacelis, Department of Obstetrics and Gynecology, Sahlgrenska University Hospital Östra (East), Gothenburg, Sweden.
Xueping Liu, Department of Epidemiology Research, Statens Serum Institut, Copenhagen, Denmark.
Lisa M. Muglia, Center for Prevention of Preterm Birth, Perinatal Institute, Cincinnati Children’s Hospital Medical Center and March of Dimes Prematurity Research Center Ohio Collaborative, Cincinnati, OH, USA.
Julius Juodakis, Department of Obstetrics and Gynecology, Institute of Clinical Sciences, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden.
Daniel E. Miller, Center for Autoimmune Genomics and Etiology, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, USA.
Nadia Litterman, 23andMe, Inc. Mountain View, CA, USA.
Pan-Pan Jiang, 23andMe, Inc. Mountain View, CA, USA.
Laura Russell, 23andMe, Inc. Mountain View, CA, USA.
David A. Hinds, 23andMe, Inc. Mountain View, CA, USA.
Youna Hu, 23andMe, Inc. Mountain View, CA, USA.
Matthew T. Weirauch, Center for Autoimmune Genomics and Etiology; Divisions of Biomedical Informatics and Developmental Biology, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, USA.
Xiaoting Chen, Center for Autoimmune Genomics and Etiology, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, USA.
Arun R. Chavan, Department of Ecology and Evolutionary Biology, Yale University, New Haven, CT, Yale Systems Biology Institute, West Haven, CT, USA.
Günter P. Wagner, Department of Ecology and Evolutionary Biology, Yale University, New Haven, CT, Yale Systems Biology Institute, West Haven, CT, Department of Obstetrics, Gynecology and Reproductive Sciences, Yale Medical School, New Haven, CT, Department of Obstetrics and Gynecology, Wayne State University, Detroit, MI, USA.
Mihaela Pavličev, Center for Prevention of Preterm Birth, Perinatal Institute, Cincinnati Children’s Hospital Medical Center and March of Dimes Prematurity Research Center Ohio Collaborative, Cincinnati, OH, USA.
Mauris C. Nnamani, Department of Ecology and Evolutionary Biology, Yale University, New Haven, CT, Yale Systems Biology Institute, West Haven, CT, USA.
Jamie Maziarz, Department of Ecology and Evolutionary Biology, Yale University, New Haven, CT, Yale Systems Biology Institute, West Haven, CT, USA.
Minna K. Karjalainen, PEDEGO Research Unit and Medical Research Center Oulu, University of Oulu and Department of Children and Adolescents, Oulu University Hospital, Oulu, Finland.
Mika Rämet, PEDEGO Research Unit and Medical Research Center Oulu, University of Oulu and Department of Children and Adolescents, Oulu University Hospital, Oulu, Finland.
Verena Sengpiel, Department of Obstetrics and Gynecology, Sahlgrenska University Hospital Östra (East), Gothenburg, Sweden.
D Frank Geller, Department of Epidemiology Research, Statens Serum Institut, Copenhagen, Denmark.
Heather A. Boyd, Department of Epidemiology Research, Statens Serum Institut, Copenhagen, Denmark.
Aarno Palotie, Analytic and Translational Genetics Unit, Department of Medicine, Massachusetts General Hospital, Boston, MA, USA.; Program in Medical and Population Genetics, The Broad Institute of MIT and Harvard, Cambridge, MA, USA; The Stanley Center for Psychiatric Research, The Broad Institute of MIT and Harvard, Cambridge, MA, USA. Institute for Molecular Medicine Finland, University of Helsinki, Helsinki, Finland. ; Psychiatric & Neurodevelopmental Genetics Unit, Department of Psychiatry, Massachusetts General Hospital, Boston, MA, USA.; Department of Neurology, Massachusetts General Hospital, Boston, MA, USA.
Allison Momany, Department of Pediatrics, Carver College of Medicine, University of Iowa, Iowa City, IA, USA.
Bruce Bedell, Department of Pediatrics, Carver College of Medicine University of Iowa, Iowa City, IA, USA.
Kelli K. Ryckman, Department of Epidemiology, College of Public Health and Department of Pediatrics, Carver College of Medicine, University of Iowa, Iowa City, IA, USA.
Johanna M. Huusko, PEDEGO Research Unit and Medical Research Center Oulu, University of Oulu and Department of Children and Adolescents, Oulu University Hospital, Oulu, Finland; Center for Prevention of Preterm Birth, Perinatal Institute, Cincinnati Children’s Hospital Medical Center and March of Dimes Prematurity Research Center Ohio Collaborative, Cincinnati, OH, USAaww.
Carmy R. Forney, Center for Autoimmune Genomics and Etiology, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, USA.
