Abstract
Genetic alterations of ARID1B have been recently recognized as one of the most common mendelian causes of intellectual disability and are associated with both syndromic and non-syndromic phenotypes. The ARID1B protein, a subunit of the chromatin remodeling complex SWI/SNF-A, is involved in the regulation of transcription and multiple downstream cellular processes. We report here the clinical, genetic and proteomic phenotypes of an individual with a unique apparent de novo mutation of ARID1B due to an intragenic duplication. His neurodevelopmental phenotype includes a severe speech/language disorder with full scale IQ scores 78–98 and scattered academic skill levels, expanding the phenotypic spectrum of ARID1B mutations. Haploinsufficiency of ARID1B was determined both by RNA sequencing and quantitative RT-PCR. Fluorescence in situ hybridization analysis supported an intragenic localization of the ARID1B copy number gain. Principal component analysis revealed marked differentiation of the subject’s lymphoblast proteome from that of controls. Of 3426 proteins quantified, 1,014 were significantly up- or down-regulated compared to controls (q<0.01). Pathway analysis revealed highly significant enrichment for canonical pathways of EIF2 and EIF4 signaling, protein ubiquitination, tRNA charging and chromosomal replication, among others. Network analyses revealed down-regulation of: (1) intracellular components involved in organization of membranes, organelles and vesicles; (2) aspects of cell cycle control, signal transduction and nuclear protein export; (3) ubiquitination and proteosomal function; and (4) aspects of mRNA synthesis/splicing. Further studies are needed to determine the detailed molecular and cellular mechanisms by which constitutional haploinsufficiency of ARID1B causes syndromic and non-syndromic developmental disabilities.
Keywords: ARID1B, SWI/SNF, SWI/SNF-A, chromatin, regulation, development, intellectual disability, proteome, proteomic
INTRODUCTION
Intellectual disability is characterized by significant limitations in cognitive functioning and adaptive behaviors [American Psychiatric Association, 2013] and affects 1–3% of the general population. Mutations of ARID1B (AT-rich interactive domain 1B) are an epidemiologically significant subset of mendelian causes of neurodevelopmental disability and are associated with non-syndromic intellectual disability as well as syndromic forms of intellectual disability such as Coffin-Siris syndrome [Santen and Clayton-Smith, 2014; Sim et al., 2015]. The product of ARID1B is a ubiquitous nuclear-localized protein that is a subunit of SWI/SNF-A, a chromatin remodeling complex that contains over 25 core subunits and that is involved in the regulation of many biological processes, including regulation of transcription [Euskirchen et al., 2012]. ARID1B mutations associated with intellectual disability include whole gene deletions, intragenic deletions, splice site, nonsense, and frameshift mutations, all of which point to haploinsufficiency as the mechanism causing the phenotype, as well as rare and less well-studied duplications [reviewed in Santen and Clayton-Smith, 2014; Sim et al., 2015]. Here, we report the clinical, genetic, and proteomic findings of an individual having a unique loss-of-function mutation of ARID1B due to an intragenic duplication.
CLINICAL REPORT
This study was approved by the Institutional Review Board of the Cleveland Clinic. The subject is a 14-year-old male born at 39 weeks of gestation to a healthy primagravida 38-year-old mother and unrelated 49-year-old father with non-contributory family histories. There were no medical concerns during infancy. Early developmental milestones were met until 1-year of age but no use of sentences occurred until about 2.5–3 years old.
Physical examinations during early childhood noted language delays, borderline macrocephaly, strabismus, dysarthria, mild hypotonia and mild gross and fine motor incoordination. Diminished physical endurance was also apparent in early childhood and has persisted. He had prolonged recovery times from illnesses, including several developmental regressions that lasted two or more months between ages 7–9 years, as well as two episodes of difficulty recovering from general anesthesia at 3 and 6 years old. Ophthalmologic exam revealed a right optic nerve pit. Growth parameters at 6.5 years included head circumference 54.8 cm (98%), weight 22.1 kg (52%) and height 116.9 cm (38%). Physical exam at 12.4 years showed a non-dysmorphic male with weight 36.6 kg (18%) and height 140.3 cm (7%); neurological exam showed slowed processing to questions or directions, abnormal gait with bilateral intoeing, mild imbalance, mild dysmetria, slow rapid alternating movements and clumsy fine motor function.
