Abstract
The steady advances in machine learning and accumulation of biomedical data have contributed to the development of numerous computational models that assess the impact of missense variants. Different methods, however, operationalize impact differently. Two common tasks in this context are the prediction of the pathogenicity of variants and the prediction of their effects on a protein’s function. These are related but distinct problems and it is unclear whether methods developed for one are optimized for the other. The Critical Assessment of Genome Interpretation (CAGI) experiment provides a means to address this question empirically. To this end, we participated in various protein-specific challenges in CAGI with two objectives in mind. First, to compare the performance of methods in the MutPred family with the state-of-the-art. Second and more importantly, to investigate the applicability of general-purpose pathogenicity predictors to the classification of specific function-altering variants without additional training or calibration. We find that our pathogenicity predictors performed competitively with other methods, outputting score distributions in agreement with experimental outcomes. Overall, we conclude that binary classifiers learned from disease-causing mutations are capable of modeling important aspects of the underlying biology and the alteration of protein function resulting from mutations.
Keywords: CAGI, pathogenicity prediction, functional effect prediction, MutPred, severity, generalization, machine learning, MutPred2
Introduction
In the era of high-throughput sequencing, computational methods that predict the impact of newly discovered variants have become integral to the studies of disease (Peterson et al., 2013; Niroula and Vihinen, 2016; Rost et al., 2016). Different methods, however, use different definitions of impact, depending on the established practices in the research community that their developers and users belong to. Broadly speaking, an impactful variant can be interpreted in an evolutionary sense as ‘deleterious’; i.e., affecting reproductive fitness, or in a biochemical and molecular sense, as ‘damaging’ to a protein’s function; i.e., function-affecting (MacArthur et al., 2014). Another term, ‘pathogenicity’, has been used to describe disease-causing variants and is usually interpreted as clinically significant (Richards et al., 2015). These terms are often used interchangeably and, consequently, predictions from one class of methods may be misused to inform decisions regarding related but different concepts. It has been pointed out, for instance, that evolutionary conservation is not sufficient to establish pathogenicity (MacArthur et al., 2014) and that many variants that affect protein function appear to be disease-neutral (Schaefer et al., 2012; Bromberg et al., 2013; Lugo-Martinez et al., 2016).
Computational methods for variant impact prediction either rely on rules derived from expert knowledge or on data-driven machine learning algorithms; see Peterson et al. (2013) for an overview. The basic idea behind supervised machine learning methods involves ‘training’ a computer to ‘learn’ a mathematical function (model) that relates observable features (e.g., protein sequence) to known outcomes (e.g., protein function), in order to make predictions on a previously unseen set of observed features. Ideally, the choice of training data and features leads to the optimization of a mathematical function that captures the underlying biology. For instance, it has been observed that a predictor of protein-protein interaction (PPI) sites tends to predict hotspots (residues critical for interactions) more strongly than other interacting residues (Ofran and Rost, 2007). However, this behavior is not guaranteed. In the context of impact prediction, there are methods, particularly for missense variants, that rely on similar training data and feature sets to predict various notions of impact. Several questions, thus, naturally arise. Do these models learn some principles that underlie what makes an amino acid substitution impactful? Are they all learning similar or different classification functions? Or how can one use available data from disparate experimental studies to improve learning of each notion of impact?
Before addressing such questions, it is important to understand these methods in the context of the aforementioned variance in terminology. Supervised or semi-supervised learning methods can be divided into those that are trained on variants demonstrated to affect protein function in vitro or in vivo; e.g., SNAP (Bromberg and Rost, 2007), SNAP2 (Hecht et al., 2015), and those that are trained directly on known pathogenic variants; e.g., PhD-SNP (Capriotti et al., 2006), MutPred (Li et al., 2009), PolyPhen-2 (Adzhubei et al., 2010), FATHMM (Shihab et al., 2013) and VEST (Carter et al., 2013). This allows us to disambiguate the two different but related binary classification tasks that these methods address: the classification of amino acid substitutions as (1) protein function-altering or not, and (2) pathogenic or benign. This distinction has been made previously (Schaefer et al., 2012) and has recently been reiterated (Hecht et al., 2015; Richards et al., 2015).
Pathogenic amino acid substitutions can be understood as those that severely affect protein function (note that we do not specifically address variants that disrupt protein structure and its dynamics, but rather group those into the broader classes of function-impacting, pathogenic, or other variants depending on their downstream consequences). Therefore, a predictor trained on functional variants should be able to predict pathogenic ones as those that score highly. Indeed, it has been shown that pathogenic amino acid substitutions can be predicted using functional effect predictors with reasonable accuracy (Schaefer et al., 2012; Reeb et al., 2016). Schaefer et al. (2012) further demonstrated that SNAP’s prediction scores for disease mutations tend to be higher than even those for the function-altering mutations that it was trained on and that PhD-SNP predicted nearly two-thirds of function-altering substitutions from SNAP’s training set as disease-causing. However, Bromberg et al. (2013) found that SIFT and PolyPhen-2 do not capture effect severity as well as SNAP does. Therefore, it remains unclear under what conditions and to what extent pathogenicity predictors can directly be transferred to infer the effects of substitutions on the specific aspects of protein function.
There are several factors to consider when investigating the generalizability of pathogenicity predictors to the functional effect prediction problem. First, the definition of functional and/or pathogenic variants varies between studies and depends on the evidence at hand. Genetic evidence is insufficient when factors such as penetrance and epistatic effects are unaccounted for. In vitro assays, on the other hand, provide evidence of functional impact in controlled settings but do not necessarily reflect what happens at the cellular or organismal level. In vivo experiments rely on model organisms and cell lines, and their findings may not necessarily translate to the clinical setting. Currently, training variants for pathogenicity predictions are obtained from databases such as the Human Gene Mutation Database (HGMD) (Stenson et al., 2014) and SwissVar (Mottaz et al., 2010), which do not necessarily annotate the lines of evidence used to establish impact and are sparse on specific experimental outcomes. This suggests that pathogenicity predictors, in particular, are exposed to a broad spectrum of functional outcomes under the umbrella of ‘pathogenic’ and ‘benign’. Second, there has been a growing debate over whether general-purpose pathogenicity prediction models trained on variants combined from multiple genes are more powerful than gene-specific models. Recently, a case has been made for the development of both as the performance advantages relate to the amount and composition of the training data available for each protein (Riera et al., 2016). Finally, in spite of substantial curation efforts, errors and biases are major issues with public databases (Schnoes et al., 2009, 2013). This has particularly been noted in the case of HGMD (George et al., 2008; Cassa et al., 2013) and it is not clear if learned models are robust to this noise in practice.
Community experiments such as the Critical Assessment of Genome Interpretation (CAGI) provide excellent opportunities to not only assess the status of the field but also address the aforementioned questions through controlled studies. To this end, we participated in multiple challenges in CAGI by primarily submitting predictions from MutPred, a random-forest based predictor that we previously developed to classify pathogenic and benign variants (Li et al., 2009). A distinguishing feature of MutPred is that it includes internal predictors for specific structural and functional properties such as secondary structure, intrinsic disorder, DNA-binding, phosphorylation, and others. For a given amino acid substitution, MutPred runs these predictors on the original and mutated sequence, models the putative change in the propensities for these properties and uses them to predict pathogenicity. Sequence-and structure-based analyses have revealed that such changes in local properties account for a substantial fraction of disease-causing mutations and can classify pathogenic variants more effectively than conservation-based approaches (Li et al., 2009; Mort et al., 2010; Lugo-Martinez et al., 2016). However, it is not clear whether such site-specific or region-specific changes in structure and function are predictive of impact beyond the protein level.
In this study, we take advantage of the numerous data sets available as part of the CAGI experiment to assess the utility of general sequence-based methods for pathogenicity prediction such as MutPred for more specific prediction tasks. In particular, we address the following questions: (1) Are pathogenicity predictors trained on mutations from multiple proteins useful for individual protein-specific prediction tasks? (2) Can predictors trained to distinguish between pathogenic and benign variants predict the outcomes of function-specific experimental assays? (3) Are pathogenicity predictors robust to heterogeneity in evidence for pathogenicity/functionality in current training and test sets? (4) Do supervised learning approaches capture the biologically intuitive relationship between pathogenicity and functionality? By addressing these questions, we aim to provide a more thorough understanding of the variant impact predictors and lay the groundwork for the development of next-generation methodologies.
Materials and Methods
Data sets
Over the four iterations of the CAGI experiment, several prediction challenges accompanied by data sets obtained through different types of experimental techniques have been archived on the CAGI website (https://genomeinterpretation.org/). In this study, only those data sets containing mutations (predominantly amino acid substitutions) from a single gene or a small set of genes were selected (Table 1). For each challenge, raw ground truth values were extracted from the official results file provided along with the data set. For four challenges, experimental results have already been published by the data providers and were obtained directly from the literature: the p16 challenge (Scaini et al., 2014), the NaV1.5 channel challenge (Calloe et al., 2013), the 2010 CBS challenge (Dimster-Denk et al., 2013) and the CHEK2 challenge (Le Calvez-Kelm et al., 2011). When numbering all the mutations in this work, the initiation codon was assumed to be codon 1.
