Abstract
Purpose
To assess the APTw-MRI features of isocitrate dehydrogenase (IDH)-wildtype and IDH-mutant grade-II gliomas and to test the hypothesis that the APTw signal is a surrogate imaging marker for identifying IDH mutation status preoperatively.
Methods
27 patients with pathologically confirmed low-grade glioma, who were previously scanned at 3T, were retrospectively analyzed. The Mann-Whitney test was used to evaluate relationships between APTw intensities for IDH-mutant and IDH-wildtype groups, and the receiver-operator-characteristic analysis was used to assess the diagnostic performance of APTw.
Results
Based on histopathology and molecular analysis, seven cases were diagnosed as IDH-wildtype grade-II gliomas, and 20 cases as IDH-mutant grade-II gliomas. The maximum and minimum APTw values, based on multiple regions of interest, as well as the whole-tumor histogram-based mean and 50th percentile APTw values, were significantly higher in the IDH-wildtype gliomas than in the IDH-mutant groups. This corresponded to the areas under the receiver-operator-characteristic curves of 0.89, 0.76, 0.75, and 0.75, respectively, for the prediction of the IDH mutation status.
Conclusions
IDH-wildtype lesions were typically associated with relatively high APTw signal intensities, compared with IDH-mutant lesions. The APTw signal could be a valuable imaging biomarker by which to identify IDH1 mutation status in grade-II gliomas.
Keywords: Glioma, IDH, CEST, amide proton transfer imaging, imaging biomarker
INTRODUCTION
Gliomas are the most common primary brain tumors, and more than 23,770 new cases are diagnosed in the USA each year (1). The recent breakthrough in the understanding of isocitrate dehydrogenase (IDH) mutations in glioma has resulted in a prompt reappraisal of the molecular oncogenesis of this group of diseases (2–5). Notably, for the first time, the most recent 2016 WHO classification of central nervous system (CNS) tumors uses molecular parameters, in addition to histology, to define tumor entities, thus formulating a basis for how CNS tumor diagnoses should be structured in the molecular era (6). The IDH enzyme catalyzes the oxidative decarboxylation of isocitrate to α-ketoglutarate (α-KG), needed for producing NADH and NADPH during cellular respiration. Mutations in IDH genes, which occur in the majority of WHO grade-II and -III gliomas and secondary glioblastomas, have been postulated to indicate a favorable clinical prognosis (2–5). Since IDH-gene-encoded enzymes are closely involved in the energy-producing Krebs cycle as catalytic isozymes, mutations in IDH genes may cause widespread disturbances of cellular metabolism, including alteration of amino acid concentrations and enzymatic activity (7, 8), and raise global DNA hypermethylation and global downregulation of protein expression as well (9, 10). Therefore, IDH-directed therapy, including IDH inhibitors and DNA methyltransferase inhibitors, have been developed and may impart a cumulative clinical benefit for the targeted patients (11).
As popularly applied in clinical practice, IDH mutation screening is generally performed with an immunohistochemical (IHC) assay on surgical samples using antibodies which detect IDH1-R132H. More than 85% of all genetic alterations in IDH1 in gliomas are heterozygous missense mutations of arginine to histidine (R132H); thus, this immunohistochemical approach will detect the majority of cases (12). For IHC-negative or IHC-equivocal cases, DNA sequencing is recommended as a complementary test (13). However, it is not uncommon that lab tests lead to false-negative results when inadequate neoplastic cells from a biopsy procedure are mixed in with the normal cell background. Thus, sufficient tumor tissue should be obtained from surgery, while balancing the benefits of removing tumor versus the potential risks for damaging normal tissue in patients with low-grade gliomas. It is these aspects of IDH-wildtype and IDH1-mutant gliomas that provide the impetus for the pre-operative determination of IDH mutational status, with methods such as MRI, which, potentially, can rapidly and noninvasively provide a comprehensive assessment. Such imaging methods could at minimum be used to better guide the biopsies needed for ICH assessment.
In recent years, researchers have evaluated the feasibility of applying several MRI techniques to determine IDH status in patients with gliomas (14–18), including gadolinium-enhanced T1-weighted (Gd-T1w) images (19), diffusion (20–22), perfusion (23), sodium MRI (24), and machine-learning algorithms (25, 26). Notably, since lab studies have proven that the oncometabolite 2-hydroxyglutarate (2-HG) holds a 100-fold increase in IDH-mutant cells compared to IDH-wildtype type cells (27), recent studies have also shown that significantly higher 2-HG detected by single-voxel MRS can identify patients with IDH-1 mutant gliomas, with high sensitivity (28–30). However, the MRS detection of 2-HG in IDH-mutant gliomas requires a large tumor volume (31), and is time-consuming, which limits the application of MRS. In addition, partial volume effects between different tumor regions may obscure the presence of 2-HG in smaller regions.
Amide proton transfer-weighted (APTw) MRI, a specific type of chemical exchange-dependent saturation transfer (CEST) imaging (32, 33), is an upcoming molecular imaging technique that can generate image contrast based on the endogenous cellular mobile proteins in tissue (34, 35). Since APT imaging was first reported in 2003 (34, 35), it has been studied as an imaging biomarker in a variety of cancers (36–41) and non-oncologic diseases (42–45). For glial neoplasms, in particular, consistent APTw MRI results have been produced across various labs, showing promise for the grading of gliomas (distinct hyperintensity in grades-III and -IV gliomas vs. iso-intensity to minimal hyperintensity in grade-II gliomas) (46–48), which demonstrate increasing protein concentrations with grade, as revealed by proteomics (49, 50) and in vivo MR spectroscopy (51). In this study, we assessed the APTw MRI features for IDH-wildtype and IDH-mutant grade-II gliomas before surgery, with the hypothesis that APTw signal intensity metrics could aid in the preoperative identification of the IDH genotype in grade-II gliomas.
METHODS
Patients
This retrospective study was approved by the local institutional review board, and informed consent was waived. Enrollment criteria were as follows: ≥18 years old; APTw and routine MRI scanning occurred within 7 days preoperatively; histopathologically confirmed as a WHO grade-II astrocytoma, oligoastrocytoma, or oligodendroglioma (according to their medical records; originally based on the 2007 WHO criteria); IDH1 mutation status and 1p/19q co-deletion status available from operative sample; and received no radiotherapy or chemotherapy before imaging. Exclusion criteria included: inferior image quality due to various reasons.
MRI Data Acquisition
MRI scanning was performed on a 3T clinical MRI scanner (Achieva; Philips Medical Systems, Best, The Netherlands). APT image data were obtained using a fat-suppressed, single-shot, fast spin-echo pulse sequence, using the following parameters: radiofrequency saturation power = 2 µT; duration = 800 ms (four block pulses; 200 ms each; inter-pulse delay, 10 ms); repetition time = 3 sec; echo time = 11 ms; matrix = 140×70 (reconstructed to be 400 × 400); sensitivity-encoding factor = 2; turbo-spin-echo factor, 37; field of view = 240×240 mm2; and slice thickness = 6 mm. A single-slice, combined APTw imaging and Z-spectrum acquisition protocol [31 offsets = 0, ±0.25, ±0.5, ±0.75, ±1, ±1.5, ±2, ±2.5, ±3 (2), ±3.25 (4), ±3.5 (8), ±3.75 (4), ±4 (2), ±4.5, ±5, ±6 ppm; the values in parentheses were the number of acquisitions, which was 1, if not specified] was applied to the maximum cross-sectional tumor area (as determined by standard MRI images). More acquisitions were obtained at and around ±3.5 ppm to obtain a sufficient signal-to-noise ratio for APTw images; more offsets were used near 0 ppm to increase the fitting accuracy of B0 maps; and more offsets were used close to ±3.5 ppm to increase the interpolation accuracy of APTw data for B0 correction (36). A saturated image at 15.6 ppm (or 2 kHz at 3 Tesla) was also acquired to calculate magnetization transfer ratio (MTR) values for conventional semisolid magnetization transfer (MT) imaging. The scanning time for this combined scan was about three minutes. Several standard MRI sequences were acquired for reference, including T2-weighted (T2w), T1w, and Gd-T1w. The Gd-T1w imaging (0.2 mL/kg body weight; Magnevist; Bayer Schering, Guangzhou, China) was the last sequence acquired.