The earliest neuropsychological assessment, at 3.5 years, used the Stanford Binet 5 tool and showed full-scale IQ 98, verbal IQ 122 and non-verbal IQ 74. At 5 years, using the WPPSI-III, he was noted to have a full-scale IQ 83, verbal IQ 78 and non-verbal IQ 96, a poor fund of general knowledge and difficulty formulating and expressing verbal concepts. At 8 years, using the WISC-4, there was a full-scale IQ 78, with verbal comprehension 85, perceptual reasoning 102, working memory 86 and processing speed 50. His strongest skills related to nonverbal visual-spatial reasoning. There was slow processing of information, deficiency of working memory and poor visual/graphomotor skills. He was diagnosed with a severe language disorder. Auditory evaluation at 10 years showed an auditory processing disorder with severe difficulty in figure-ground discrimination, integration of words and sentences, temporal integration and phonological processing, and low average auditory comprehension and average auditory short-term memory. At 12.5 years, using the Wide Range Achievement Test-4, he scored 101, 104, 91 and 67 in word reading, sentence comprehension, spelling and math computation, respectively.
Cranial MRIs at 6 months and 7 years of age showed a mildly dysmorphic corpus callosum. A 48-hour EEG at 5 years was unremarkable. Routine blood tests and urinalysis were normal. Normal metabolic testing included thyroid function tests, plasma amino acids, blood ammonia, and urinary organic acids, acylglycines, guanidinoacetate and creatine. Newborn screening, including for galactosemia, was negative. A fasting global plasma metabolomic analysis was unremarkable. The blood lactate and plasma butyrylcarnitine levels were intermittently increased. The fibroblast lactate:pyruvate ratio was normal, as were activities of fibroblast electron transport chain complexes II, III and IV and pyruvate dehydrogenase. A fibroblast loading study for defects of mitochondrial fatty acid beta-oxidation was negative. A lymphocyte cytogenetic analysis showed a normal 46,XY karyotype at the 550 band resolution. Whole exome sequencing did not reveal pathogenic variants that were likely or definitely related to the clinical phenotype. A separate sequencing of the mitochondrial genome was unremarkable.
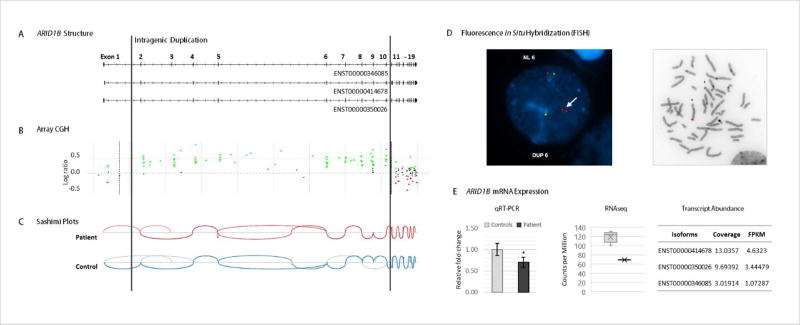
Array CGH showed a copy number gain within chromosome band 6q25.3 of approximately 0.361 Mb in size, arr [GRCh37] 6q25.3(157133792–157495187)×3 dn (Figure 1B). The duplication, which was not observed in either parent, involves a segment containing exons 2–10 (ENST00000346085) of ARID1B. Array analysis also showed heterozygosity for an approximately 0.003 Mb maternally inherited copy number loss, arr [GRCh37] 9p13.3(34647598–34650608)×1 mat, including part of GALT. There was no clinical or biochemical evidence for galactosemia, nor was a mutation of the other GALT allele detected.
FIGURE 1.