Table 1.
Summary of data sets selected for this study
| Gene symbol |
Protein name | Challenge name |
CAGI year |
Number of protein variants |
Number of variants with amino acid substitutions |
|---|---|---|---|---|---|
| NAGLU | Alpha-N-acetylglucosaminidase | NAGLU | 2015 | 165 | 165 |
| PKLR | Pyruvate kinase PKLR | Pyruvate kinase | 2015 | 543 | 543 |
| UBE2I | SUMO-conjugating enzyme UBC9 | SUMO ligase | 2015 | 5,109 | 5,109a |
| NPM-ALK | Nucleophosmin - Anaplastic Lymphoma Kinase fusion protein | NPM-ALK | 2015 | 23 | 19a |
| BRCA1 | Breast cancer type 1 susceptibility protein | BRCA | 2013 | 36 | 19 |
| BRCA2 | Breast cancer type 2 susceptibility protein | 64 | 42 | ||
| CDKN2A | Cyclin-dependent kinase inhibitor 2A | p16 | 2013 | 10 | 10 |
| MRE11A | Double-strand break repair protein MRE11A | MRN | 2013 | 51 | 24 |
| NBN | Nibrin | 40 | 24 | ||
| RAD50 | DNA repair protein RAD50 | RAD50 | 2011 | 69 | 35 |
| SCN5A | Sodium channel protein type 5 subunit alpha | NaV1.5 channel | 2011 | 3 | 3 |
| CBS | Cystathionine beta-synthase | CBS | 2011 | 84 | 84 |
| 2010 | 51 | 51 | |||
| CHEK2 | Serine/threonine-protein kinase Chk2 | CHEK2 | 2010 | 41 | 34a |
Includes protein variants with more than one substitution
Data processing
For all challenges, only missense variants were extracted from the original data sets. Variants without experimental values and/or predictions (due to mismatch between reference residue in the variant and that in the reference sequence) were excluded. For challenges where experimental quantities were expressed as percentages, the values were mapped to the [0, 1] interval. In challenges where the combined effects of multiple mutations were to be predicted, the predictors were run on each mutation individually and the maximum value was chosen as the final prediction. This approach was chosen over other potential approaches in a systematic analysis on the SUMO ligase challenge and applied to the other data sets without further experimentation. The underlying assumption of this approach was that the combined effects of multiple substitutions would not exceed the effect of the most deleterious variant. Other challenge-specific aspects of data set construction are outlined below.
The MRN, RAD50 and CHEK2 challenges contained data sets derived from case-control sequencing studies. In these special cases, the ground truth for each variant was available through the numbers of case and control individuals harboring it. When counts for different sub-populations were available, they were summed together to create single pooled count sets. To establish ground truth values, the probability of pathogenicity for a variant was estimated in two ways: (1) as the fraction of its occurrence in case individuals among all individuals carrying the variant, and (2) as the same fraction when the imbalance between case and controls was accounted for. This case-control-balanced frequency was calculated as
where ‘carrier’ indicates that the variant is carried by an individual. A frequency value of 0.5 indicates that a variant is equally likely to occur in the case and control cohorts. Any variant with a value greater than this was treated as a pathogenic variant. To assess the effect of this frequency threshold on predictor performance, it was varied between 0.1 and 0.9 in increments of 0.1 and AUC values were calculated at each threshold.
In the case of CBS, binary class labels (non-functional or other) and raw experimental values for the 2010 and 2011 challenges were obtained from the work of Dimster-Denk et al. (2013) and the CAGI repository, respectively. ‘Sensitive’ mutations were included in the ‘non-functional’ class and ‘Heme rescue’ mutations were excluded. Only those raw experimental values that corresponded to the assay conditions defined in the original challenge description were extracted.
For the SUMO ligase challenge, experimental values could be negative and were set to zero to fall within our predictors’ score distributions. Apart from evaluating predictions in terms of raw experimental values, all variants were classified into four groups, based on thresholds defined by the official CAGI assessors. If a value for a variant was < 0.3, it was considered to be ‘deleterious’. If its value was ≥ 0.3 and < 0.7, it was treated as ‘unsure’. If the variant had a value ≥ 0.7 and < 1.3, it was considered to be ‘wild-type’ and any variant with a value ≥ 1.3 was treated as ‘advantageous’.
Predictors
Pathogenicity predictions were made on all missense mutations from these data sets using two different predictors. First, MutPred, an existing random forest-based method that relies on sequence, conservation, predicted structural and functional features to make pathogenicity predictions, was used (Li et al., 2009). Second, a neural network ensemble using an expanded feature set, was trained on a much larger and more heterogeneous data set obtained from HGMD (Stenson et al., 2014), SwissVar (Mottaz et al., 2010), dbSNP (Sherry et al., 2001), and others (details of this approach will be described elsewhere). This predictor will be referred to as MutPred2 (Pejaver, et al., 2017). The predictor was run in two modes, with or without accounting for gene families in training. These features simply enumerate proteins in the human and mouse genomes at various levels of sequence identity to the protein in which the variant is observed. We informally refer to these features as ‘homolog counts’. Both MutPred and MutPred2 require only a protein sequence and an amino acid substitution as input, and output scores between zero (benign) and one (pathogenic). For challenges where experimental assay scores were expressed as a fraction of the wild-type function, prediction scores were ‘inverted’ by subtracting them from one. Thus, a score of zero would indicate a complete loss of function and a score of one would indicate similar function levels as the wild-type protein.
Evaluation
For prediction tasks that were treated as soft (unthresholded) classification problems, the predictors were assessed as rankers. The area under the receiver operating characteristic (ROC) curve (AUC) was therefore chosen as an evaluation metric (Fawcett, 2006). In the case of regression tasks, to measure the concordance between prediction scores and actual experimental values, one or more of the following measures was used: Pearson’s correlation, Spearman’s rank correlation and the root-mean-square deviation (RMSD) (Rosner, 2010). The choice of metric varied depending on how evaluations were undertaken by the official CAGI assessors for each challenge. Performance values for other methods were obtained from the assessors’ materials on the CAGI website. For more systematic comparisons, the reader is referred to the assessors’ publications.
Results
The CAGI experiment has resulted in a valuable repository of data sets associated with a variety of prediction tasks. For the purposes of this study, we concentrated on protein-specific data sets rich in amino acid substitutions. This resulted in a collection of 11 data sets covering 13 proteins, with the number of protein variants ranging from three to 5,109 (Table 1). Depending on the experimental means used to determine ground truth values, this collection can be grouped into three broad categories: (a) challenges with genetic evidence from large sequencing studies (BRCA1, BRCA2, CHEK2, MRE11A, NBN and RAD50), (b) challenges with biochemical functional assays (NAGLU, NPM-ALK, PKLR) and, (c) challenges that measure effects such as growth rate at the cellular level (CBS, CDKN2A, SCN5A, UBE2I). We note that these proteins can also be grouped along the lines of their phenotypic roles. For instance, BRCA1, BRCA2, CDKN2A, CHEK2, the MRN complex proteins, and the NPM-ALK fusion protein have all been implicated in a variety of cancers while CBS, NAGLU, PKLR and SCN5A have been implicated in relatively rare genetic disorders. In the following sections, we describe the nuances associated with the specific prediction tasks within each of the three aforementioned groups and their evaluation.
Prediction of the effects of pathogenic amino acid substitutions identified through sequencing studies
In this section, we describe results for prediction challenges where the true values for impact were assigned based solely on genetic evidence with no additional biochemical or molecular experiments. For the four challenges covered here, frequency of occurrence or the mere presence of variants in patient and control cohorts largely influenced their categorization as being pathogenic.
BRCA challenge
Myriad Genetics, a molecular diagnostic company, has created the proprietary BRACAnalysis® test for the assessment of a woman’s risk of developing hereditary breast or ovarian cancer. This test is based on the detection of mutations in BRCA1 and BRCA2 and their classification into one of four classes: (1) Deleterious, (2) Benign, (3) Genetic Variant - Favor Polymorphism (VFP), and (4) Variant of Unknown Significance (VUS). The basis for these designations is proprietary but is thought to incorporate information from patient testing, population-level variant frequencies and segregation of variants in families with disease. In the BRCA challenge, the goal was to predict the probability that Myriad Genetics classified each of 100 BRCA variants into one of these four classes. Apart from this multiclass classification task, a more straight-forward task was to predict the probability that Myriad Genetics classified a given variant as deleterious (a binary classification problem). Since our predictors were directly amenable to this task, we concentrated on the prediction of the deleteriousness of variants, particularly missense variants. As a result, of the 19 and 42 missense variants in BRCA1 and BRCA2, respectively, only four and seven were considered for evaluation. The remaining missense variants had been classified as VUS by Myriad Genetics and any quantitative evaluation was infeasible.