APTw and MT Image Processing
The image analysis was performed by the interactive data language (IDL, Version 7; Exelis Visual Information Solutions, Inc., Boulder, CO, USA). To reduce possible motion artifacts during the scanning, the acquired APTw image or Z-spectrum series was registered to the saturated image at 3.5 ppm (52). The acquired APTw image or Z-spectrum series was further corrected for the B0 inhomogeneity effect on a voxel-by-voxel basis, as described previously (36). The APTw image was constructed with the so-called magnetization transfer-ratio asymmetry at the offsets of ±3.5 ppm (34, 35): MTRasym(3.5 ppm) = Ssat(−3.5 ppm)/S0 − Ssat(+3.5 ppm)/S0, where Ssat and S0 are the imaging signal intensities with and without selective radiofrequency irradiation, respectively. To account for the contribution of the possible nuclear Overhauser enhancement (NOE) effect of protons in mobile and relatively mobile proteins at −3.5 ppm on MTRasym(3.5ppm) (53–56), as well as possible asymmetries in the conventional semisolid-based MT effect (57), the calculated MTRasym(3.5ppm) image is best described as the APTw image (46). In addition, for conventional MT imaging, MTR(2 kHz) = 1 − Ssat(2 kHz)/S0.
APTw and MT Image Analysis
To perform quantitative APTw and MTR analyses, the acquired conventional MR images from each case were co-registered to the corresponding saturated Ssat image at 3.5 ppm, which was co-registered with the APTw image, as described previously (52). Primarily based on the T2w image, regions of interest (ROIs) were carefully drawn by radiologists (S.J. and X.W., who have had ten and seven years of experience in brain imaging, respectively), together with MRI physicists. One large ROI covering the whole area of abnormal intensity on the T2w image was first drawn. Similar to some previous reports (38, 39), five small tumor ROIs were further chosen in the T2w-hyperintense lesion area. Each ROI encompassed around 100–130 voxels (reconstructed). ROIs excluded the large cyst, hemorrhage, or vessels evident on standard MRI sequences. The contralateral normal-appearing white matter (CNAWM) was also analyzed, and relative APTw values were reported (ROI APTw - CNAWM APTw). For each case, the maximum and minimum APTw values among the five small tumor ROIs, as well as the Z-spectrum data and APTw histogram data corresponding to the ROI with maximum APTw (“tumor core”), were recorded. For the large whole-tumor ROI, the Z-spectrum data, APTw histogram data, and average MTR value were recorded. The histogram data were analyzed for mean, variance, skewness, kurtosis, slope, 10th percentile, 50th percentile, 90th percentile, and peak values, as defined before (58).
Histopathological Evaluation
Operative tissue samples were processed using standard clinical techniques. IDH1-R132H evaluation was performed by IHC and DNA sequencing, as described previously (13, 59). Paraffin-embedded slices of operative specimens were stained with IDH1-R132H-specific antibody (1:50; H09 clone, Dianova; Hamburg, Germany). To amplify exon 4 (codon R132) of the IDH1 gene, we designed the forward primer (5′-ACC AAA TGG CAC CAT ACG A-3′) and reverse primer (5′-GCA AAA TCA CAT TAT TGC CAA C-3′). Fluorescent in situ hybridization testing was applied to assess the 1p/19q codeletion. An experienced pathologist (Y.W.), blinded to the imaging findings, evaluated and re-classified all the cases according to the 2016 WHO classification of central nervous system tumors (6). Tumor grading documented in the clinical pathological reports (originally based on the 2007 WHO criteria and reported by multiple pathologists) was confirmed for all patients enrolled. The molecular pathological reports (IDH, 1p/19q) were taken into account.
Statistical Analysis
The data were reported as means and standard deviations. After normality testing, the Mann-Whitney U test was used to analyze the statistical differences between quantitative imaging parameters for the two IDH mutation statuses. We further employed receiver operating characteristic curve (ROC) analysis to assess the diagnostic performance of APTw metrics. All statistical analyses were performed using statistical software (SPSS, Version 23; Chicago, IL, USA). Generated P-values were two-tailed, and P < 0.05 was considered statistically significant.
RESULTS
Patient Demographics
Between 2014 and 2016, 31 patients who fulfilled the eligibility criteria according to their medical records were retrospectively analyzed. Patients typically showed some neurological deficit or seizures. 27 patients (15 males, 12 females) were finally recruited for this study, after four patients were excluded (inferior image quality duo to the patient movement, n = 3; or irremovable artifacts caused by a large intratumoral hemorrhage, n = 1). The clinical characteristics of the patient population are summarized in Table 1. Tissue samples were available from all subjects either by stereotactic biopsy (n = 3); gross total resection (n = 21); or subtotal resection (n = 3). IDH mutations were found in 20 cases (74.1%), 15 of which were histopathologically classified as oligodendrogliomas, IDH mutant and 1p/19q codeleted, and five as diffuse astrocytomas, IDH mutant. In the seven patients (25.9%) with IDH-wildtype glioma, all were classified as diffuse astrocytomas, IDH wild-type.
Table 1.
Clinical Characteristics of Patient Population
| IDH wild-type (n = 7) | IDH mutant (n = 20) | |
|---|---|---|
| Clinical parameters: | ||
| Gender (male/female) | 4/3 | 11/9 |
| Ages (y)* | 37.1±7.9 | 40.5±13.7 |
| Seizures (yes/no) | 5/2 | 14/6 |
| KPS score* | 82.6±9.7 | 85.4±8.9 |
| Imaging parameters: | ||
| Cerebral lobes involved (<2/≥2) | 4/3 | 15/5 |
| Contrast enhancement (yes/no) | 0/7 | 1/19 |
| Histopathological Diagnosis: | ||
| Oligodendroglioma | 0 | 15 |
| Diffuse astrocytoma | 7 | 5 |
Data are mean ± standard deviation.
Comparison between MTRasym Spectra for IDH-wildtype and IDH-mutant Gliomas
The average MTRasym spectra for the groups of patients with IDH-wildtype and IDH-mutant gliomas were compared to explore the specific characteristics of the APT effect at an offset of ~3.5 ppm downfield from water (Fig. 1). Both glioma types demonstrated stronger protein-based APT and other CEST effects in the offset range of 1.5–4 ppm, compared to the CNAWM. As reported previously (46), all MTRasym spectra were negative at the higher frequency (>4.5 ppm) due to the upfield NOE effect and the conventional MT asymmetry. The CEST signal intensities in the offset range of 1.5–4 ppm (including the APT signal at 3.5 ppm downfield from water) were relatively higher in IDH-wildtype than in IDH-mutant gliomas, except for the CEST effect at 2.0 ppm (35, 60), which showed no difference in absolute MTRasym between the two glioma types, but stood out in intensity relative to the other frequencies in the mutant type itself. Particularly in the IDH-wildtype tumor core, a relatively high APT effect was clearly observed at 3.5 ppm offset from water, at which the amide protons of endogenous mobile proteins and peptides resonate (61). The APTw-MRI metrics are associated with relevant proteomic characteristics that may provide valuable information for the non-invasive discrimination of IDH mutation status.