Molecular and genetic characterization of the ARID1B microduplication. (a) Representation of the ARID1B transcripts expressed in the patient with exon numbering based on an Ensembl transcript ENST00000346085. (b)Array CGH shows the duplicated region comprising exons 2 to 10 between the vertical lines. (Green dots represent probes with log ratio above 0.2 and red dots those with log ratio below −0.2. Duplication or deletion is considered when probes are ≥0.4 or ≤−0.6, respectively.) (c) Sashimi plots generated from the RNAseq datasets depict the splice junctions that have a minimum of 3 reads supporting each junction.No novel junctions are observed in the patient. (d) FISH analysis (performed with a BAC clone, RP11-680A17, labeled in red and a control probe, RP11-719E16, labeled in green) confirms the duplication in interphase and metaphase cells, showing the duplication on chromosome 6 (as indicated by the white arrow) and not inserted into another chromosome. (e) (left to right) Decreased expression levels of ARID1B measured by qRT-PCR (using primers on exon 9 and on the junction of exons 10 and 11), in comparison to 7 age- and gender-matched controls (p-val 0.02), and by RNAseq in comparison to 5 age-matched controls. Error bars represent standard deviation. Transcript abundance analysis in the patient shows the 3 ARID1B expressed transcripts, measured from the RNAseq dataset using Cufflinks. [Color figure can be viewed at wileyonlinelibrary.com].
MATERIALS AND METHODS
Molecular Cytogenomic Analysis
Array CGH was performed on a 400K oligonucleotide microarray (version 10.2) designed by the Medical Genetics Laboratories at Baylor College of Medicine and manufactured by Agilent Technologies (Santa Clara, CA). It includes exonic coverage of over 4000 candidate and disease genes at an average resolution of 30 kb with 60,000 SNP probes and 670 probes for the mitochondrial genome. Data was extracted using Agilent’s Feature Extraction software (version 9.5.3.1) and was analyzed using a web-based software platform [Cheung et al., 2005; Lu et al., 2007].
Transcriptome Analysis
Gene expression was measured by RNAseq and quantitative RT-PCR. Total RNA was extracted from a patient-derived EBV-transformed lymphoblastoid cell line (LCL) and control EBV-transformed LCLs using TRIzol® (Invitrogen) followed by RNeasy Mini Kit (Qiagen) column purification. cDNA was synthetized from 1µg of extracted RNA using SuperScript® II Reverse Transcriptase (ThermoFisher Scientific with oligo(dT), random hexamers, and RNase inhibitor). The RNAseq library was prepared using the Illumina TruSeq kit and manufacturer’s instructions. Libraries were multiplexed, pooled and sequenced on multiple lanes of an Illumina HiSeq2500, generating an average of 30 million paired-end reads of 76 bp. Quality assessment of sequence reads was performed using fastQC (v. 0.10.1) (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Sequence reads were then aligned to human reference genome Ensembl GRCh37 (v. 71) using GSNAP (v. 12-19-2014) [Wu and Nacu, 2010] at its default parameter setting. Quality checking of alignments was assessed by a custom script utilizing Picard Tools (http://broadinstitute.github.io/picard/), RNASeQC [DeLuca et al., 2012], RSeQC [Wang et al., 2012] and SamTools [Li et al., 2009]. Novel transcript analysis was performed using Cufflinks, and visualized on Integrative Genomics Viewer [Robinson et al., 2011; Thorvaldsdóttir et al., 2013]. Counts per Million, generated from gene level counts which were tabulated using BedTools’s multibamcov algorithm (v. 2.17.0) [Quinlan and Hall, 2010] on unique alignments for each library at all Ensembl genes (GRCh37 v.71), were calculated to compare expression levels with control samples from five healthy individuals.