Leah C. Kottyan, Center for Autoimmune Genomics and Etiology, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, USA.
Mikko Hallman, PEDEGO Research Unit and Medical Research Center Oulu, University of Oulu and Department of Children and Adolescents, Oulu University Hospital, Oulu, Finland.
Kari Teramo, Obstetrics and Gynecology, University of Helsinki and Helsinki University Hospital, Helsinki, Finland.
Ellen A. Nohr, Research Unit of Gynaecology & Obstetrics, Institute of Clinical Research, University of Southern Denmark, Odense, Denmark.
George Davey-Smith, Medical Research Council Integrative Epidemiology Unit (IEU) at the University of Bristol, School of Social and Community Medicine, University of Bristol, Oakfield House, Oakfield Grove, Bristol, United Kingdom.
Mads Melbye, Department of Epidemiology Research, Statens Serum Institut, Copenhagen, Denmark; Department of Clinical Medicine, University of Copenhagen, Copenhagen, Denmark; Department of Medicine, Stanford University School of Medicine, Stanford, CA, USA.
Bo Jacobsson, Department of Obstetrics and Gynecology, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden; Department of Genetics and Bioinformatics, Area of Health Data and Digitalisation, Norwegian Institute of Public Health, Oslo, Norway.
Louis J. Muglia, Division of Human Genetics, Center for Prevention of Preterm Birth, Perinatal Institute, Cincinnati Children’s Hospital Medical Center and March of Dimes Prematurity Research Center Ohio Collaborative, Cincinnati, OH, USA.
References
- 1.Martin JA, Hamilton BE, Osterman MJ. Births in the United States, 2014. NCHS data brief 2015:1-8. [PubMed] [Google Scholar]
- 2.Yoshida S, Martines J, Lawn JE, et al. Setting research priorities to improve global newborn health and prevent stillbirths by 2025. J Glob Health 2016;6:010508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu L, Oza S, Hogan D, et al. Global, regional, and national causes of child mortality in 2000-13, with projections to inform post-2015 priorities: an updated systematic analysis. Lancet 2015;385:430-40. [DOI] [PubMed] [Google Scholar]
- 4.Butler AS, Behrman RE. Preterm Birth:: Causes, Consequences, and Prevention: National Academies Press; 2007. [PubMed] [Google Scholar]
- 5.Muglia LJ, Katz M. The enigma of spontaneous preterm birth. N Engl J Med 2010;362:529-35. [DOI] [PubMed] [Google Scholar]
- 6.Romero R, Dey SK, Fisher SJ. Preterm labor: one syndrome, many causes. Science 2014;345:760-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bezold KY, Karjalainen MK, Hallman M, Teramo K, Muglia LJ. The genomics of preterm birth: from animal models to human studies. Genome Med 2013;5:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clausson B, Lichtenstein P, Cnattingius S. Genetic influence on birthweight and gestational length determined by studies in offspring of twins. BJOG : an international journal of obstetrics and gynaecology 2000;107:375-81. [DOI] [PubMed] [Google Scholar]
- 9.York TP, Eaves LJ, Lichtenstein P, et al. Fetal and maternal genes' influence on gestational age in a quantitative genetic analysis of 244,000 Swedish births. American journal of epidemiology 2013;178:543-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plunkett J, Feitosa MF, Trusgnich M, et al. Mother's genome or maternally-inherited genes acting in the fetus influence gestational age in familial preterm birth. Hum Hered 2009;68:209-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kistka ZA, DeFranco EA, Ligthart L, et al. Heritability of parturition timing: an extended twin design analysis. American journal of obstetrics and gynecology 2008;199:43 e1-5. [DOI] [PubMed] [Google Scholar]
- 12.Boyd HA, Poulsen G, Wohlfahrt J, Murray JC, Feenstra B, Melbye M. Maternal contributions to preterm delivery. American journal of epidemiology 2009;170:1358-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altman DG, Royston P. The cost of dichotomising continuous variables. Bmj 2006;332:1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirschhorn JN. Genomewide association studies—illuminating biologic pathways. N Engl J Med 2009;360:1699-701. [DOI] [PubMed] [Google Scholar]
- 15.McCarthy MI, Abecasis GR, Cardon LR, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nature reviews Genetics 2008;9:356-69. [DOI] [PubMed] [Google Scholar]
- 16.Monangi NK, Brockway HM, House M, Zhang G, Muglia LJ. The genetics of preterm birth: Progress and promise. Semin Perinatol 2015;39:574-83. [DOI] [PubMed] [Google Scholar]
- 17.Wu W, Clark E, Manuck T, Esplin M, Varner M, Jorde L. A Genome-Wide Association Study of spontaneous preterm birth in a European population [version 1; referees: 2 approved with reservations]2013. [Google Scholar]
- 18.Zhang H, Baldwin DA, Bukowski RK, et al. A genome-wide association study of early spontaneous preterm delivery. Genetic epidemiology 2015;39:217-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Durand EY, Do CB, Mountain JL, Macpherson JM. Ancestry Composition: A Novel, Efficient Pipeline for Ancestry Deconvolution. bioRxiv 2014. [Google Scholar]