Fig. 1.
a,b: Comparison of the average MTRasym spectra of whole tumor, tumor core, and CNAWM for IDH-wildtype (a) and IDH-mutant (b) grade-II gliomas. The CEST effects were clearly visible at multiple frequencies in the frequency offset range to 1.5–4ppm. c,d: MTRasym spectra relative to CNAWM providing the relative APTw intensities. The most significant CEST effect was the APT at 3.5ppm in the IDH-wildtype tumor core.
Comparison between APTw Images for IDH-wildtype and IDH-mutant Gliomas
The APTw features of the IDH-wildtype and IDH-mutant gliomas (WHO grade-II) were further accessed, using several standard MRI (T2w, T1w, and Gd-T1w) sequences. Of seven IDH-wildtype gliomas (WHO grade-II), four lesions were located in the temporal lobe, and the remaining three lesions mainly involved in the frontotemporal lobe. Figure 2 shows one example of standard and APTw MR images for an IDH-wildtype glioma (WHO grade-II). All seven lesions showed no visible enhancement on the post-contrast T1w images. The APTw images showed mildly increased hyperintensity in T2w-hyperintense areas (at least some portions), compared to the CNAWM, demonstrating the IDH-wildtype lesions as relatively heterogeneous masses with scattered punctate or pitchy high APTw signals.
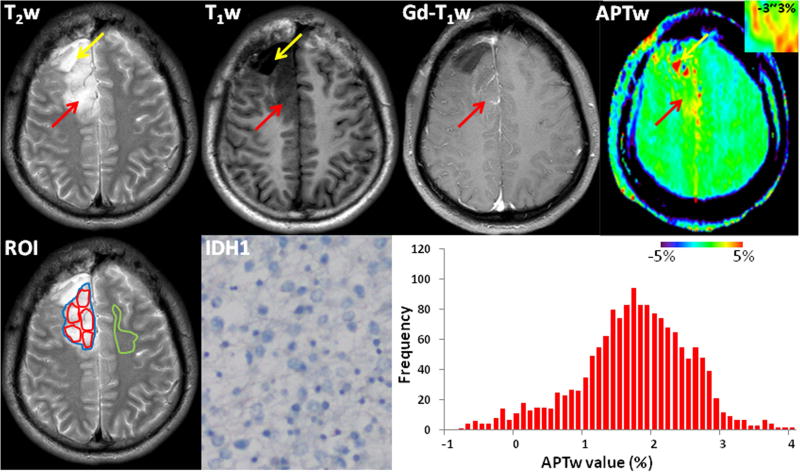
Fig. 2.
Conventional and APTw MR images, a mutant IDH1-R132H-stained section, and a whole-tumor histogram of an IDH-wildtype, WHO grade-II diffuse astrocytoma (male; 33y), who underwent partial tumor resection and left a visible surgical cavity and measurable residual tumor. The tumor (red arrows) involved the right frontal lobe and showed heterogeneous hyperintensity on the T2w image, hypointensity on the T1w image, and no obvious Gd enhancement. Compared to the CNAWM, the well-defined lesion showed scattered punctate and pitchy hyperintensity on the APTw image (inset, using a small window of −3~3%). The operative sample demonstrated IDH1-negative expression on the IHC-stained slice. The whole-tumor APTw histogram had a mean APTw value of 1.75% and a 50th percentile APTw value of 1.80%. Yellow arrows were a large cyst. ROIs for quantitative analysis were placed on the T2w image: red-solid ROIs for five small tumor ROIs; blue-solid ROI for the whole tumor; and green-solid ROI for the CNAWM.
Of 20 patients with IDH-mutant gliomas (WHO grade-II), 11 lesions were located in the frontal lobe, five were located in the temporal lobe, and five had the major lesion located in the frontotemporal lobe. Figure 3 shows one example of the standard and APTw MR images for a patient with an IDH-mutant oligodendroglioma. These IDH-mutant gliomas showed no (n = 19) or slightly central faint (n = 1) enhancement on the post-contrast T1w images. All lesions showed visually homogeneous iso-intensity to minimal APTw hyperintensity in T2w-hyperintense areas, compared to the CNAWM.
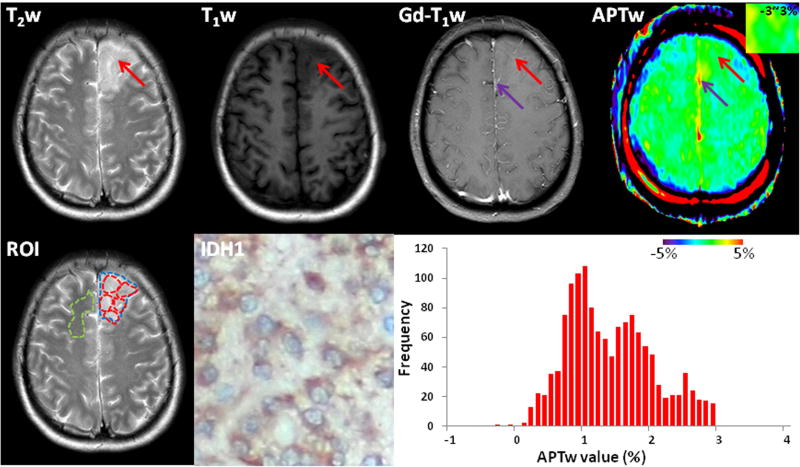
Fig. 3.
Standard and APTw MR images, a mutant IDH1-R132H-stained section, and a whole-tumor histogram of an IDH1-mutant, WHO grade-II oligodendroglioma (female; 52y). The tumor (red arrows) mainly involved the left frontal lobe, showing homogeneous hyperintensity on the T2w image, hypointensity on the T1w image, and no obvious enhancement. Compared to the CNAWM, the ill-defined lesion showed iso-intensity to minimal hyperintensity on the APTw image (inset, using a small window of −3~3%). The operative sample demonstrated diffusely IDH1-positive expression in the tumor cytoplasm on the IHC-stained slice. The whole-tumor APTw histogram had a mean APTw value of 1.37% and a 50th percentile APTw value of 1.30%. Purple arrows were a large vessel. ROIs for quantitative analysis were placed on the T2w image: red-dashed ROIs for five small tumor ROIs; blue-dashed ROI for the whole tumor; and green-dashed ROI for the CNAWM.
Quantitative APTw and MTR Analyses
Figure 4 shows histograms of APTw values obtained from whole tumor, tumor core (corresponding to the ROI with maximum APTw), and CNAWM for IDH-wildtype and IDH-mutant grade-II gliomas. The multi-ROI-based (maximum and minimum) and whole-tumor histogram-based (mean, variance, skewness, kurtosis, slop, 10th percentile, 50th percentile, 90th percentile, and peak) APTw metrics for IDH-wildtype and IDH-mutant gliomas, together with the MTR value, are summarized in Table 2. Differences between APTw values for IDH-wildtype and IDH-mutant gliomas, based on histogram analysis that corresponded to the ROI with maximum APTw, are summarized in Supporting Table S1.