Quantitative RT-PCR was performed for ARID1B using custom designed primers and ACTB, GAPDH and POLR2A were used as endogenous controls. Primers were as described: ARID1B (forward - tgcgtcccctcatctctcca, reverse - aggcatctgactacctggga), ACTB (forward - tgaagtgtgacgtggacatc, reverse - ggaggagcaatgatcttgat), GAPDH (forward - ggacctgacctgccgtctag, reverse - gtagcccaggatgcccttga), POLR2A (forward - gcaccacgtccaatgacat, reverse - gtgcggctgcttccataa). Primers (0.75 µM final), cDNA (1:100 final) and nuclease-free water were added to the LightCycler® 480 SYBR Green I Master Mix (Roche) for a final 10µL reaction volume. A LightCycler® 480 (Roche) was used for data acquisition. Values for the subject and seven age-and gender-matched controls were obtained in at least three technical replicates. Results of technical replicates for ARID1B were normalized and compared against the average of the three endogenous gene controls using the ΔΔCt method. Results are expressed as fold-change relative to the averaged control individuals. A two-tailed T-test was used to assess statistical significance.
Fluorescence in situ hybridization
Peripheral blood from the proband was collected in a sodium heparin vacutainer tube, cultured for 72 hours with the mitogen phytohemagglutinin, and harvested by standard cytogenetic methods. Slides containing both interphase and metaphase cells were hybridized according to standard protocol with fluorescently labeled BAC clones, RP11-680A17 and RP11-719E16, localized to 6q25.3 and 6q13, respectively. The BAC clones had been grown in broth medium with 20 µg/mL of chloramphenicol followed by DNA extraction using a Plasmid Miniprep kit (Qiagen). The target clone, RP11-680A17, was labeled directly with Spectrum GreenTM dUTP by nick translation with the Abbott Molecular kit. After hybridization and in accordance with the laboratory’s standard confirmation protocol for duplications less than 1 Mb, 50 interphase cells were manually scored using a fluorescent microscope to confirm the presence of the duplication and a metaphase cell was examined to confirm the localization of the duplicated segment.
Proteome analysis
LCLs from the subject, obtained at 13 years of age, and 5 male controls ages 31–40 years were used. 15 µg of protein from each sample was digested with LysC for 1 hour and trypsin overnight at 37°C. Reverse phase LC-MS/MS was performed as described [Tomechko et al., 2015], except that 600 ng of peptide digests was loaded on the HPLC column and the gradient of solvent B ranged from 2 to 40% over 210 min.
Raw LC-MS/MS data files for each sample were processed using Rosetta Elucidator (Rosetta Biosoftware, Seattle, WA) (Version 3.3.01 SP4 25). Automated differential quantification of peptides was performed as previously described [Neubert et al., 2008; Schlatzer et al., 2012; Azzam et al., 2016]. Briefly, LC-MS/MS raw data were imported, and for each MS spectrum profile of each LC-MS/MS run, chromatographic peaks and monoisotopic masses were extracted and aligned. Chromatographic peaks were first aligned by retention time and monoisotopic mass. Peaklists with the monoisotopic mass and corresponding MS/MS data were then generated for each sample and searched using Mascot. Resultant peptide identifications were imported into Elucidator and monoisotopic masses annotated with peptide identifications. The false discovery rate for protein identifications was calculated to be 0.02%. The MS/MS peak lists were searched by Mascot (version 2.4.1) (Matrix Science, London, UK) using the human UniProt database. Search settings were as follows: trypsin enzyme specificity; mass accuracy window for precursor ion, 25 ppm; mass accuracy window for fragment ions, 0.8Da; variable modifications including carbamidomethlylation of cysteines, 1 missed cleavage and oxidation of methionine.
Statistical Methods and Bioinformatic Analyses
Raw MS data were obtained for each region in a .csv file; this file contained intensity values, with rows corresponding to peptides and columns corresponding to the sample. Missing values were imputed using a weighted k-nearest neighbors algorithm [Troyanskaya et al., 2001]. Next, data were log2-transformed to achieve normality. Data were visualized with principal component analysis and complete linkage hierarchical clustering. These preprocessing steps were performed using InfernoRDN (formerly DanTE) [Polpitiya et al., 2008]. Data were imported into the R statistical programming environment for subsequent analyses.