The predictions for these 11 variants are summarized in Table 2 along with their class labels. In the case of BRCA1, both predictors returned perfect predictions with AUC values of one. For BRCA2, the AUC values of MutPred and MutPred2 were 0.90 and 0.67, respectively. However, when the two variants classified as ‘probably benign’ were excluded, both methods resulted in AUCs of one. This suggests that the difference between the methods lies in how such ‘probably benign’ variants were ranked relative to other variants in BRCA2. In fact, the MutPred2 score distribution was such that a low score threshold was sufficient to classify variants as deleterious (Table 2). This was probably due to the selection of the MutPred2 model that included protein-level homolog counts as features. Although this model performed better than the one without these features, it typically rescales scores based on the number of homologs that a given protein has in human and mouse. Overall, while it appears that MutPred performed better than MutPred2, the results were inconclusive, due to the small data set size, class imbalance and the lack of additional experimental evidence for deleteriousness. We note that only MutPred predictions were included in our official submission to the challenge and it was found to be the best-performing method by the assessors.
Table 2.
Predictions on missense variants classified by Myriad Genetics
| (i) BRCA1 | |||
| Variant | True classification | MutPred score | MutPred2 score |
|
| |||
| p.Cys39Phe | deleterious | 0.99 | 0.76 |
| p.Tyr777Asp | benign | 0.54 | 0.54 |
| p.Ile1275Val | benign | 0.20 | 0.02 |
| p.Asp1546Glu | benign | 0.11 | 0.03 |
| (ii) BRCA2 | |||
| Variant | True classification | MutPred score | MutPred2 score |
|
| |||
| p.Val211Leu | deleterious | 0.24 | 0.29 |
| p.Ile975Ser | deleterious | 0.37 | 0.34 |
| p.Arg2336Hisa | deleterious | 0.69 | 0.15 |
| p.Val3079Ile | deleterious | 0.58 | 0.09 |
| p.Leu2368Val | benign | 0.19 | 0.09 |
| p.Glu2956Ala | probably benign | 0.23 | 0.18 |
| p.Pro3243Leu | probably benign | 0.35 | 0.21 |
Present in both predictors’ training sets as a pathogenic variant
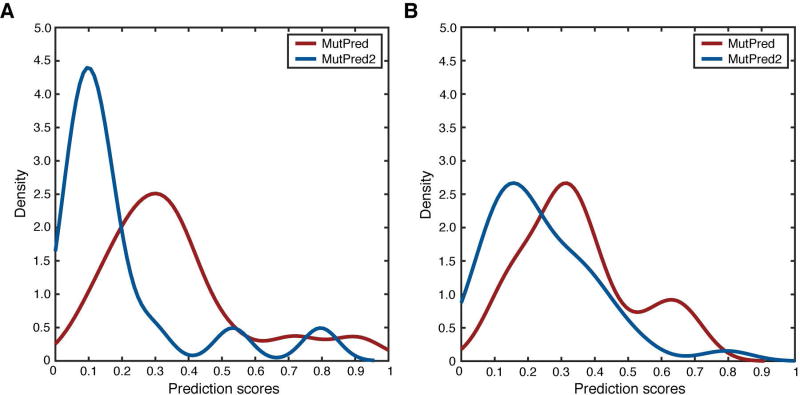
We then performed a qualitative analysis of the scores from both predictors for the 50 variants designated as VUS. We found that VUSs from both genes were dominated by benign predictions for both predictors (Figure 1). This is in general agreement with the majority of the official CAGI submissions. However, the score distributions of MutPred and MutPred2 differed from each other significantly for both BRCA1 (Kolmogorov-Smirnov P = 1.1 × 10−3) and BRCA2 (Kolmogorov-Smirnov P = 9.7 × 10−3). In the case of MutPred, the score distribution peaked in the interval [0.3, 0.4] for both BRCA1 and BRCA2 and was slightly bimodal for BRCA2. On the other hand, MutPred2 favored lower scores, peaking in the interval [0.1, 0.2] for both genes with a shorter right tail for BRCA2. This trend agrees with that in Table 2. We speculate that, in addition to the homolog count features, the different score distributions for the two predictors largely arise due to an expanded feature set and the use of neural networks in MutPred2.
Figure 1.
MRN and CHEK2 challenges
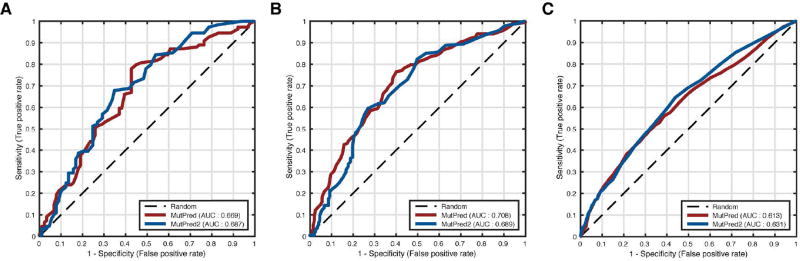
In addition to the BRCA genes, there were four other genes with only genetic evidence for pathogenicity of their variants in the CAGI experiment. These were MRE11A, NBN, RAD50 and CHEK2 and their variants were provided in three distinct challenges: the MRN challenge (for the first two genes), the RAD50 challenge and the CHEK2 challenge. However, all of these challenges were structured similarly. Variants in these four genes were obtained from breast cancer case and control individuals through population-level sequencing at the respective loci; for details see (Le Calvez-Kelm et al., 2011; Damiola et al., 2014). The goal of these challenges was to predict the probability of a given variant occurring in an individual from the case group. Since the challenge focused primarily on rare variants, these probabilities (derived from frequencies) were very similar to each other and evaluating the task as a regression problem would be too stringent. Therefore, we treated this task as a binary classification problem, applied our predictors to only missense variants and evaluated them using AUC. However, it is important to note that the definition of whether a variant was truly pathogenic or not depended on its frequency in cases and controls. Therefore, in addition to the default threshold of 0.5, we investigated the effects of varying class label thresholds on AUCs as well.
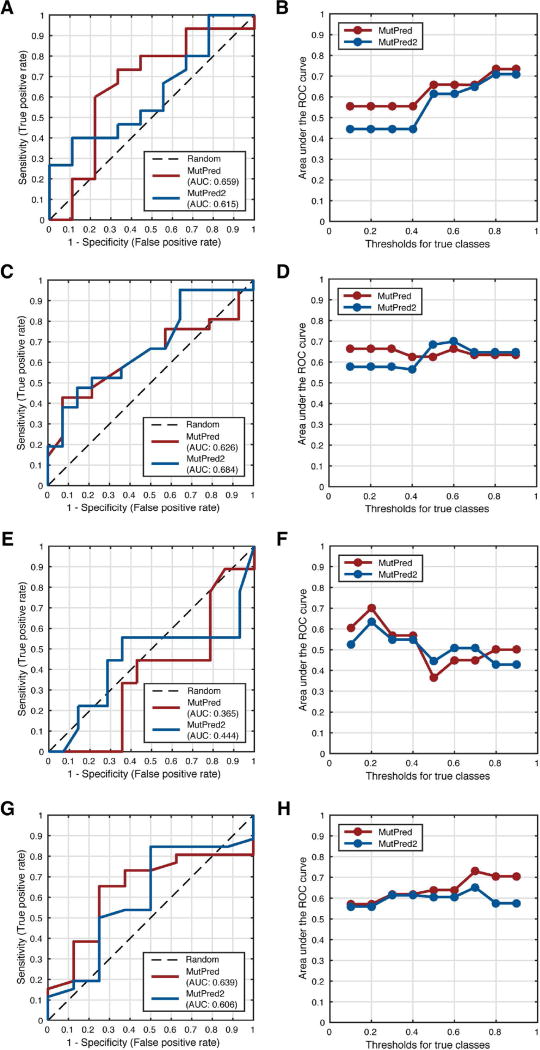
Evaluation results for all four genes are summarized in Figure 2. In all four cases, the MutPred2 model with homolog count features was used as it performed comparably or better than the model without these features. In the case of MRE11A, the AUC for MutPred was higher than that for MutPred2 (0.66 vs. 0.62). However, at low fpr values, MutPred2 identified more true positives than MutPred (Figure 2A). As the cutoff for pathogenicity was increased, both predictors improved performance and converged to similar AUC values (Figure 2B). For RAD50, although MutPred had a smaller AUC value than MutPred2 (0.63 vs. 0.68), the performances were very similar, differing only at high fpr values (Figure 2C). Again, both predictors showed similar performance at higher frequency thresholds, with MutPred being more stable (Figure 2D). NBN deviated from the other members of the MRN complex in that both predictors performed worse than random at the default frequency threshold, with MutPred2 doing slightly better (Figure 2E). Interestingly, this performance was recovered for both predictors when the threshold was decreased; i.e., unlike the previous case, more relaxed thresholds ([0.2, 0.3]) yielded better performances, with MutPred reaching AUC values up to 0.7 (Figure 2F). Finally, in Figures 2G and 2H, we describe the results for CHEK2. Although the AUC for MutPred was higher than that for MutPred2 (0.64 vs. 0.61), their true positive rates (tpr) in the [0, 0.1] fpr interval were comparable. Furthermore, AUC values for both predictors at lower frequency thresholds were equal, with MutPred’s performance improving at more stringent thresholds. These results held even when the true pathogenicity labels were assigned based on case-control-balanced frequencies (Supp. Tables S1–S4).
Figure 2.