Fig. 4.
a,b: Average histograms of APTw values from the whole tumor, the tumor core, and the CNAWM of IDH-wildtype (a) and IDH-mutant (b) grade-II gliomas. The red lines for tumor cores in (a) and (b) were amplified five-fold. c,d: Comparison of APTw histograms for two IDH mutation types for whole-tumor (c) and tumor core (d).
Table 2.
Differences between APTw and MTR Values for IDH-wildtype and IDH-mutant Gliomas
| Parameters | IDH wild-type (n = 7) | IDH mutant (n = 20) | P values |
|---|---|---|---|
| APTw multi-ROI analysis: | |||
| Maximum | 2.03±0.72 | 0.99±0.33 | <0.001 |
| Minimum | 0.99±0.47 | 0.59±0.32 | 0.02 |
| Whole-tumor APTw histogram analysis: | |||
| Mean | 1.39±0.49 | 0.93±0.44 | 0.03 |
| Variance | 0.61±0.36 | 0.97±0.73 | 0.23 |
| Skewness | −0.13±0.28 | −0.35±0.83 | 0.50 |
| Kurtosis | 0.57±0.67 | 1.82±3.32 | 0.34 |
| Slope | 2.27±0.77 | 2.65±0.91 | 0.33 |
| 10th percentile | 0.48±0.54 | −0.14±0.76 | 0.06 |
| 50th percentile | 1.39±0.46 | 0.96±0.36 | 0.02 |
| 90th percentile | 2.30±0.64 | 1.98±0.49 | 0.18 |
| Peak | 1.33±0.52 | 1.02±0.37 | 0.09 |
| Whole-tumor MTR: | |||
| Mean | 14.9±2.1 | 16.3±5.3 | 0.63 |
Note: Data are mean ±standard deviation. Significant results are printed in bold.
Based on the multi-ROI analysis, the IDH-wildtype gliomas had higher maximum (2.03 ± 0.72 vs. 0.99 ± 0.33) and minimum (0.99 ± 0.47 vs. 0.59 ± 0.32) APTw values than the IDH-mutant gliomas, both of which were statistically significant (P < 0.001 and P = 0.02, respectively). Based on the histogram-based analysis, the IDH-wildtype gliomas had significantly higher mean (1.39 ± 0.49 vs. 0.93 ± 0.44; P = 0.03) and 50th percentile (1.39 ± 0.46 vs. 0.96 ± 0.36; P = 0.02) APTw values than the IDH-mutant group. Three other histogram parameters, 10th percentile, 90th percentile, and peak values, showed a higher trend in the IDH-wildtype group, compared to the IDH-mutant group (P = 0.06–0.18; insignificantly). For variance, skewness, kurtosis, and slope of APTw values, there were no significant differences between the two glioma groups (P > 0.2). Further, the whole-tumor MTR values showed no significant difference between the IDH-wildtype and IDH-mutant groups (14.9 ± 2.1 vs. 16.3 ± 2.5.3; P = 0.63).
Prediction of IDH Mutation with APTw Metrics
Among 11 APTw metrics, the multi-ROI-based maximum and minimum APTw values, as well as the histogram-based mean and 50th percentile APTw values, differed significantly between the IDH-wildtype and IDH-mutant groups. Based on ROC curve analyses (Supporting Fig. S1), the multi-ROI-based maximum values showed the highest area under the ROC curve (0.89), and the areas under the ROC curves for the multi-ROI-based minimum, histogram-based mean, and histogram-based 50th percentile APTw metrics in predicting the IDH mutation status were 0.76, 0.75, and 0.75, respectively (Table 3). IDH mutation status was thus predictable with APTw imaging, non-invasively.
Table 3.
Diagnostic Performance of Whole-tumor Histogram-based(Mean, 50th percentile) and Multi-ROI-based (Maximum, Minimum)APTw Parameters in Predicting IDH Genotypes
| Parameters | AUC (95% CI) |
Sensitivity (95% CI) |
Specificity (95% CI) |
PPV (95%CI) |
NPV (95%CI) |
Cut-off value (%) |
|---|---|---|---|---|---|---|
| Mean | 0.75 (0.52–1) | 0.57 (0.20–0.94) | 1 | 1 | 0.87 (0.73–1.01) | 1.58 |
| 50th percentile | 0.75 (0.49–1) | 0.71 (0.38–1.05) | 0.95 (0.85–1.05) | 0.83 (0.54–1.13) | 0.91 (0.78–1.03) | 1.45 |
| Maximum | 0.89 (0.73–1) | 0.57 (0.18–0.90) | 1 | 1 | 0.87 (0.74–0.94) | 1.67 |
| Minimum | 0.76 (0.51–1) | 0.43 (0.10–0.82) | 1 | 1 | 0.83 (0.72–0.90) | 1.12 |
Note: AUC, area under the ROC curve; 95 %CI, 95 % confidence interval; PPV, positive predictive value; NPV, negative predictive value.
Discussion
Low-grade gliomas yield widely different clinical outcomes. The modernized understanding of the somatic mutations of the IDH 1 and 2 genes in gliomas allows us to better sub-classify this group of entities which have distinguished prognosis, opening up a new era in the treatment of brain tumors. In spite of the frequency of IDH wild-type is relatively rare in the population with WHO grade-II gliomas, around 20% (62), IDH-wildtype gliomas are prone to have poorer prognosis (63). Thus, predicting the IDH mutant status preoperatively, preferably with noninvasive imaging modalities, is becoming a focus of the oncology community. Although promising, most previous studies have been experimental, and results have been mixed thus far, which has hampered consistent clinical application. Therefore, most clinicians agree that more specific imaging modalities are urgently needed to help distinguish IDH-wildtype from IDH-mutant gliomas, especially in ambiguous cases where biopsy could potentially be avoided. CEST imaging provides an important contrast mechanism for molecular MRI (32, 33), and this study is the first to evaluate the ability of CEST-MRI to discriminate IDH-wildtype and IDH-mutant low-grade gliomas preoperatively. Based on the entire MTRasym spectra (Fig. 1), we found that the CEST signal intensities in the offset range of 1.5–4 ppm (including the protein-based APTw signal at 3.5 ppm downfield from water) were relatively higher in IDH-wildtype than in IDH-mutant grade-II gliomas, except for the CEST effect at 2.0 ppm, which showed no difference between the two glioma types. Interestingly, IDH-mutant gliomas showed an obviously decreased CEST effect at 2.5ppm besides 3.5ppm (APT effect), compared to IDH-wildtype gliomas. The exact mechanism for this needs to be further explored in the future.
In this study, we applied two ROI drawing approaches for APTw value analysis: histogram analysis and multi-ROI-based analysis, and multiple APTw metrics were then obtained. Both APTw analysis methods are able to yield discriminated indexes. Generally, histogram analysis has priority over multi-ROI-based analysis in terms of the operation simplicity, since the latter should be used with caution by excluding large cysts, hemorrhage, or vessels. However, multi-ROI-based analysis is useful for finding tumor hotspots. We found that multi-ROI-based maximum and minimum APTw values, as well as the histogram-based mean and 50th percentile APTw values, were significantly higher in IDH-wildtype gliomas than in the IDH-mutant gliomas (Table 2), corresponding to areas under the ROC curves of 0.89, 0.76, 0.75, and 0.75, respectively, for the prediction of the IDH mutation status (Table 3). Although the APTw difference between IDH-wildtype and IDH-mutant was very small, it was enough to help making discrimination, showing moderate areas under the ROC curves for these APTw metrics. Therefore, the APTw signal could be a valuable imaging biomarker by which to identify IDH1 mutation status in grade-II gliomas.