Because there was one case and multiple controls, we treated individual peptides as observations of a given protein. Therefore, proteins with only one quantified peptide were excluded from our analysis. To compare protein abundance between the case and controls, we constructed a linear mixed effects model that adjusted for subject to account for the non-independence of peptides derived from the same sample. Proteins with q < 0.01 were imported into DAVID version 6.8 (https://david.ncifcrf.gov/tools.jsp) for ontology analysis. EASE score threshold was set at a value of 1.0 and minimum count for an annotation term was set to 3. The entire set of proteins with more than one peptide (n = 2,351) was used as background for enrichment analysis. Network and pathway analyses were performed in Ingenuity Pathway Analysis (IPA®, www.qiagen.com/ingenuity); proteins with q-value < 0.01 were used for this purpose. Network connections were curated based on data from all species and all cell lines and tissues. Enrichment scores and p-values for canonical pathway and network analysis were determined by a one-tailed Fisher’s exact test.
RESULTS
Measurement of ARID1B expression by quantitative RT-PCR showed a significant decrease of mRNA levels in the patient in comparison to control subjects (p-value 0.02) and a similar result was observed using RNAseq (Figure 1E). Allele-specific expression could not be assessed since the subject was not heterozygous for SNPs located in coding exons or in the untranslated regions. The most abundant mRNA transcript observed in the RNAseq dataset was ENST00000414678, consistent with the GTEx database for EBV-transformed lymphoblasts; ENST00000350026 and ENST00000346085 were also detected in the patient sample (Figure 1E). IGV Sashimi plots did not reveal novel junctions or transcripts for this gene (Figure 1C), providing no evidence that duplication of this exonic region resulted in alterations in splicing.
Follow-up fluorescence in situ hybridization analyses showed the presence of duplicated ARID1B genome in interphase cells (Figure 1D). The latter, in turn, was localized to chromosome 6q25.3 on analysis of a metaphase cell, the site of the ARID1B gene (Figure 1D), confirming that the microduplication occurred within the same chromosome and excluding the hypothesis of complex rearrangements.
Proteomic analyses of the LCLs resulted in the quantification of 15,792 peptides, corresponding to 3,426 proteins. There were 1,014 proteins that were differentially expressed between the case and controls (q < 0.01). The differentially expressed proteins and related data are noted in Supplementary Table 1. Principal component analysis and hierarchical clustering analysis revealed marked separation of the subject’s proteome from that of controls (Supplementary Figure 1).
Gene ontology analysis in DAVID revealed enrichment of several annotation clusters with terms relating to ATP binding, mitochondrion and flavin adenine dinucleotide (Supplementary Table 2). Bioinformatic analysis with IPA® revealed enrichment for canonical pathways of EIF2 signaling (p-value 6.31 E-17), protein ubiquitination (p-value 1.58 E-16), regulation of eIF4 and p70S6K signaling (p-value 1.00 E-14), tRNA charging (p-value 5.01 E-13) and cell cycle control of chromosomal replication (p-value 6.31 E-11) (Supplementary Table 3). Down-regulation of: (1) intracellular components involved in intracellular organization of membranes, organelles and vesicles; (2) aspects of cell cycle control, signal transduction and nuclear protein export; (3) ubiquitination and proteosomal function; and (4) aspects of mRNA synthesis/splicing are noted in IPA® network analyses (Supplementary Figures 2A–D).
DISCUSSION
The subject has a unique ARID1B mutation, a copy number gain involving part of that gene that was initially determined by a chromosomal microarray analysis and which was thought likely to cause a pathologic reduction of ARID1B gene expression. Subsequent gene expression data, by both qRT-PCR and RNAseq, indicated haploinsufficiency of ARID1B and was consistent with localization of the duplication within the ARID1B gene, disrupting expression of the affected allele. Confirmation that there is a large intragenic ARID1B duplication was established by a fluorescence in situ hybridization analysis. Haploinsufficiency for ARID1B is likely to be the cause of developmental delay in this individual as no additional genetic or metabolic defects were identified. While the measurement of RNA in LCLs does not necessarily reflect the effect of the genetic lesion in the brain, the association of heterozygous inactivating mutations in ARID1B with neurodevelopmental phenotypes in other subjects suggests that reduced gene expression also occurs in the central nervous system. As ARID1B is expressed at different levels in the brain, namely higher in the cerebellum than in many peripheral tissues and other measured areas of the brain (http://www.gtexportal.org/), the consequences of its reduced expression may be even more pronounced there, possibly accounting for aspects of subject’s phenotype.