We note that the official CAGI assessors used measures other than AUC. Although our AUC values for these four genes were modest at best, the official assessment concluded that MutPred was the best-performing method for the RAD50 challenge (MutPred2 had not been developed at the time). To the best of our knowledge, all submitted predictors performed poorly on MRE11A and NBN and our predictors were among the better-performing ones. Although some of the variants in these data sets were included in our predictors’ training sets (more so for MutPred2; data not shown), the comparable performances of both predictors indicates no significant advantage to either of our methods. This suggests that either variants from these genes are difficult to predict on with current approaches or that the frequency-based annotations of pathogenicity are generally unreliable.
Prediction of the effects of amino acid substitutions on protein activity and function
In these challenges, the ground truth values were established through in vitro biochemical assays. Typically, these experiments first involved expression of proteins of interest in cells followed by their extraction from cell lysates and purification. Then specific functions such as enzyme activity or binding affinity were assayed against known substrates or analogs. Although several mutations, and almost all of the proteins (NAGLU, NPM-ALK and PKLR), covered in this section are medically important, the prediction tasks here focused mostly on the specific functional consequences of these mutations, without regard to cellular or organismal phenotype.
NAGLU challenge
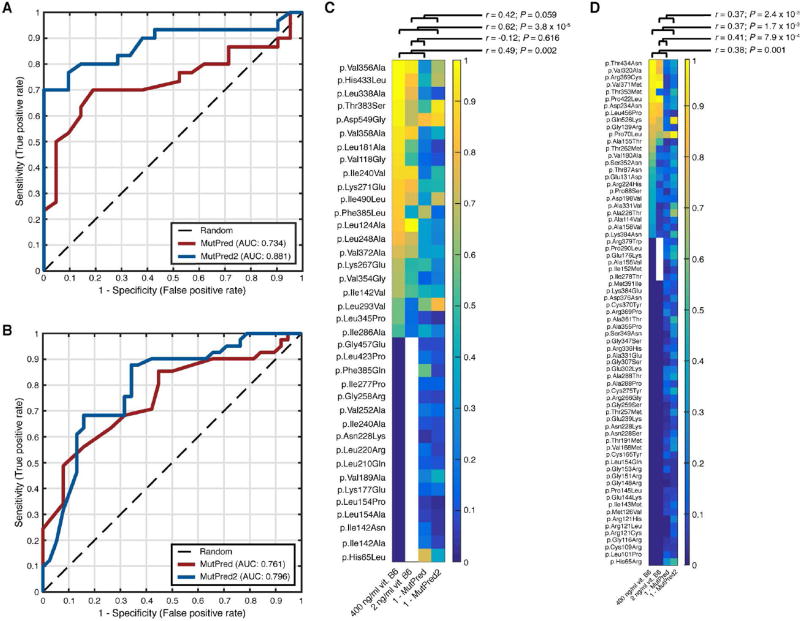
The most recent challenge in this category was the prediction of the effect of naturally-occurring missense mutations on the enzymatic activity of N-acetyl-glucosaminidase (NAGLU), a lysosomal glycohydrolyase. Deficiency of NAGLU and/or its mutations cause Mucopolysaccharidosis IIIB or Sanfillipo B disease. The data set consisted of 165 missense mutations extracted from the Exome Aggregation Consortium (ExAC) data resource (Lek et al., 2016). The effects of these mutations on NAGLU’s hydrolysis activity were assayed and quantified relative to the wild-type to establish ground truth values. An activity value of zero indicated complete abolition of activity, a value of one indicated wild-type levels of activity and any value greater than one indicated activity greater than the wild-type value. We evaluated our predictions in two ways. First, we treated every mutation with activity levels > 0.1 (Wyatt T. Clark, personal communication) as wild-type-like and evaluated the task as a binary classification problem. Second, we treated the task as a regression problem and used Pearson’s correlation coefficients to measure the agreement between our predicted activity values and actual experimental values.
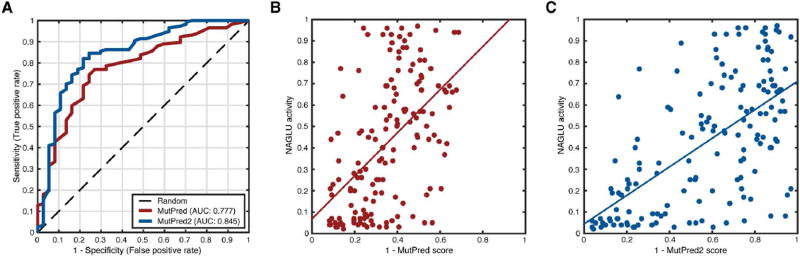
We found that the inclusion of homolog count features had little impact on the predictive performance of MutPred2 and, therefore, considered the model without these features. Irrespective of whether mutations with activity levels above 1 were included or not, the AUCs of MutPred and MutPred2 were 0.78 and 0.85, respectively (Figure 3A). This was the case even when considering Pearson correlation coefficients. MutPred was less correlated with activity values (r = 0.54; P = 3.4 × 10−13; t-test) than MutPred2 was (r = 0.62; P = 1.1 × 10−17; t-test) when mutations with activity levels above 1 were excluded. When these mutations were included, the corresponding correlation coefficients were 0.54 (P = 1.6 × 10−13; t-test) and 0.61 (P = 6.7 × 10−18; t-test). Overall, MutPred2 outperformed MutPred and was, in fact, judged to be the best-performing method among all submissions for this challenge.
Figure 3.
Interestingly, as in the case of the BRCA VUS, the distributions of scores output by MutPred and MutPred2 were very different. Unlike MutPred2, MutPred predicted no activity values to be greater than 0.7 and the spread of scores appeared more ‘compressed’ (Figure 3B and 3C). One possible explanation for this is that MutPred2 was trained on 22 more NAGLU variants than MutPred, about half of which were not pathogenic. On the other hand, MutPred was only presented with pathogenic NAGLU variants during training and may have tended to favor pathogenic predictions in this protein. However, it is unlikely that the inclusion of such a small set of training variants (relative to the whole training set) would substantially affect performance. For instance, seven non-pathogenic mutations from the NAGLU data set overlapped with MutPred2’s training set. However, three of these have activities < 0.5 and are more likely to be damaging mutations. Therefore, any advantages gained here by MutPred2 would be nullified. An alternative explanation for the difference in score distributions is that MutPred2 uses a neural network ensemble, resulting in a smoother classification function that scales scores more evenly than the random forest model in MutPred.
NPM-ALK challenge
NPM-ALK is a protein formed through the fusion of nucleophosmin (NPM1) and Anaplastic lymphoma kinase (ALK). This fusion results in the ALK tyrosine kinase being constitutively activated and contributing to cancer. NPM-ALK is dependent on the molecular chaperone Hsp90 for its stability and activity. In this challenge, the goal was to predict the effects of 23 mutations (19 missense) on two distinct and separately-assayed functions of NPM-ALK relative to the wild-type protein, kinase activity and Hsp90-binding affinity. In both cases, the experimental values were set to zero if the given function was completely lost, 0.5 if the activity or binding was less than that of the wild-type protein, one if the function was at wild-type levels and two if the function was above wild-type levels. We ran and evaluated our predictors solely for this study and did not officially participate in this challenge. We note that MutPred2 uses predicted losses and gains of specific protein properties (as a consequence of mutation) such as catalytic activity, stability and protein-protein interactions (PPIs) as features. Therefore, in addition to the general scores output by our predictors, we evaluated scores from these individual predictors as well.
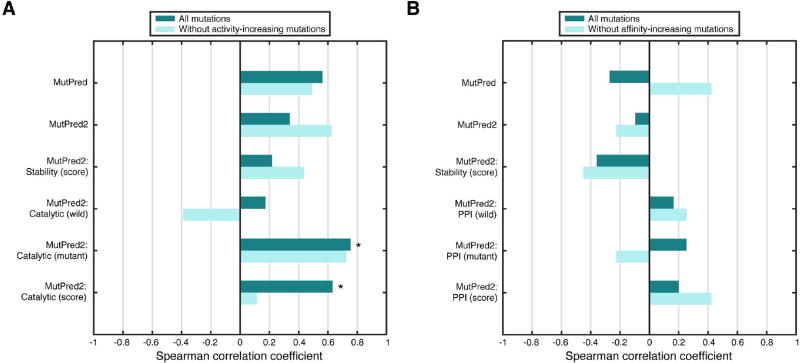
We note that all mutations in the data set were mappable to the ALK protein and predictions could be made on either NPM-ALK or ALK. We chose ALK, as our performance on this sequence was better overall. For the first task, MutPred correlated better with kinase activities than MutPred2 when all mutations were considered. However, the trend reversed when only those mutations with activity values ≤ 1 were considered (Figure 4A). To put these results in context, r of the best-performing method at CAGI was 0.88. However, we note that these methods were developed and/or customized for this challenge. When the assessors used general pathogenicity predictors such as SIFT (Ng and Henikoff, 2001), PolyPhen-2 (Adzhubei et al., 2010) and PROVEAN (Choi et al., 2012), the best-performing method (PolyPhen-2) had a coefficient of 0.47. Furthermore, when MutPred2’s internal catalytic site predictors were considered, correlation levels increased to those comparable to the top CAGI submissions.