According to the CEST theory (64), APTw imaging can generate contrast that, to a large extent, depends on the concentration of endogenous cellular proteins in tissue and the exchange properties of their amide protons with water protons (pH dependent), while other parameters (tissue water content, T1 of water, saturation efficiency) affect the contrast. However, it is imperative to realize that the effect of an increasing water T1 on the measured APTw signal is mostly canceled out by the effect of the increasing water content in the tumor (34, 65). Our further results (66) have shown that it may not be necessary to correct for the influence of water T1 on APT imaging of gliomas, at least at 2 µT, as was used in this study, although this remains an interesting research topic in the field. Based on previous 31P NMR spectroscopy studies in experimental brain tumors and in patients with brain tumors (67, 68), the intracellular pH of untreated malignant gliomas was near neutrality, or a little alkaline (with a slight increase of 0.05–0.08 pH unit reported in the literature). Thus, the observed APT effect in tumor is mostly dominated by the increased amide proton concentration related to mobile cellular proteins (such as cytosolic proteins, many endoplasmic reticulum proteins, and secreted proteins) (69), but an extra contribution from the possibly alkaline intracellular pH may exist, as discussed in our early papers (65). Notably, lab research results have shown global downregulation of protein expression in mutant IDH1-driven glioma cells, compared to oncogenic HRAS IDH1-wild type glioma cells (10), which is consistent with our findings that significantly lower APTw signal intensities were observed in IDH-mutant grade-II gliomas. Conventional MT imaging generates unique contrast in MRI that is related to semi-solid macromolecules, such as those in membranes and nuclei (70). Our research did not find any significant difference in MTR between IDH-mutant and IDH-wildtype grade-II gliomas, suggesting a negligible difference in the more solid environment of the cell for these two tumor types. In addition, our results in this study are consistent with a recent study by Xiong et al. (22), who reported that IDH mutations significantly correlated with a lower cell proliferation in grade-II oligodendroglial tumors, thus leading to a lower APTw signal.
There are some limitations or weaknesses in this study. A limitation of our study was the relatively small sample size, especially for the numbers in the IDH-wildtype group due to the inherent IDH mutation distribution ratio in the general population (63). A large-scale study would be required to obtain more conclusive results. Another limitation is that different experimental saturation settings will give different results, and the contrast needs to be standardized. The final potential limitation was the semi-quantitative characteristic of APTw signal intensity. It means that the APTw intensity has multiple contributions including the upfield NOE effect (53–56) and some conventional semisolid MT asymmetry that are mixed in when performing MTRasym analysis. Fortunately, our recent studies showed that, for the saturation parameters we used, the APT effect (rather than the NOE) would be the major contributor to the APTw image contrast between the tumor and normal brain tissue (or the relative APTw intensity value in the tumor, compared to normal brain tissue, as used in this study) (54, 65, 71). Thus, we believe that our results would be very close to those based on the absolute quantitative APT metrics. Several alternative APTw imaging analysis or acquisition approaches have been proposed to quantify a pure APT effect (72–75). Notably, the extrapolated semisolid magnetization transfer reference model that is being developed by our group (71) should achieve more pure APT quantification, and we will apply this promising imaging analysis method to our future research.
CONCLUSIONS
The present study represents the first analysis of the ability to use APTw MRI to differentiate between IDH-wildtype and IDH-mutant grade-II gliomas. IDH mutation status is associated with distinct APTw signatures, and IDH1-wildtype lesions are typically correlated with relatively high APTw signal intensities, compared with IDH1-mutant lesions. Our early results highlight the potential future of APTw imaging to provide a more precise genotypic diagnostic workup of gliomas. Non-invasive prediction of IDH1 mutation status could provide more valuable supplementary information about the prognosis of grade-II gliomas before surgery.
Supplementary Material
Acknowledgments
The authors thank Ms. Mary McAllister for editorial assistance. This work was supported in part by grants from the National Institutes of Health (R01EB009731, R01CA166171, R01NS083435, R01 EB015032, and P41EB015909), the National Natural Science Foundation of China (81171322), the Guangdong Provincial Natural Science Foundation (2014A030313271, S2012010009114), the Guangdong Provincial Science and Technology Project (2014A020212726), and the Southern Medical University clinical research project (LC2016ZD028).
Footnotes
Additional Supporting Information may be found in the online version of this article.
Supporting Table S1. Differences between APTw Values Based on Histogram Analysis corresponding to the ROI with maximum APTw (“tumor core”) for IDH-wildtype and IDH-mutant Gliomas
Supporting Fig. S1. ROC diagnostic performance analyses of whole-tumor histogram-based (mean, 50th percentile) and multi-ROI-based (maximum, minimum) APTw parameters in predicting IDH genotypes. The maximum APTw value showed the most accurate diagnostic ability.
References
- 1.American Cancer Society. Cancer facts and figures. 2016 http://www.cancer.org.
- 2.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA, Jr, Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pollard PJ, Ratcliffe PJ. Cancer. Puzzling patterns of predisposition. Science. 2009;324:192–194. doi: 10.1126/science.1173362. [DOI] [PubMed] [Google Scholar]
- 5.Kloosterhof NK, Bralten LB, Dubbink HJ, French PJ, van den Bent MJ. Isocitrate dehydrogenase-1 mutations: a fundamentally new understanding of diffuse glioma? Lancet Oncol. 2011;12:83–91. doi: 10.1016/S1470-2045(10)70053-X. [DOI] [PubMed] [Google Scholar]