There are two reports of smaller microduplications of ARID1B. The clinical and genetic phenotypes of these two individuals and our case are summarized in Supplementary Table 4. Our case has several unique aspects to his phenotype. Few individuals with ARIB1B haploinsufficiency are reported with low-normal intellectual function [Santen et al., 2014]. Our patient, while having a significant speech/language disorder and cognitive disability, has had several IQ determinations within the normal range but there was considerable (and reproducible) variability in selected skills. In addition, his clinical course has been characterized by easy fatigability and episodes of developmental regression, prompting evaluation for a possible metabolic underpinning of these clinical concerns. Apart from intermittent elevated blood lactate levels, extensive metabolic testing was unrevealing. To our knowledge, other cases of individuals with ARID1B haploinsufficiency do not have histories of episodic regression, although the episodic memory dysfunction in one case may be relevant [Yu et al., 2015].
Our results support the view that ARID1B is a dosage sensitive gene whose expression can be affected by deletions or duplications. Indeed, the evolutionary constraint on this gene shows that it is highly intolerant to loss-of-function mutations according to its ExAC pLI score of 1.00 (http://exac.broadinstitute.org/). As we did not identify novel transcripts, the duplicated allele either was not transcribed or was transcribed but its RNA was degraded by surveillance mechanisms. Other chromatin regulators have also been noted to be dosage sensitive causes of neurodevelopmental phenotypes, including MBD5, EHMT1, CHD8 and SATB2 [Talkowski et al., 2012].
The lymphoblast proteomic data are striking: statistically highly significant dysregulation of pathways related to initiation of translation were noted, in addition to dysregulation of pathways and networks relating to protein ubiquitination, mTOR signaling, apoptosis, major metabolic pathways (e.g., TCA cycle, mitochondrial oxidative phosphorylation), cell cycle control, mRNA synthesis/processing and intracellular vesicular transport. A number of the differentially expressed pathways and networks relate to described roles of ARID1B and the SWI/SNF-A chromatin remodeling complex in different tissues such as cell cycle regulation, ubiquitination, mRNA synthesis/processing, apoptosis and intracellular signaling pathways [Nagl et al., 2007; Xi et al., 2008; Li et al., 2010; Sim et al., 2014; Raab et al., 2015]; involvement of other pathways, including many metabolic pathways and intracellular vesicular trafficking, is newly described. Our data do not specifically implicate dysregulation of Wnt/β-catenin signaling as was noted in another recent study [Vasileiou et al., 2015]. Yet, although our proteomic analysis indicates widespread and marked differential expression of proteins in the ARID1B haploinsufficient lymphoblasts, there are three important potential limitations of these findings: first, one cannot generalize these results until proteomic analysis of additional cases of ARID1B haploinsufficiency are carried out; second, the proteomic findings, while predictive of marked dysregulation of multiple pathways and networks, do not reveal which dysregulated pathways are of greatest pathophysiological significance; and, third, the relevance of lymphoblast findings to brain biology (and that of other tissues) is uncertain.
The roles of ARID1B in brain development are starting to be understood. Recent studies reveal that it may be critical in cell proliferation and differentiation and is important in dendritic arborization and synapse formation [Nagl et al., 2007; Yan et al., 2008; Tuoc et al., 2013; Harmacek et al., 2014]. Consequently, ARID1B deficiency may induce neurodevelopmental defects via abnormal brain wiring induced by the defective differentiation of neurons [Ka et al., 2016]. This report provides additional perspectives regarding ARID1B haploinsufficiency and its associated developmental disabilities. Further studies are needed to understand the precise mechanisms whereby altered chromatin remodeling leads to changed brain development and cognitive function.