Figure 4.
Next, we considered the Hsp90-binding affinity predictions. This task turned out to be much harder with the only positive correlation value arising from predictions made by MutPred when affinity-increasing mutations were excluded (Figure 4B). Interestingly, the PPI property predictors resulted in positive correlations in all but one case. However, none of these were statistically significant and were only better than MutPred when all mutations were considered. In the official assessment, all predictions were virtually random with the top r being 0.05, much below those of a majority of our approaches. These trends held even when considering Pearson’s correlation coefficients, except in the case of PPI mutant scores; a positive correlation was observed when affinity-increasing mutations were excluded, suggesting a strong linear relationship between the predicted and true values (Supp. Figure S1).
Pyruvate kinase challenge
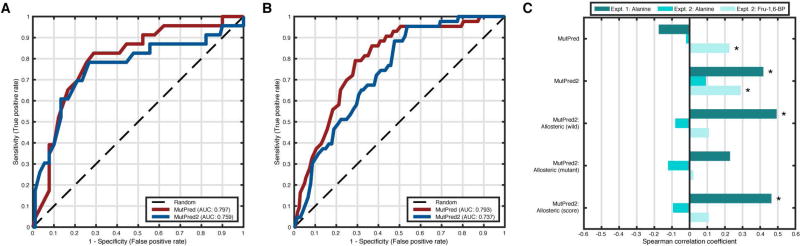
Pyruvate kinase is an enzyme that catalyzes the last step in glycolysis and is regulated by two allosteric effectors, alanine and fructose 1,6 bisphosphate (Fru-1,6-BP). Of the four isoforms expressed from the pklr gene in mammals, the one specific to the liver, L-PYK, is of particular interest. Variants in this gene have been associated with pyruvate kinase deficiency and, in L-PYK, several variants fall in or around allosteric effector-binding sites, suggesting a link between allosteric regulation and disease. In the pyruvate kinase challenge, there were two major goals. The first goal was to predict the effect of missense mutations on kinase activity defined simply as the presence or absence of activity. The second goal involved the prediction of the real-valued effects of these mutations on allosteric regulation by the two effectors. The values between zero and one indicated allosteric inhibition, values above one indicated allosteric activation and values of one indicated no allosteric effects. As in the case of the NPM-ALK challenge, in addition to our general impact scores, we also evaluated MutPred2’s predicted allosteric site-related features for the second task.
There were two data sets in this challenge that were derived from two experiments. The first data set consisted of 113 amino acid substitutions at nine different positions close to the alanine-binding site. On this data set, the AUC for MutPred was greater than that for the MutPred2 model with homolog count features (0.80 vs. 0.76) al-though the latter identified more activity-disrupting mutations at low fpr values (Figure 5A). The second data set consisted of 430 mutations generated through an alanine scan on all non-alanine/glycine positions in L-PYK. The trends here were similar (AUC of MutPred = 0.79 vs. AUC of MutPred2 = 0.74), with MutPred also doing slightly better than MutPred2 at low fpr values (Figure 5B). Although we did not officially participate in this challenge, it is instructive to contextualize our performance in relation to the state-of-the-art. The official assessors used balanced accuracy, calculated as the average of sensitivity and specificity (Fawcett, 2006), as the primary measure and found the best-performing methods to have values of 0.77 and 0.75 for the first and second data sets, respectively. We calculated MutPred’s balanced accuracy to be 0.54 and 0.61 at a score threshold of 0.5. The corresponding values for MutPred2 were 0.59 and 0.67. When score thresholds corresponding to the maximum possible balanced accuracy were considered, MutPred had values of 0.77 and 0.75 (score threshold: 0.80 and 0.81, respectively), and MutPred2 had values of 0.76 and 0.71 (score threshold: 0.74 and 0.59, respectively) for the two data sets.
Figure 5.
Next, we evaluated the ability of our methods to predict effects on allosteric regulation by alanine and Fru-1,6-BP. These were measured in both experiments for alanine and only in the second experiment for Fru-1,6-BP. Unlike other challenges, all mutations were retained here because none of the these values exceeded one for alanine and all but one were below one for Fru-1,6-BP. We found that MutPred2 was the only approach with positive correlations in all three cases (Figure 5C). Interestingly, the predicted allosteric site features in MutPred2 performed better than the original MutPred model, achieving positive correlations in two out of the three data sets. To put these results in perspective, Spearman’s rank correlation coefficients of the best-performing methods at CAGI were 0.35, 0.01 and 0.05 for the first and second alanine data sets and the Fru-1,6-BP data, respectively. Since none of these mutations were in our training sets, we interpret these results as follows. The score distribution returned by MutPred is optimized to perform better (and perhaps the best) when assessing kinase activity. However, MutPred2’s score distribution generalizes better to the different definitions of function in this challenge at the cost of a modest loss in accuracy when considering kinase activity (the primary function of L-PYK).
Prediction of the effects of amino acid substitutions measured at the cellular level
In order to establish the causal roles of variants in disease, it is often insufficient to demonstrate their effects on an isolated protein’s function. A common approach is to measure the effects of such mutations on measurable cellular parameters such as growth rate, detection of signals in or outside the cell, protein turnover, among others. The CAGI experiment included four such challenges: the SUMO ligase challenge, the p16 challenge, the SCN5A challenge and the CBS challenge (with two different data sets). Each of these used different experimental end-points to assign variant impact and are described in this section along with the evaluation of our predictions. It is important to note that these experiments are often carried out on cell lines and may or may not reflect actual phenotypes at the organism level.
SUMO ligase challenge
This challenge constitutes the largest data set of mutations in our collection and was generated through a high-throughput yeast complementation assay. UBE2I is the only human SUMO-conjugating protein (SUMO E2 ligase). It regulates the function of a multitude of proteins through the covalent attachment of small ubiquitin-like modifier proteins (SUMOs) to them (Geiss-Friedlander and Melchior, 2007). The goal here was to predict the effect of missense mutations on the function of UBE2I defined as the change in fraction of mutant SUMO ligase clones, relative to wild-type clones, in a competitive yeast growth assay. This set of variants was divided into three subsets. The first subset consisted of 219 amino acid substitutions whose experimental values were robust to multiple internal replicates. The second subset consisted of 463 amino acid substitutions whose experimental values were assigned with lower confidence than those in subset 1. Finally, the third subset consisted of 4,427 combinations of two or more amino acid substitutions occurring together. The predicted values were expected to be between zero (no growth) and one (wild-type growth), or greater than one (more growth than wild-type). We note that for this challenge, a score distribution was provided to calibrate raw prediction scores. While our official submissions included this calibration step, in this study, we aimed to investigate the direct applicability of our methods and, therefore, used the raw scores. Calibration only provided modest improvements over the results from the raw scores.
Based on thresholds established by the CAGI assessors, we divided the data set into two classes (deleterious or wild-type) and obtained ROC curves for both predictors (Figure 6). In subsets 1 and 2, we found that both predictors had comparable performances with slight advantages belonging to MutPred2 in subset 1 and MutPred in subset 2. This trend remained even when the ‘unsure’ variants were included in the ‘deleterious’ class (Table 3(i) and 3(ii)). However, both predictors’ performance improved on subset 1 and dropped on subset 2 when the ‘unsure’ variants were re-assigned. In this case, the trend for subset 1 agreed with the trend for subset 3 and we concluded that subset 2 deviated from the other subsets due to the noise introduced by a larger fraction of low-confidence growth values. As can be seen in Table 3, Spearman correlation coefficients between predicted and experimental growth rates followed a similar pattern as the AUC values with modest differences between the two predictors. We note that the exclusion of ‘unsure’ variants made both subsets easier to predict on and that the performance improved for both predictors (data not shown).
Figure 6.
Table 3.