- 6.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 7.Reitman ZJ, Jin G, Karoly ED, Spasojevic I, Yang J, Kinzler KW, He Y, Bigner DD, Vogelstein B, Yan H. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc Natl Acad Sci U S A. 2011;108:3270–3275. doi: 10.1073/pnas.1019393108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ichimura K, Pearson DM, Kocialkowski S, Backlund LM, Chan R, Jones DT, Collins VP. IDH1 mutations are present in the majority of common adult gliomas but rare in primary glioblastomas. Neuro Oncol. 2009;11:341–347. doi: 10.1215/15228517-2009-025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, Verhaak RG, Hoadley KA, Hayes DN, Perou CM, Schmidt HK, Ding L, Wilson RK, Van Den Berg D, Shen H, Bengtsson H, Neuvial P, Cope LM, Buckley J, Herman JG, Baylin SB, Laird PW, Aldape K. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doll S, Urisman A, Oses-Prieto JA, Arnott D, Burlingame AL. Quantitative Proteomics Reveals Fundamental Regulatory Differences in Oncogenic HRAS and Isocitrate Dehydrogenase (IDH1) Driven Astrocytoma. Mol Cell Proteomics. 2017;16:39–56. doi: 10.1074/mcp.M116.063883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turkalp Z, Karamchandani J, Das S. IDH mutation in glioma: new insights and promises for the future. JAMA Neurol. 2014;71:1319–1325. doi: 10.1001/jamaneurol.2014.1205. [DOI] [PubMed] [Google Scholar]
- 12.Watanabe T, Vital A, Nobusawa S, Kleihues P, Ohgaki H. Selective acquisition of IDH1 R132C mutations in astrocytomas associated with Li-Fraumeni syndrome. Acta Neuropathol. 2009;117:653–656. doi: 10.1007/s00401-009-0528-x. [DOI] [PubMed] [Google Scholar]
- 13.Agarwal S, Sharma MC, Jha P, Pathak P, Suri V, Sarkar C, Chosdol K, Suri A, Kale SS, Mahapatra AK. Comparative study of IDH1 mutations in gliomas by immunohistochemistry and DNA sequencing. Neuro Oncol. 2013;15:718–726. doi: 10.1093/neuonc/not015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Metellus P, Coulibaly B, Colin C, de Paula AM, Vasiljevic A, Taieb D, Barlier A, Boisselier B, Mokhtari K, Wang XW, Loundou A, Chapon F, Pineau S, Ouafik L, Chinot O, Figarella-Branger D. Absence of IDH mutation identifies a novel radiologic and molecular subtype of WHO grade II gliomas with dismal prognosis. Acta Neuropathol. 2010;120:719–729. doi: 10.1007/s00401-010-0777-8. [DOI] [PubMed] [Google Scholar]
- 15.Carrillo JA, Lai A, Nghiemphu PL, Kim HJ, Phillips HS, Kharbanda S, Moftakhar P, Lalaezari S, Yong W, Ellingson BM, Cloughesy TF, Pope WB. Relationship between tumor enhancement, edema, IDH1 mutational status, MGMT promoter methylation, and survival in glioblastoma. AJNR Am J Neuroradiol. 2012;33:1349–1355. doi: 10.3174/ajnr.A2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qi S, Yu L, Li H, Ou Y, Qiu X, Ding Y, Han H, Zhang X. Isocitrate dehydrogenase mutation is associated with tumor location and magnetic resonance imaging characteristics in astrocytic neoplasms. Oncol Lett. 2014;7:1895–1902. doi: 10.3892/ol.2014.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reyes-Botero G, Dehais C, Idbaih A, Martin-Duverneuil N, Lahutte M, Carpentier C, Letouze E, Chinot O, Loiseau H, Honnorat J, Ramirez C, Moyal E, Figarella-Branger D, Ducray F. Contrast enhancement in 1p/19q-codeleted anaplastic oligodendrogliomas is associated with 9p loss, genomic instability, and angiogenic gene expression. Neuro-Oncol. 2014;16:662–670. doi: 10.1093/neuonc/not235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sonoda Y, Shibahara I, Kawaguchi T, Saito R, Kanamori M, Watanabe M, Suzuki H, Kumabe T, Tominaga T. Association between molecular alterations and tumor location and MRI characteristics in anaplastic gliomas. Brain Tumor Pathol. 2015;32:99–104. doi: 10.1007/s10014-014-0211-3. [DOI] [PubMed] [Google Scholar]
- 19.Yamashita K, Hiwatashi A, Togao O, Kikuchi K, Hatae R, Yoshimoto K, Mizoguchi M, Suzuki SO, Yoshiura T, Honda H. MR Imaging-Based Analysis of Glioblastoma Multiforme: Estimation of IDH1 Mutation Status. AJNR American journal of neuroradiology. 2016;37:58–65. doi: 10.3174/ajnr.A4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee S, Choi SH, Ryoo I, Yoon TJ, Kim TM, Lee SH, Park CK, Kim JH, Sohn CH, Park SH, Kim IH. Evaluation of the microenvironmental heterogeneity in high-grade gliomas with IDH1/2 gene mutation using histogram analysis of diffusion-weighted imaging and dynamic-susceptibility contrast perfusion imaging. J Neurooncol. 2015;121:141–150. doi: 10.1007/s11060-014-1614-z. [DOI] [PubMed] [Google Scholar]
- 21.Xiong J, Tan WL, Pan JW, Wang Y, Yin B, Zhang J, Geng DY. Detecting isocitrate dehydrogenase gene mutations in oligodendroglial tumors using diffusion tensor imaging metrics and their correlations with proliferation and microvascular density. J Magn Reson Imaging. 2016;43:45–54. doi: 10.1002/jmri.24958. [DOI] [PubMed] [Google Scholar]
- 22.Xiong J, Tan W, Wen J, Pan J, Wang Y, Zhang J, Geng D. Combination of diffusion tensor imaging and conventional MRI correlates with isocitrate dehydrogenase 1/2 mutations but not 1p/19q genotyping in oligodendroglial tumours. Eur Radiol. 2016;26:1705–1715. doi: 10.1007/s00330-015-4025-4. [DOI] [PubMed] [Google Scholar]
- 23.Tan W, Xiong J, Huang W, Wu J, Zhan S, Geng D. Noninvasively detecting Isocitrate dehydrogenase 1 gene status in astrocytoma by dynamic susceptibility contrast MRI. J Magn Reson Imaging. 2017;45:492–499. doi: 10.1002/jmri.25358. [DOI] [PubMed] [Google Scholar]
- 24.Biller A, Badde S, Nagel A, Neumann JO, Wick W, Hertenstein A, Bendszus M, Sahm F, Benkhedah N, Kleesiek J. Improved brain tumor classification by sodium MR imaging: Prediction of IDH mutation status and tumor progression. AJNR Am J Neuroradiol. 2016;37:66–73. doi: 10.3174/ajnr.A4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macyszyn L, Akbari H, Pisapia JM, Da X, Attiah M, Pigrish V, Bi Y, Pal S, Davuluri RV, Roccograndi L, Dahmane N, Martinez-Lage M, Biros G, Wolf RL, Bilello M, O'Rourke DM, Davatzikos C. Imaging patterns predict patient survival and molecular subtype in glioblastoma via machine learning techniques. Neuro Oncol. 2016;18:417–425. doi: 10.1093/neuonc/nov127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang B, Chang K, Ramkissoon S, Tanguturi S, Bi WL, Reardon DA, Ligon KL, Alexander BM, Wen PY, Huang RY. Multimodal MRI features predict isocitrate dehydrogenase genotype in high-grade gliomas. Neuro Oncol. 2017;19:109–117. doi: 10.1093/neuonc/now121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Vander Heiden MG, Su SM. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andronesi OC, Kim GS, Gerstner E, Batchelor T, Tzika AA, Fantin VR, Vander Heiden MG, Sorensen AG. Detection of 2-hydroxyglutarate in IDH-mutated glioma patients by in vivo spectral-editing and 2D correlation magnetic resonance spectroscopy. Sci Transl Med. 2012;4:116ra114. doi: 10.1126/scitranslmed.3002693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pope WB, Prins RM, Albert Thomas M, Nagarajan R, Yen KE, Bittinger MA, Salamon N, Chou AP, Yong WH, Soto H, Wilson N, Driggers E, Jang HG, Su SM, Schenkein DP, Lai A, Cloughesy TF, Kornblum HI, Wu H, Fantin VR, Liau LM. Non-invasive detection of 2-hydroxyglutarate and other metabolites in IDH1 mutant glioma patients using magnetic resonance spectroscopy. J Neurooncol. 