Supplementary Material
Acknowledgments
We thank the family for participating in this case report. Funding was provided by FCT Fellowship SFRH/BD/52049/2012 to CMS, NIH grant GM061354 to JFG and MET, SFARI grant 308955 to JFG and R00MH095867 to MET and Autism Research Institute grant to MRN.
Footnotes
The authors have no conflicts of interest regarding this work.
References
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders (DSM-5) 5. 2013. [DOI] [PubMed] [Google Scholar]
- Azzam S, Schlatzer D, Maxwell S, Li X, Bazdar D, Chen Y, Asaad R, Barnholtz-Sloan J, Chance MR, Sieg SF. Proteome and protein network analyses of memory T cells find altered translation and cell stress signaling in treated human immunodeficiency virus patients exhibiting poor CD4 recovery. Open Forum Infect Dis. 2016;3 doi: 10.1093/ofid/ofw037. ofw037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung SW, Shaw CA, Yu W, Li J, Ou Z, Patel A, Yatsenko SA, Cooper ML, Furman P, Stankiewicz P, Lupski JR, Chinault AC, Beaudet AL. Development and validation of a CHG microarray for clinical cytogenetic diagnosis. Genet Med. 2005;7:422–432. doi: 10.1097/01.gim.0000170992.63691.32. [DOI] [PubMed] [Google Scholar]
- DeLuca DS, Levin JZ, Sivachenko A, Fennell T, Nazaire M-D, Williams C, Reich M, Winckler W, Getz G. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012;28:1530–1532. doi: 10.1093/bioinformatics/bts196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euskirchen G, Auerbach RK, Snyder M. SWI/SNF Chromatin-remodeling factors: multiscale analyses and diverse fnctions. J Biol Chem. 2012;287:30897–30905. doi: 10.1074/jbc.R111.309302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmacek L, Watkins-Chow DE, Chen J, Jones KL, Pavan WJ, Salbaum JM, Niswander L. A unique missense allele of BAF155, a core BAF chromatin remodeling complex protein, causes neural tube closure defects in mice. Dev Neurobiol. 2014;74:483–497. doi: 10.1002/dneu.22142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer J, Ekici AB, Endele S, Popp B, Zweier C, Wiesener A, Wohlleber E, Dufke A, Rossier E, Petsch C, Zweier M, Göhring I, Zink AM, Rappold G, Schröck E, Wieczorek D, Riess O, Engels H, Rauch A, Reis A. Haploinsufficiency of ARID1B, a member of the SWI/SNF-A chromatin-remodeling complex, is a frequent cause of intellectual disability. Am J Hum Genet. 2012;90:565–572. doi: 10.1016/j.ajhg.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ka M, Chopra DA, Dravid SM, Kim W-Y. Essential roles for ARID1B in dendritic arborization and spine morphology of developing pyramidal neurons. J Neurosci. 2016;36:2723–2742. doi: 10.1523/JNEUROSCI.2321-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence alignment/map format and SAM tools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XS, Trojer P, Matsumura T, Treisman JE, Tanese N. Mammalian SWI/SNF-A subunit BAF250/ARID1 is an E3 ubiquitin ligase that targets histone H2B. Molec Cell Biol. 2010;30:1673–1688. doi: 10.1128/MCB.00540-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Shaw CA, Patel A, Li J, Cooper ML, Wells Wr, Sullivan CM, Sahoo T, Yatsenko SA, Bacino CA, Stankiewicz P, Ou Z, Chinault AC, Beaudet AL, Lupski JR, Cheung SW, Ward PA. Clinical implementation of chromosomal microarray analysis: summary of 2513 postnatal cases. PLoS One. 2007;2:e327. doi: 10.1371/journal.pone.0000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagl NG, Wang X, Patsialou A, Van Scoy M, Moran E. Distinct mammalian SWI/SNF chromatin remodeling complexes with opposing roles in cell-cycle control. EMBO J. 2007;26:752–763. doi: 10.1038/sj.emboj.7601541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubert H, Bonnert TP, Rumpel K, Hunt BT, Henle ES, James IT. Label-free detection of differential protein expression by LC/MALDI mass spectrometry. J Proteome Res. 2008;7:2270–2279. doi: 10.1021/pr700705u. [DOI] [PubMed] [Google Scholar]