Performance measures for the prediction of the deleteriousness of amino acid substitutions in SUMO ligase. All correlations are significant at a P-value threshold of 0.05.
| (i) Subset 1 | ||||
| Method | Processing | AUC | Spearman correlation coefficient | |
|
| ||||
| Unsure as "Wild-type" | Unsure as "Deleterious" | |||
|
| ||||
| MutPred | NA | 0.669 | 0.694 | 0.380 |
| MutPred2 | NA | 0.687 | 0.703 | 0.408 |
| (ii) Subset 2 | ||||
| Method | Processing | AUC | Spearman correlation coefficient | |
|
| ||||
| Unsure as "Wild-type" | Unsure as "Deleterious" | |||
|
| ||||
| MutPred | NA | 0.708 | 0.671 | 0.389 |
| MutPred2 | NA | 0.689 | 0.653 | 0.360 |
| (iii) Subset 3 | ||||
| Method | Processing | AUC | Spearman correlation coefficient | |
|
| ||||
| Unsure as "Wild-type" | Unsure as "Deleterious" | |||
|
| ||||
| MutPred | Meana | 0.615 | 0.625 | 0.197 |
| MutPred2 | 0.608 | 0.628 | 0.184 | |
| MutPred | Max | 0.613 | 0.624 | 0.189 |
| MutPred2 | 0.631 | 0.649 | 0.226 | |
| MutPred | Product | 0.601 | 0.624 | 0.182 |
| MutPred2 | 0.609 | 0.638 | 0.170 | |
Official submission to CAGI
Subset 3 was a harder task for both predictors as neither of them were designed to predict the combined effects of multiple substitutions. We therefore decided to run the predictors on individual mutations and perform post-processing to assign a single value to each multi-mutant protein variant. Table 3(iii) shows the three different post-processing methods that we explored and, clearly, taking the maximum predicted value among individual predictions for a variant worked the best for MutPred2 and the average predicted value worked slightly better for MutPred. Both predictors showed modest correlations with experimental values. We note that the official assessors for this particular challenge used multiple sophisticated metrics to identify the best method. Although our methods were not the best-performing, their performances were very similar to the state-of-the-art, suggesting that UBE2I is a hard target to predict on with current methods.
p16 challenge
The CDKN2A gene codes for two proteins through alternate splicing of its first exons, both of which have tumor suppressor functions. Among these, the p16 isoform inhibits cyclin dependent kinase (CDK4/6) and, thus, promotes cell cycle arrest and affects cell proliferation. Mutations that affect the inhibitory function of p16 cause cells to grow rapidly and contribute to malignant melanoma. The p16 challenge involved the prediction of cell proliferation rates as a consequence of 10 different mutations at four positions in p16. These rates were set by the data providers to be 0.5 for wild-type cells (negative controls) and one for tumor-like cells (positive controls). Since a mutation resulting in a growth rate closer to 0.5 could be interpreted as being ‘benign’ and that with a rate closer to one could be interpreted as ‘pathogenic’, we directly applied our predictors to this data set. Our predictions were evaluated in two ways. First, we treated every mutation with growth rate above 0.75 as tumor-like and evaluated the task as a binary classification problem. Second, we treated the task as a regression problem and calculated the RMSD to determine how close our predicted proliferation rates were to actual experimental values.
The results for the p16 data set are summarized in Table 4. When evaluated as a classification task, both models in MutPred2 (with or without homolog counts) outperformed MutPred (AUC values of 0.80 and 0.84, respectively, vs. 0.68). This is the case even for RMSD values except that the MutPred2 model with homolog counts had the lowest RMSD (0.1397), followed by the other MutPred2 model (0.1506) and MutPred (0.2061). As suggested before, the better performance of the homolog count model can be explained by the fact that the inclusion of these features rescales scores, which better mimic actual proliferation rate distributions. Since we did not participate in this challenge, we directly compared our performance values to other methods that were officially assessed. MutPred2 had an AUC value comparable to the top-performing methods and the second-best RMSD value among all methods submitted.
Table 4.
Predictions of the effects of 10 mutations in p16 on cell proliferation rates. Bold predictions represent those where the absolute difference between the real and predicted values < 0.20.
| Mutation | Proliferation rate |
Tumor- like?a |
MutPred | MutPred2b | ||
|---|---|---|---|---|---|---|
| Score | Abs. diff. |
Score | Abs. diff. |
|||
| p.Gly23Arg | 0.54 | No | 0.54 | 0.00 | 0.68 | 0.14 |
| p.Gly23Cysc | 0.87 | Yes | 0.83 | 0.03 | 0.89 | 0.02 |
| p.Gly23Arg | 0.92 | Yes | 0.56 | 0.35 | 0.79 | 0.13 |
| p.Gly23Ser | 0.69 | No | 0.52 | 0.17 | 0.66 | 0.04 |
| p.Gly23Val | 0.90 | Yes | 0.62 | 0.29 | 0.83 | 0.07 |
| p.Gly35Gluc | 0.60 | No | 0.92 | 0.32 | 0.80 | 0.20 |
| p.Gly35Arg | 0.54 | No | 0.80 | 0.26 | 0.84 | 0.30 |
| p.Gly35Trp | 0.86 | Yes | 0.81 | 0.05 | 0.82 | 0.04 |
| p.Leu65Proc | 0.66 | No | 0.78 | 0.12 | 0.59 | 0.08 |
| p.Leu94Proc | 0.94 | Yes | 0.95 | 0.01 | 0.83 | 0.11 |
Using a proliferation rate threshold of 0.75, as defined by CAGI assessors
Model with homolog count features shown here
Present in both predictors’ training sets as pathogenic mutations
NaV 1.5 challenge
This challenge involves the smallest data set among all the challenges described in this study. SCN5A encodes the NaV 1.5 integral membrane protein, primarily found in cardiac muscle cells and is responsible for mediating the fast influx of Na+ ions across the cell membrane. This influx, in turn, results in the fast depolarization of the cardiac action potential. Thus, this protein plays an important role in impulse propagation through the heart. Although mutations in SCN5A have been associated with multiple heart diseases, this challenge concentrates on those that have been linked to Brugada Syndrome (Calloe et al., 2013). The data set contains three missense mutations obtained by sequencing two independent families and screening the resulting mutations against a control group. The effects of these mutations on current flow were measured by patch clamp experiments and expressed as the fraction of the reduction in current density due to the mutation, when compared to the wild type protein. The goal of this challenge was to predict these current density fraction values. We did not officially participate in this challenge and ran our predictors on this data set solely for this study.
Table 5 shows the prediction scores for the three mutations along with actual current density values obtained from Calloe et al. (2013). Although the data set was too small to assign statistical significance to our results, we found the Spearman correlation coefficients to be one for both prediction methods. The Pearson correlation coefficients were 0.83 and 0.90 for MutPred and MutPred2, respectively. This suggests that while scores from both predictors were monotonically related to current density fractions, MutPred2 scores were linearly related to these fractions and could directly be applied to this prediction task. Interestingly, although both function-altering mutations were present in MutPred2’s training set as pathogenic, it was able to reasonably predict actual outcomes from the patch clamp experiments.
Table 5.
Predictions of the change in cellular current densities due to SCN5A mutations associated with Brugada syndrome
| Mutation | Fraction of current density reduction as compared with wild-type |
1 - Prediction score | |
|---|---|---|---|
|
| |||
| MutPred | MutPred2 | ||
| p.Arg620His | 1.00 | 0.64 | 0.93 |
| p.Arg811His | 0.57 | 0.14 | 0.22 |
| p.Ser1218Ile | 0.00 | 0.12 | 0.07 |
CBS challenge
One of the earliest challenges included in the CAGI experiment was the prediction of the effects of mutations in cystathionine beta-synthase (CBS) on its function. CBS plays a role in cysteine biosynthesis via the transsulfuration pathway and has been implicated in homocystinuria. There have been two CBS challenges in CAGI with different data sets for each: one in 2010 and the other in 2011. The 2010 data set contained 51 synthetically generated amino acid substitutions and the 2011 data set contained 84 substitutions observed in patients with homocystinuria. The experimental setup in both situations was the same and involved the expression of mutated human CBS clones in yeast cells with the orthologous gene CYS4 removed. Thus, in this in vivo yeast complementation assay, the growth of yeast cells was directly dependent upon the level of mutant human CBS and could be used to infer functionality for each mutation. Since human CBS requires vitamin B6 (supplemented in the soluble form of pyridoxine) as a co-factor for its function, experiments were carried out with high (400 ng/ml) and low (2 ng/ml) co-factor concentrations. The goal was to predict cell growth rates expressed as a fraction relative to the wild-type with the same amount of pyridoxine supplementation, where zero indicates complete loss of function and one indicates wild-type levels of function. Since the data sets also provided explicit binary information on whether a variant was non-functional or not, we directly ran our predictors on these data sets and evaluated them as both classification and regression problems.
We found that MutPred2 (without homolog counts) had greater AUC values than MutPred in both data sets (Figures 7A and 7B). However, in the 2011 data set, MutPred was more sensitive in the low fpr interval; 79 of the 84 mutations in the 2011 data set were present in MutPred’s training set (78 pathogenic and one benign) and 81 were present in MutPred2’s training set (79 pathogenic and two benign). Of these pathogenic mutations, only p.Asn228Lys occurred in the 2010 data set as well. Since our predictors were trained in a binary classification scenario similar to this challenge (albeit with different class definitions), we calculated Spearman’s correlation coefficients as an additional measure of performance. When mutations with relative growth rates above one were excluded, MutPred2’s scores correlated better with experimental values than MutPred’s for the 2010 data set but the converse was true for the 2011 data set (Figures 7C and 7D). Predictions from both methods correlated better with the rates at high pyridoxine concentrations in the 2010 set but were similar in both experimental conditions in the 2011 data set. This was perhaps due to the lack of null mutant growth rates for the low concentration experiment in the 2010 data set (Figure 7C; second column). When the full set of mutations was considered for each data set, MutPred2 outperformed MutPred on all data sets in all experimental conditions (Supp. Table S5), suggesting that its scores scaled better even when mutations outside its prediction range were included. We note that neither of these methods were officially submitted to CAGI. The best-performing method in 2010 had an AUC value of 0.90 and Spearman coefficients of 0.62 and 0.65 for high and low concentrations of pyridoxine, respectively. Since our training sets substantially overlapped with the 2011 data set, we did not compare our performance to the official assessors’ results.