2012;107:197–205. doi: 10.1007/s11060-011-0737-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi C, Ganji SK, DeBerardinis RJ, Hatanpaa KJ, Rakheja D, Kovacs Z, Yang XL, Mashimo T, Raisanen JM, Marin-Valencia I, Pascual JM, Madden CJ, Mickey BE, Malloy CR, Bachoo RM, Maher EA. 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat Med. 2012;18:624–629. doi: 10.1038/nm.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de la Fuente MI, Young RJ, Rubel J, Rosenblum M, Tisnado J, Briggs S, Arevalo-Perez J, Cross JR, Campos C, Straley K, Zhu D, Dong C, Thomas A, Omuro AA, Nolan CP, Pentsova E, Kaley TJ, Oh JH, Noeske R, Maher E, Choi C, Gutin PH, Holodny AI, Yen K, DeAngelis LM, Mellinghoff IK, Thakur SB. Integration of 2-hydroxyglutarate-proton magnetic resonance spectroscopy into clinical practice for disease monitoring in isocitrate dehydrogenase-mutant glioma. Neuro Oncol. 2016;18:283–290. doi: 10.1093/neuonc/nov307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ward KM, Aletras AH, Balaban RS. A new class of contrast agents for MRI based on proton chemical exchange dependent saturation transfer (CEST) Journal of Magnetic Resonance. 2000;143:79–87. doi: 10.1006/jmre.1999.1956. [DOI] [PubMed] [Google Scholar]
- 33.Zhou J, van Zijl PC. Chemical exchange saturation transfer imaging and spectroscopy. Progr NMR Spectr. 2006;48:109–136. [Google Scholar]
- 34.Zhou J, Payen J, Wilson DA, Traystman RJ, van Zijl PCM. Using the amide proton signals of intracellular proteins and peptides to detect pH effects in MRI. Nature Med. 2003;9:1085–1090. doi: 10.1038/nm907. [DOI] [PubMed] [Google Scholar]
- 35.Zhou J, Lal B, Wilson DA, Laterra J, van Zijl PCM. Amide proton transfer (APT) contrast for imaging of brain tumors. Magn Reson Med. 2003;50:1120–1126. doi: 10.1002/mrm.10651. [DOI] [PubMed] [Google Scholar]
- 36.Wen Z, Hu S, Huang F, Wang X, Guo L, Quan X, Wang S, Zhou J. MR imaging of high-grade brain tumors using endogenous protein and peptide-based contrast. NeuroImage. 2010;51:616–622. doi: 10.1016/j.neuroimage.2010.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou J, Zhu H, Lim M, Blair L, Quinones-Hinojosa A, Messina AA, Eberhart CG, Pomper MG, Laterra J, Barker PB, van Zijl PCM, Blakeley JO. Three-dimensional amide proton transfer MR imaging of gliomas: Initial experience and comparison with gadolinium enhancement. J Magn Reson Imaging. 2013;38:1119–1128. doi: 10.1002/jmri.24067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Togao O, Yoshiura T, Keupp J, Hiwatashi A, Yamashita K, Kikuchi K, Suzuki Y, Suzuki SO, Iwaki T, Hata N, Mizoguchi M, Yoshimoto K, Sagiyama K, Takahashi M, Honda H. Amide proton transfer imaging of adult diffuse gliomas: correlation with histopathological grades. Neuro-Oncology. 2014;16:441–448. doi: 10.1093/neuonc/not158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang S, Yu H, Wang X, Lu S, Li Y, Feng L, Zhang Y, Heo HY, Lee DH, Zhou J, Wen Z. Molecular MRI differentiation between primary central nervous system lymphomas and high-grade gliomas using endogenous protein-based amide proton transfer MR imaging at 3 Tesla. Eur Radiol. 2016;26:64–71. doi: 10.1007/s00330-015-3805-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jia G, Abaza R, Williams JD, Zynger DL, Zhou JY, Shah ZK, Patel M, Sammet S, Wei L, Bahnson RR, Knopp MV. Amide proton transfer MR imaging of prostate cancer: A preliminary study. J Magn Reson Imaging. 2011;33:647–654. doi: 10.1002/jmri.22480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dula AN, Arlinghaus LR, Dortch RD, Dewey BE, Whisenant JG, Ayers GD, Yankeelov TE, Smith SA. Amide proton transfer imaging of the breast at 3 T: Establishing reproducibility and possible feasibility assessing chemotherapy response. Magn Reson Med. 2013;70:216–224. doi: 10.1002/mrm.24450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou JY, van Zijl PCM. Defining an acidosis-based ischemic penumbra from pH-weighted MRI. Transl Stroke Res. 2012;3:76–83. doi: 10.1007/s12975-011-0110-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harston GW, Tee YK, Blockley N, Okell TW, Thandeswaran S, Shaya G, Sheerin F, Cellerini M, Payne S, Jezzard P, Chappell M, Kennedy J. Identifying the ischaemic penumbra using pH-weighted magnetic resonance imaging. Brain. 2015;138:36–42. doi: 10.1093/brain/awu374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li C, Peng S, Wang R, Chen H, Su W, Zhao X, Zhou J, Chen M. Chemical exchange saturation transfer MR imaging of Parkinson's disease at 3 Tesla. Eur Radiol. 2014;24:2631–2639. doi: 10.1007/s00330-014-3241-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang H, Kang H, Zhao X, Jiang S, Zhang Y, Zhou J, Peng Y. Amide proton transfer (APT) MR imaging and magnetization transfer (MT) MR imaging of pediatric brain development. Eur Radiol. 2016;26:3368–3376. doi: 10.1007/s00330-015-4188-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou J, Blakeley JO, Hua J, Kim M, Laterra J, Pomper MG, van Zijl PCM. Practical data acquisition method for human brain tumor amide proton transfer (APT) imaging. Magn Reson Med. 2008;60:842–849. doi: 10.1002/mrm.21712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sakata A, Okada T, Yamamoto A, Kanagaki M, Fushimi Y, Dodo T, Arakawa Y, Schmitt B, Miyamoto S, Togashi K. Grading glial tumors with amide proton transfer MR imaging: different analytical approaches. J Neuro-Oncol. 2015;122:339–348. doi: 10.1007/s11060-014-1715-8. [DOI] [PubMed] [Google Scholar]
- 48.Choi YS, Ahn SS, Lee SK, Chang JH, Kang SG, Kim SH, Zhou J. Amide proton transfer imaging to discriminate between low- and high-grade gliomas: added value to apparent diffusion coefficient and relative cerebral blood volume. Eur Radiol. 2017 doi: 10.1007/s00330-00017-04732-00330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hobbs SK, Shi G, Homer R, Harsh G, Altlas SW, Bednarski MD. Magnetic resonance imaging-guided proteomics of human glioblastoma multiforme. J Magn Reson Imag. 2003;18:530–536. doi: 10.1002/jmri.10395. [DOI] [PubMed] [Google Scholar]
- 50.Li J, Zhuang Z, Okamoto H, Vortmeyer AO, Park DM, Furata M, Lee Y-S, Oldfield EH, Zeng W, Weil RJ. Proteomic profiling distinguishes astrocytomas and identifies differential tumor markers. Neurology. 2006;66:733–736. doi: 10.1212/01.wnl.0000201270.90502.d0. [DOI] [PubMed] [Google Scholar]