- Polpitiya AD, Qian W, Jaitly N, Petyuk VA, Adkins JN, Camp DG, 2nd, Anderson GA, Smith RD. DAnTE : a statistical tool for quantitative analysis of -omics data. Bioinformatics. 2008;24:1556–1558. doi: 10.1093/bioinformatics/btn217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab JR, Resnick S, Magnuson T. Genome-wide transcriptional regulation mediated by biochemically distinct SWI/SNF complexes. PLoS Genetics. 2015;11:e1005748. doi: 10.1371/journal.pgen.1005748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santen GWE, Clayton-Smith J. The ARID1B phenotype: what we have learned so far. Am J Med Genet Part C. 2014;166C:276–289. doi: 10.1002/ajmg.c.31414. [DOI] [PubMed] [Google Scholar]
- Schlatzer DM, Sugalski J, Dazard J-E, Chance MR, Anthony DD. A quantitative proteomic approach for detecting protein profiles of activated human myeloid dendritic cells. J Immunol Methods. 2012;375:39–45. doi: 10.1016/j.jim.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim JCH, White SM, Lockhart PJ. ARID1B-mediated disorders: mutations and possible mechanisms. Intractable Rare Dis Res. 2015;4:17–23. doi: 10.5582/irdr.2014.01021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkowski ME, Rosenfeld J, Blumenthal I, Pillalamarri V, Chiang C, Heilbut A, Ernst C, Hanscom C, Rossin E, Lindgren AM, Pereira S, Ruderfer D, Kirby A, Ripke S, Harris DJ, Lee J-H, Ha K, Kim H-G, Solomon BD, Gropman AL, Lucente D, Sims K, Ohsumi TK, Borowsky ML, Loranger S, Quade B, Lage K, Miles J, Wu B-L, Shen Y, Neale B, Shaffer LG, Daly MJ, Morton CC, Gusella JF. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 2012;149:525–37. doi: 10.1016/j.cell.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomechko SE, Lundberg KC, Jarvela J, Bebek G, Chesnokov NG, Schlatzer D, Ewing RM, Boom WH, Chance MR, Silver RF. Proteomics. 2015;15:3797–3805. doi: 10.1002/pmic.201400496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyanskaya O, Cantor M, Sherlock G, et al. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17:520–525. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- Tuoc TC, Boretius S, Sansom SN, Pitulescu M-E, Frahm J, Livesey FJ, Stoykova A. Chromatin regulation by BAF170 controls cerebral cortical size and thickness. Dev Cell. 2013;25:256–269. doi: 10.1016/j.devcel.2013.04.005. [DOI] [PubMed] [Google Scholar]
- Vasileiou G, Ekici AB, Uebe S, et al. Chromatin remodeling factor ARID1B represses Wnt/β-catenin signaling. Am J Hum Genet. 2015;97:445–456. doi: 10.1016/j.ajhg.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28:2184–2185. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Q, He W, Zhang XHF, Le H-v, Massagué Genome-wide impact of the BRG1 SWI/SNF chromatin remodeler on the transforming growth factor β transcriptional program. J Biol Chem. 2008;283:1146–1155. doi: 10.1074/jbc.M707479200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Wang Z, Sharova L, Sharov AA, Ling C, Piao Y, Aiba K, Matoba R, Wang W, Ko MSH. BAF250B-associated SWI/SNF chromatin-remodeling complex is required to maintain undifferentiated mouse embryonic stem cells. Stem Cells. 2008;26:1155–1165. doi: 10.1634/stemcells.2007-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Yao R, Wang L, Fan Y, Huang X, Hirschhorn J, Dauber A, Shen Y. De novo mutations in ARID1B associated with both syndromic and non-syndromic short stature. BMC Genomics. 2015;16:701. doi: 10.1186/s12864-015-1898-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.