Figure 7.
Discussion
In this study, we applied two pathogenicity predictors, MutPred and MutPred2, to various single-gene prediction challenges related to impact of missense mutations. This impact was measured through genetic sequencing in case-control settings (BRCA1, BRCA2, MRE11A, RAD50, NBN, CHEK2), biochemical assays for protein function (NAGLU, NPM-ALK, PKLR) and assays measuring cellular variables (UBE2I/SUMO ligase, CDKN2A/p16, SCN5A/NaV 1.5, CBS). Both methods were trained on large sets consisting of amino acid substitutions from thousands of genes and were not specifically tuned for any particular gene or task at hand. They performed comparably or better than the top-performing methods identified by the independent assessors. In some cases, these methods only utilized information specific to the given gene, implicitly suggesting that generic models trained on larger data sets, benefit from information learned from other genes. This is in general agreement with a recent systematic study that concluded that the best-performing generic models frequently outperform gene-specific ones (Riera et al., 2016). In addition, our results provide insights on the transferability of pathogenicity predictors to the prediction of functional effects of missense mutations, the robustness of these predictors to heterogeneity in evidence types in current test sets, and the interpretability of these models in a biological context; i.e., the relationship between the problem of predicting pathogenic variants and that of predicting function-altering variants.
MutPred and MutPred2 models generalize to different prediction tasks
It has been previously argued that functional effect predictors can predict disease-related mutations with reasonable accuracy and that prediction scores correlate with effect severity (Schaefer et al., 2012). We inverted this question and asked whether pathogenicity predictors can similarly capture different specific functional effects of amino acid substitutions. We used a variety of CAGI prediction tasks to investigate this and found that pathogenicity predictors performed well on almost all of the functional effect prediction tasks that this study covers. Furthermore, prediction scores (especially MutPred2) often correlated with actual experimental values without any calibration or pre-processing. Although the direct use of these prediction scores without any calibration can be context-dependent, the evidence suggests that one can generally utilize the rankings to prioritize variants of interest. Another interesting observation has to do with mutations that overlap with our methods’ training sets. For example, nearly all of the variants in the 2011 CBS data set were present in MutPred’s and MutPred2’s training sets as pathogenic variants. Despite being trained to identify all of these mutations as one class, our methods generated distributions of scores that agreed with actual experimental measurements. Moreover, we found that MutPred2 generally outperformed MutPred. In the case of NPM-ALK and pyruvate kinase, we demonstrated that the new functional site predictors (allosteric site and PPI-binding) in MutPred2 provide alternative ways to infer the impact of amino acid substitutions. Furthermore, the prediction and inclusion of such residue-level functional impact (a distinguishing attribute of MutPred and MutPred2) improved the prediction of both pathogenic and function-altering substitutions. We conclude that our methods can be directly used to predict experimental values without any calibration and, at the very least, can serve as a baseline approach or a feature for customized functional impact prediction tasks. We recommend that a set of models similar to (and potentially including) ours, be selected by the CAGI organizers and used as a baseline in each relevant prediction challenge.
Predictor score distributions matter
When considering metrics that measure the concordance between experimental and predicted values, MutPred2 performed better than MutPred; e.g., the NAGLU, pyruvate kinase, p16 and the CBS challenges. A major factor contributing to this are the differences between the score distributions of the two methods. MutPred2 had a spikier score distribution on the BRCA VUS data set and was more spread out in the NAGLU challenge. The learning algorithm in MutPred2 is a neural network, which can approximate any discriminant function (Cybenko, 1989) as well as posterior probabilities of the underlying distribution of inputs and outputs (Rojas, 1996). Score distributions of predictors are often viewed and optimized in terms of the separation between different classes. However, our results support the attempts to approximate posterior distributions as closely as possible (Rost et al., 2016). While random forests (MutPred) performed comparably or even better than MutPred2 on classification tasks, they were frequently outperformed on the prediction of functional effect severity. We speculate that this may be due to the differences between the individual components of the two ensemble models: neural networks and classification trees. Although both neural network ensembles and random forests involve averaging the outputs of these individual predictors, the smoother approximation of posterior distributions by the neural networks seems to better capture the intermediate levels of functional effect severity. Our arguments are further supported by the fact that SNAP (which has been demonstrated to correlate with the magnitude of functional effects) also relies on neural networks.
The relationship between predictive performance and the evidence for pathogenicity and functional impact
As illustrated through the different CAGI prediction tasks, the strength of the annotation of a pathogenic variant depends on the operating definition of a pathogenic variant. Our results suggest better performance of the pathogenicity predictors when information on biochemical, molecular or functional impact is available. For instance, in the cases of MRE11A, RAD50, NBN and CHEK2, AUC values for our predictors seldom exceeded 0.7 and yet our predictors were among the top-performing methods. Despite being trained for the purpose of distinguishing between pathogenic and benign variants, state-of-the-art methods did not perform as well as expected. However, in the case of challenges with evidence of functional impact (NAGLU, CBS and p16), our predictors and the community itself fared much better. Based on these observations, we reason that evaluations against disease variants annotated simply based on their frequencies in case-control studies are confounded by several external factors and these likely contribute to the underestimation of predictive performance (Jain et al., 2017).
Predictors trained on variants from multiple genes in HGMD are robust
Public databases provide valuable data for the systematic hypotheses testing and the development of predictive models. However, issues of uncertainty (overlap between class-conditional distributions in a given feature space), sample selection bias, and noise will always have to be overcome either through best practices in data processing or by the models themselves (Rost et al., 2016). We found that supervised learning methods that rely on large resources for training, were actually useful on tasks specific to individual proteins. Our predictors were competitive with or in some cases better than methods customized for the given protein and/or task; e.g., the p16, CBS and NPM-ALK challenges.
Interestingly, all pathogenic mutations in the MutPred training set and a substantial fraction in the MutPred2 training set derive from HGMD. From our experience going beyond this specific study, both MutPred models heavily benefited from the use of this database. Results from recent large-scale studies on the development of meta-predictors for pathogenicity prediction support this view; predictors trained on HGMD mutations tended to perform better than other methods (Dong et al., 2015; Ioannidis et al., 2016). This can be explained by the fact that the specialization of HGMD and its expert curators for the task of collecting and storing disease variants reduces noise, especially when the evidence for pathogenicity is variable and/or sparse.
Limitations and future work
Our work is not without limitations and we discuss them here. First, unlike in many previous studies (Schaefer et al., 2012; Bromberg et al., 2013), the CAGI data sets covered a dozen or so proteins with significant variability in data set sizes. While our study serves as a proof-of-concept for the direct applicability of our pathogenicity predictors to functional effect prediction tasks, more systematic analyses on larger data sets will be needed to fully disentangle the relationship between the two prediction problems. Second, the CAGI data sets likely contained some biases. Amino acid substitutions predominantly came from proteins that were of medical interest. Moreover, several data sets were obtained from ongoing studies with different objectives. For example, in the 2010 CBS data set, nearly half of all mutations were selected for their mild to severe impact on protein stability (Dimster-Denk et al., 2013), as predicted by Rosetta (Kellogg et al., 2011). Third, as of now, our methods do not account for activating mutations and this not only introduces complications for evaluation (see below) but also overestimates the direct applicability of our methods. However, since the overall ranking of amino acid substitutions were often consistent with real values, our methods would still be effective in the prioritization of interesting variants. Assigning directionality to predicted functional effects is a potential avenue for future research that should rely on predictions of specific functional activities (Lugo-Martinez et al., 2016). Finally, while our choice of evaluation criteria for a given task was influenced by the respective CAGI assessors, our analyses were not as comprehensive. Moreover, our data handling and pre-processing protocols deviated from theirs; e.g., exclusion of frameshifting indels and nonsense mutations. As a result, in some cases our performance values did not agree with CAGI assessments. For instance, in our hands, we found the AUC values for subset 1 of the SUMO ligase challenge ranged between 0.65 and 0.71 (with varying data selection criteria), but in the official assessments our methods performed consistently around 0.75. Therefore, any direct comparisons between actual performance evaluations must be interpreted cautiously. However, our general conclusions still hold as the overall trends remained the same; e.g., in the SUMO ligase challenge MutPred and MutPred2 did cluster together with other methods, in terms of performance.
Final remarks
The CAGI experiment has enabled us to assess the utility of general machine learning models for multiple function-specific tasks and draw meaningful conclusions both on a case-by-case basis and on the aggregate. Although our results may not generalize to other predictive methods, they provide empirical support that models trained on pathogenic variants with the incorporation of local structural and functional features, transfer well to the related problem of functional effect prediction. Disambiguating specific functional effects from pathogenicity, and understanding their mutual relationship, may be of interest in the future iterations of CAGI.
Supplementary Material
Acknowledgments
We thank Binghuang Cai for discussions on specific challenges. We thank Biao Li and Janita Thusberg for submitting MutPred predictions during the early iterations of the CAGI experiment. This work was funded by the National Institutes of Health (NIH) grants R01 LM009722 and R01 MH105524, and the Indiana University Precision Health Initiative. The CAGI experiment coordination is supported by NIH U41 HG007446 and the CAGI conference by NIH R13 HG006650.