- 51.Howe FA, Barton SJ, Cudlip SA, Stubbs M, Saunders DE, Murphy M, Wilkins P, Opstad KS, Doyle VL, McLean MA, Bell BA, Griffiths JR. Metabolic profiles of human brain tumors using quantitative in vivo 1H magnetic resonance spectroscopy. Magn Reson Med. 2003;49:223–232. doi: 10.1002/mrm.10367. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Y, Heo HY, Lee DH, Zhao X, Jiang S, Zhang K, Li H, Zhou J. Selecting the reference image for registration of CEST series. J Magn Reson Imaging. 2016;43:756–761. doi: 10.1002/jmri.25027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ling W, Regatte RR, Navon G, Jerschow A. Assessment of glycosaminoglycan concentration in vivo by chemical exchange-dependent saturation transfer (gagCEST) Proc Natl Acad Sci (USA) 2008;105:2266–2270. doi: 10.1073/pnas.0707666105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou J, Hong X, Zhao X, Gao J-H, Yuan J. APT-weighted and NOE-weighted image contrasts in glioma with different RF saturation powers based on magnetization transfer ratio asymmetry analyses. Magn Reson Med. 2013;70:320–327. doi: 10.1002/mrm.24784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jones CK, Huang A, Xu J, Edden RA, Schar M, Hua J, Oskolkov N, Zaca D, Zhou J, McMahon MT, Pillai JJ, van Zijl PC. Nuclear Overhauser enhancement (NOE) imaging in the human brain at 7T. Neuroimage. 2013;77:114–124. doi: 10.1016/j.neuroimage.2013.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heo H-Y, Zhang Y, Lee D-H, Hong X, Zhou J. Quantitative assessment of amide proton transfer (APT) and nuclear Overhauser enhancement (NOE) imaging with extrapolated semi-solid magnetization transfer reference (EMR) signals: Application to a rat glioma model at 4.7 T. Magn Reson Med. 2016;75:137–138. doi: 10.1002/mrm.25581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hua J, Jones CK, Blakeley J, Smith SA, van Zijl PCM, Zhou J. Quantitative description of the asymmetry in magnetization transfer effects around the water resonance in the human brain. Magn Reson Med. 2007;58:786–793. doi: 10.1002/mrm.21387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liang HY, Huang YQ, Yang ZX, Ying D, Zeng MS, Rao SX. Potential of MR histogram analyses for prediction of response to chemotherapy in patients with colorectal hepatic metastases. Eur Radiol. 2016;26:2009–2018. doi: 10.1007/s00330-015-4043-2. [DOI] [PubMed] [Google Scholar]
- 59.Yip S, Butterfield YS, Morozova O, Chittaranjan S, Blough MD, An J, Birol I, Chesnelong C, Chiu R, Chuah E, Corbett R, Docking R, Firme M, Hirst M, Jackman S, Karsan A, Li H, Louis DN, Maslova A, Moore R, Moradian A, Mungall KL, Perizzolo M, Qian J, Roldan G, Smith EE, Tamura-Wells J, Thiessen N, Varhol R, Weiss S, Wu W, Young S, Zhao Y, Mungall AJ, Jones SJ, Morin GB, Chan JA, Cairncross JG, Marra MA. Concurrent CIC mutations, IDH mutations, and 1p/19q loss distinguish oligodendrogliomas from other cancers. J Pathol. 2012;226:7–16. doi: 10.1002/path.2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cai K, Singh A, Poptani H, Li W, Yang S, Lu Y, Hariharan H, Zhou XJ, Reddy R. CEST signal at 2ppm (CEST@2ppm) from Z-spectral fitting correlates with creatine distribution in brain tumor. NMR Biomed. 2015;28:1–8. doi: 10.1002/nbm.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wuthrich K. NMR of proteins and nucleic acids. 2. New York: John Wiley & Sons; 1986. [Google Scholar]
- 62.Hartmann C, Meyer J, Balss J, Capper D, Mueller W, Christians A, Felsberg J, Wolter M, Mawrin C, Wick W, Weller M, Herold-Mende C, Unterberg A, Jeuken JW, Wesseling P, Reifenberger G, von Deimling A. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta neuropathologica. 2009;118:469–474. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 63.Cohen AL, Holmen SL, Colman H. IDH1 and IDH2 mutations in gliomas. Curr Neurol Neurosci Rep. 2013;13:345. doi: 10.1007/s11910-013-0345-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou J, Wilson DA, Sun PZ, Klaus JA, van Zijl PCM. Quantitative description of proton exchange processes between water and endogenous and exogenous agents for WEX, CEST, and APT experiments. Magn Reson Med. 2004;51:945–952. doi: 10.1002/mrm.20048. [DOI] [PubMed] [Google Scholar]
- 65.Lee DH, Heo HY, Zhang K, Zhang Y, Jiang S, Zhao X, Zhou J. Quantitative assessment of the effects of water proton concentration and water T1 changes on amide proton transfer (APT) and nuclear overhauser enhancement (NOE) MRI: The origin of the APT imaging signal in brain tumor. Magn Reson Med. 2016 doi: 10.1002/mrm.26131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Heo HY, Lee DH, Zhang Y, Zhao X, Jiang S, Chen M, Zhou J. Insight into the quantitative metrics of chemical exchange saturation transfer (CEST) imaging. Magn Reson Med. 2016 doi: 10.1002/mrm.26264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ross BD, Higgins RJ, Boggan JE, Knittel B, Garwood M. 31P NMR spectroscopy of the in vivo metabolism of an intracerebral glioma in the rat. Magn Reson Med. 1988;6:403–417. doi: 10.1002/mrm.1910060406. [DOI] [PubMed] [Google Scholar]
- 68.Maintz D, Heindel W, Kugel H, Jaeger R, Lackner KJ. Phosphorus-31 MR spectroscopy of normal adult human brain and brain tumours. Nmr in Biomedicine. 2002;15:18–27. doi: 10.1002/nbm.735. [DOI] [PubMed] [Google Scholar]
- 69.Yan K, Fu Z, Yang C, Zhang K, Jiang S, Lee DH, Heo HY, Zhang Y, Cole RN, Van Eyk JE, Zhou J. Assessing amide proton transfer (APT) MRI contrast origins in 9L gliosarcoma in the rat brain using proteomic analysis. Mol Imaging Biol. 2015;17:479–487. doi: 10.1007/s11307-015-0828-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Henkelman RM, Stanisz GJ, Graham SJ. Magnetization transfer in MRI: a review. NMR Biomed. 2001;14:57–64. doi: 10.1002/nbm.683. [DOI] [PubMed] [Google Scholar]
- 71.Heo HY, Zhang Y, Jiang S, Lee DH, Zhou J. Quantitative assessment of amide proton transfer (APT) and nuclear overhauser enhancement (NOE) imaging with extrapolated semisolid magnetization transfer reference (EMR) signals: II. Comparison of three EMR models and application to human brain glioma at 3 Tesla. Magn Reson Med. 2016;75:1630–1639. doi: 10.1002/mrm.25795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zaiss M, Schmitt B, Bachert P. Quantitative separation of CEST effect from magnetization transfer and spillover effects by Lorentzian-line-fit analysis of z-spectra. J Magn Reson. 2011;211:149–155. doi: 10.1016/j.jmr.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 73.Jin T, Wang P, Zong X, Kim S-G. MR imaging of the amide-proton transfer effect and the pH-insensitive nuclear overhauser effect at 9.4 T. Magn Reson Med. 2013;69:760–770. doi: 10.1002/mrm.24315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zu Z, Janve VA, Xu J, Does MD, Gore JC, Gochberg DF. A new method for detecting exchanging amide protons using chemical exchange rotation transfer. Magn Reson Med. 2013;69:637–647. doi: 10.1002/mrm.24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee JS, Xia D, Ge Y, Jerschow A, Regatte RR. Concurrent saturation transfer contrast in in vivo brain by a uniform magnetization transfer MRI. Neuroimage. 2014;95:22–28. doi: 10.1016/j.neuroimage.2014.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.