- 20.Genomes Project C, Abecasis GR, Auton A, et al. An integrated map of genetic variation from 1,092 human genomes. Nature 2012;491:56-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magnus P, Birke C, Vejrup K, et al. Cohort Profile Update: The Norwegian Mother and Child Cohort Study (MoBa). International journal of epidemiology 2016;45:382-8. [DOI] [PubMed] [Google Scholar]
- 22.Myking S, Boyd HA, Myhre R, et al. X-chromosomal maternal and fetal SNPs and the risk of spontaneous preterm delivery in a Danish/Norwegian genome-wide association study. PLoS One 2013;8:e61781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olsen J, Melbye M, Olsen SF, et al. The Danish National Birth Cohort—its background, structure and aim. Scandinavian journal of public health 2001;29:300-7. [DOI] [PubMed] [Google Scholar]
- 24.24. Ryckman KK, Feenstra B, Shaffer JR, et al. Replication of a genome-wide association study of birth weight in preterm neonates. The Journal of pediatrics 2012;160:19-24 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nohr EA, Timpson NJ, Andersen CS, Davey Smith G, Olsen J, Sorensen TI. Severe obesity in young women and reproductive health: the Danish National Birth Cohort. PLoS One 2009;4:e8444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paternoster L, Evans DM, Nohr EA, et al. Genome-wide population-based association study of extremely overweight young adults—the GOYA study. PLoS One 2011;6:e24303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang G, Bacelis J, Lengyel C, et al. Assessing the Causal Relationship of Maternal Height on Birth Size and Gestational Age at Birth: A Mendelian Randomization Analysis. PLoS medicine 2015;12:e1001865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Welter D, MacArthur J, Morales J, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res 2014;42:D1001-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nature genetics 2013;45:580-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang J, Ferreira T, Morris AP, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nature genetics 2012;44:369-75, S1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang J, Benyamin B, McEvoy BP, et al. Common SNPs explain a large proportion of the heritability for human height. Nature genetics 2010;42:565-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. American journal of human genetics 2011;88:76-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang G, Karns R, Sun G, et al. Extent of height variability explained by known heightassociated genetic variants in an isolated population of the Adriatic coast of Croatia. PLoS One 2011;6:e29475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gudmundsson J, Sulem P, Gudbjartsson DF, et al. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nature genetics 2009;41:1122-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geller F, Feenstra B, Carstensen L, et al. Genome-wide association analyses identify variants in developmental genes associated with hypospadias. Nature genetics 2014;46:957-63. [DOI] [PubMed] [Google Scholar]
- 36.Perry JR, Day F, Elks CE, et al. Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature 2014;514:92-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elks CE, Perry JR, Sulem P, et al. Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nature genetics 2010;42:1077-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Albertsen HM, Chettier R, Farrington P, Ward K. Genome-wide association study link novel loci to endometriosis. PLoS One 2013;8:e58257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuchenbaecker KB, Ramus SJ, Tyrer J, et al. Identification of six new susceptibility loci for invasive epithelial ovarian cancer. Nature genetics 2015;47:164-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kemp JP, Medina-Gomez C, Estrada K, et al. Phenotypic dissection of bone mineral density reveals skeletal site specificity and facilitates the identification of novel loci in the genetic regulation of bone mass attainment. PLoS genetics 2014;10:e1004423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Freathy RM, Mook-Kanamori DO, Sovio U, et al. Variants in ADCY5 and near CCNL1 are associated with fetal growth and birth weight. Nature genetics 2010;42:430-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dupuis J, Langenberg C, Prokopenko I, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nature genetics 2010;42:105-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horikoshi M, Beaumont RN, Day FR, et al. Genome-wide associations for birth weight and correlations with adult disease. Nature 2016;538:248-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sonderegger S, Pollheimer J, Knofler M. Wnt signalling in implantation, decidualisation and placental differentiation—review. Placenta 2010;31:839-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nyholt DR, Low SK, Anderson CA, et al. Genome-wide association meta-analysis identifies new endometriosis risk loci. Nature genetics 2012;44:1355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Q, Kannan A, Das A, et al. WNT4 acts downstream of BMP2 and functions via betacatenin signaling pathway to regulate human endometrial stromal cell differentiation. Endocrinology 