Footnotes
The authors declare no conflict of interest.
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg Y, Kahn PC, Rost B. Neutral and weakly nonneutral sequence variants may define individuality. Proc Natl Acad Sci USA. 2013;110:14255–14260. doi: 10.1073/pnas.1216613110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg Y, Rost B. SNAP: predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 2007;35:3823–3835. doi: 10.1093/nar/gkm238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calloe K, Refaat MM, Grubb S, Wojciak J, Campagna J, Thomsen NM, Nussbaum RL, Scheinman MM, Schmitt N. Characterization and mechanisms of action of novel NaV 1.5 channel mutations associated with Brugada syndrome. Circ Arrhythm Electrophysiol. 2013;6:177–184. doi: 10.1161/CIRCEP.112.974220. [DOI] [PubMed] [Google Scholar]
- Capriotti E, Calabrese R, Casadio R. Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics. 2006;22:2729–2734. doi: 10.1093/bioinformatics/btl423. [DOI] [PubMed] [Google Scholar]
- Carter H, Douville C, Stenson PD, Cooper DN, Karchin R. Identifying Mendelian disease genes with the variant effect scoring tool. BMC Genomics. 2013;14(Suppl 3):S3. doi: 10.1186/1471-2164-14-S3-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassa CA, Tong MY, Jordan DM. Large numbers of genetic variants considered to be pathogenic are common in asymptomatic individuals. Hum Mutat. 2013;34:1216–1220. doi: 10.1002/humu.22375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS ONE. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cybenko G. Approximation by superpositions of a sigmoidal function. Math Control Signals Syst. 1989;2:303–314. [Google Scholar]
- Damiola F, Pertesi M, Oliver J, Le Calvez-Kelm F, Voegele C, Young EL, Robinot N, Forey N, Durand G, Vallee MP, Tao K, Roane TC, et al. Rare key functional domain missense substitutions in MRE11A, RAD50, and NBN contribute to breast cancer susceptibility: results from a Breast Cancer Family Registry case-control mutation-screening study. Breast Cancer Res. 2014;16:R58. doi: 10.1186/bcr3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimster-Denk D, Tripp KW, Marini NJ, Marqusee S, Rine J. Mono and dual cofactor dependence of human cystathionine β-synthase enzyme variants in vivo and in vitro. G3 (Bethesda) 2013;3:1619–1628. doi: 10.1534/g3.113.006916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Wei P, Jian X, Gibbs R, Boerwinkle E, Wang K, Liu X. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum Mol Genet. 2015;24:2125–2137. doi: 10.1093/hmg/ddu733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett T. An introduction to ROC analysis. Pattern Recognit Lett. 2006;27:861–874. [Google Scholar]
- Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- George RA, Smith TD, Callaghan S, Hardman L, Pierides C, Horaitis O, Wouters MA, Cotton RG. General mutation databases: analysis and review. J Med Genet. 2008;45:65–70. doi: 10.1136/jmg.2007.052639. [DOI] [PubMed] [Google Scholar]
- Hecht M, Bromberg Y, Rost B. Better prediction of functional effects for sequence variants. BMC Genomics. 2015;16(Suppl 8):S1. doi: 10.1186/1471-2164-16-S8-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis NM, Rothstein JH, Pejaver V, Middha S, McDonnell SK, Baheti S, Musolf A, Li Q, Holzinger E, Karyadi D, Cannon-Albright LA, Teerlink CC, et al. REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am J Hum Genet. 2016;99:877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, White M, Radivojac P. Recovering true classifier performance in positive-unlabeled learning; Proceedings of the 31st AAAI Conference on Artificial Intelligence, AAAI 2017; 2017. pp. 2066–2072. [Google Scholar]
- Kellogg EH, Leaver-Fay A, Baker D. Role of conformational sampling in computing mutation-induced changes in protein structure and stability. Proteins. 2011;79:830–838. doi: 10.1002/prot.22921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Calvez-Kelm F, Lesueur F, Damiola F, Vallee M, Voegele C, Babikyan D, Durand G, Forey N, McKay-Chopin S, Robinot N, Nguyen-Dumont T, Thomas A, et al. Rare, evolutionarily unlikely missense substitutions in CHEK2 contribute to breast cancer susceptibility: results from a breast cancer family registry case-control mutation-screening study. Breast Cancer Res. 2011;13:R6. doi: 10.1186/bcr2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Krishnan VG, Mort ME, Xin F, Kamati KK, Cooper DN, Mooney SD, Radivojac P. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics. 2009;25:2744–2750. doi: 10.1093/bioinformatics/btp528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugo-Martinez J, Pejaver V, Pagel KA, Jain S, Mort M, Cooper DN, Mooney SD, Radivojac P. The loss and gain of functional amino acid residues is a common mechanism causing human inherited disease. PLoS Comput Biol. 2016;12:e1005091. doi: 10.1371/journal.pcbi.1005091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur DG, Manolio TA, Dimmock DP, Rehm HL, Shendure J, Abecasis GR, Adams DR, Altman RB, Antonarakis SE, Ashley EA, Barrett JC, Biesecker LG, et al. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mort M, Evani US, Krishnan VG, Kamati KK, Baenziger PH, Bagchi A, Peters BJ, Sathyesh R, Li B, Sun Y, Xue B, Shah NH, et al. In silico functional profiling of human disease-associated and polymorphic amino acid substitutions. Hum Mutat. 2010;31:335–346. doi: 10.1002/humu.21192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottaz A, David FP, Veuthey AL, Yip YL. Easy retrieval of single amino-acid polymorphisms and phenotype information using SwissVar. Bioinformatics. 2010;26:851–852. doi: 10.1093/bioinformatics/btq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niroula A, Vihinen M. Variation interpretation predictors: principles, types, performance, and choice. Hum Mutat. 2016;37:579–597. doi: 10.1002/humu.22987. [DOI] [PubMed] [Google Scholar]
- Ofran Y, Rost B. Protein-protein interaction hotspots carved into sequences. PLoS Comput Biol. 2007;3:e119. doi: 10.1371/journal.pcbi.0030119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pejaver V, Urresti J, Lugo-Martinez J, Pagel KA, Lin GN, Nam HJ, Mort M, Cooper DN, Sebat J, Iakoucheva LM, Mooney SD, Radivojac P. MutPred2: inferring the molecular and phenotypic impact of amino acid variants. bioRxiv. 2017 doi: 10.1038/s41467-020-19669-x. doi: https://doi.org/10.1101/134981. [DOI] [PMC free article] [PubMed]
- Peterson TA, Doughty E, Kann MG. Towards precision medicine: advances in computational approaches for the analysis of human variants. J Mol Biol. 2013;425:4047–4063. doi: 10.1016/j.jmb.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeb J, Hecht M, Mahlich Y, Bromberg Y, Rost B. Predicted molecular effects of sequence variants link to system level of disease. PLoS Comput Biol. 2016;12:e1005047. doi: 10.1371/journal.pcbi.1005047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riera C, Padilla N, de la Cruz X. The complementarity between protein-specific and general pathogenicity predictors for amino acid substitutions. Hum Mutat. 2016;37:1013–1024. doi: 10.1002/humu.23048. [DOI] [PubMed] [Google Scholar]
- Rojas R. A short proof of the posterior probability property of classifier neural networks. Neural Comput. 1996;8:41–43. [Google Scholar]
- Rosner B. Fundamentals of Biostatistics. Cengage Learning 2010 [Google Scholar]
- Rost B, Radivojac P, Bromberg Y. Protein function in precision medicine: deep understanding with machine learning. FEBS Lett. 2016;590:2327–2341. doi: 10.1002/1873-3468.12307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaini MC, Minervini G, Elefanti L, Ghiorzo P, Pastorino L, Tognazzo S, Agata S, Quaggio M, Zullato D, Bianchi-Scarra G, Montagna M, D’Andrea E, et al. CDKN2A unclassified variants in familial malignant melanoma: combining functional and computational approaches for their assessment. Hum Mutat. 2014;35:828–840. doi: 10.1002/humu.22550. [DOI] [PubMed] [Google Scholar]
- Schaefer C, Bromberg Y, Achten D, Rost B. Disease-related mutations predicted to impact protein function. BMC Genomics. 2012;13(Suppl 4):S11. doi: 10.1186/1471-2164-13-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnoes AM, Brown SD, Dodevski I, Babbitt PC. Annotation error in public databases: misannotation of molecular function in enzyme superfamilies. PLoS Comput Biol. 2009;5:e1000605. doi: 10.1371/journal.pcbi.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnoes AM, Ream DC, Thorman AW, Babbitt PC, Friedberg I. Biases in the experimental annotations of protein function and their effect on our understanding of protein function space. PLoS Comput Biol. 2013;9:e1003063. doi: 10.1371/journal.pcbi.1003063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shihab HA, Gough J, Cooper DN, Stenson PD, Barker GL, Edwards KJ, Day IN, Gaunt TR. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum Mutat. 2013;34:57–65. doi: 10.1002/humu.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenson PD, Mort M, Ball EV, Shaw K, Phillips A, Cooper DN. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet. 2014;133:1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.