2013;154:446-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weirauch MT, Yang A, Albu M, et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 2014;158:1431-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weirauch MT, Cote A, Norel R, et al. Evaluation of methods for modeling transcription factor sequence specificity. Nat Biotechnol 2013;31:126-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang Q, Chen Y, Meyer C, et al. A comprehensive view of nuclear receptor cancer cistromes. Cancer Res 2011;71:6940-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Griffon A, Barbier Q, Dalino J, van Helden J, Spicuglia S, Ballester B. Integrative analysis of public ChIP-seq experiments reveals a complex multi-cell regulatory landscape. Nucleic Acids Res 2015;43:e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gyory I, Boller S, Nechanitzky R, et al. Transcription factor EBF1 regulates differentiation stage-specific signaling, proliferation, and survival of B cells. Genes Dev 2012;26:668-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.International Consortium for Blood Pressure Genome-Wide Association S, Ehret GB, Munroe PB, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 2011;478:103-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wain LV, Verwoert GC, O'Reilly PF, et al. Genome-wide association study identifies six new loci influencing pulse pressure and mean arterial pressure. Nature genetics 2011;43:1005- 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xie G, Myint PK, Voora D, et al. Genome-wide association study on progression of carotid artery intima media thickness over 10 years in a Chinese cohort. Atherosclerosis 2015;243:30-7. [DOI] [PubMed] [Google Scholar]
- 55.Singh A, Babyak MA, Nolan DK, et al. Gene by stress genome-wide interaction analysis and path analysis identify EBF1 as a cardiovascular and metabolic risk gene. Eur J Hum Genet 2015;23:854-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Labunskyy VM, Hatfield DL, Gladyshev VN. Selenoproteins: molecular pathways and physiological roles. Physiol Rev 2014;94:739-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burnum KE, Hirota Y, Baker ES, et al. Uterine deletion of Trp53 compromises antioxidant responses in the mouse decidua. Endocrinology 2012;153:4568-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goldenberg RL, Culhane JF, Iams JD, Romero R. Epidemiology and causes of preterm birth. Lancet 2008;371:75-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haider BA, Bhutta ZA. Multiple-micronutrient supplementation for women during pregnancy. Cochrane Database Syst Rev 2015;11:CD004905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rayman MP, Wijnen H, Vader H, Kooistra L, Pop V. Maternal selenium status during early gestation and risk for preterm birth. CMAJ 2011;183:549-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Organization WH. Born too soon: the global action report on preterm birth. 2012. [Google Scholar]
- 62.Hurst R, Siyame EW, Young SD, et al. Soil-type influences human selenium status and underlies widespread selenium deficiency risks in Malawi. Sci Rep 2013;3:1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Akbar SA, Khawaja NP, Brown PR, Tayyeb R, Bamfo J, Nicolaides KH. Angiotensin II type 1 and 2 receptors gene polymorphisms in pre-eclampsia and normal pregnancy in three different populations. Acta obstetricia et gynecologica Scandinavica 2009;88:606-11. [DOI] [PubMed] [Google Scholar]
- 64.Zhou A, Dekker GA, Lumbers ER, et al. The association of AGTR2 polymorphisms with preeclampsia and uterine artery bilateral notching is modulated by maternal BMI. Placenta 2013;34:75-81. [DOI] [PubMed] [Google Scholar]
- 65.Mistry HD, Kurlak LO, Broughton Pipkin F. The placental renin-angiotensin system and oxidative stress in pre-eclampsia. Placenta 2013;34:182-6. [DOI] [PubMed] [Google Scholar]
- 66.Philibert P, Biason-Lauber A, Gueorguieva I, et al. Molecular analysis of WNT4 gene in four adolescent girls with mullerian duct abnormality and hyperandrogenism (atypical Mayer- Rokitansky-Kuster-Hauser syndrome). Fertil Steril 2011;95:2683-6. [DOI] [PubMed] [Google Scholar]
- 67.Maeda D, Shih Ie M. Pathogenesis and the role of ARID1A mutation in endometriosisrelated ovarian neoplasms. Adv Anat Pathol 2013;20:45-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim TH, Yoo JY, Wang Z, et al. ARID1A Is Essential for Endometrial Function during Early Pregnancy. PLoS genetics 2015;11:e1005537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Missmer SA, Hankinson SE, Spiegelman D, Barbieri RL, Marshall LM, Hunter DJ. Incidence of laparoscopically confirmed endometriosis by demographic, anthropometric, and lifestyle factors. American journal of epidemiology 2004;160:784-96. [DOI] [PubMed] [Google Scholar]
- 70.Mangtani P, Booth M. Epidemiology of endometriosis. Journal of epidemiology and community health 1993;47:84-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Little RE. Birthweight and gestational age: mothers' estimates compared with state and hospital records. American Journal of Public Health 1986;76:1350-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.