Abstract
Topoisomerase II beta (Top2b) is an enzyme that alters the topologic states of DNA during transcription. Top2b deletion in early retinal progenitor cells causes severe defects in neural differentiation and affects cell survival in all retinal cell types. However, it is unclear whether the observed severe phenotypes are the result of cell-autonomous/primary defects or non-cell-autonomous/secondary defects caused by alterations of other retinal cells. Using photoreceptor cells as a model, we first characterized the phenotypes in Top2b conditional knockout (cKO). Top2b deletion leads to malformation of photoreceptor outer segments (OS) and synapses accompanied with dramatic cell loss at late-stage photoreceptor differentiation. Then, we performed mosaic analysis with shRNA-mediated Top2b knockdown in neonatal retina using in vivo electroportation to target rod photoreceptors in neonatal retina. Top2b knockdown causes defective OS without causing a dramatic cell loss, suggesting a Top2b cell-autonomous function. Furthermore, RNA-seq analysis reveals that Top2b controls the expression of key genes in photoreceptor gene-regulatory network, e.g., Crx, Nr2e3, Opn1sw, and Vsx2, and retinopathy-related genes, e.g., Abca4, Bbs7, and Pde6b. Together, our data establish a combinatorial cell-autonomous and non-cell-autonomous role for Top2b in late-stage of photoreceptor differentiation and maturation.
Keywords: neural development, neural differentiation, retina photoreceptors, transcriptional regulatory network, gene expression, GSE86187
Graphical Abstract
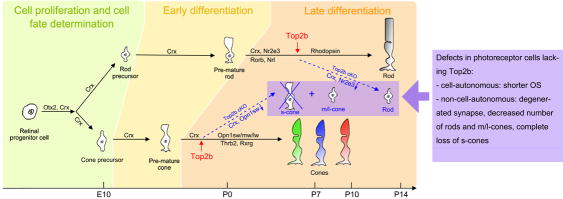
Three major stages are well defined in photoreceptor cell development: 1) cell proliferation and cell fate determination, during which the multipotent retinal progenitors proliferate and their competence being restricted as photoreceptor cell precursors; 2) early differentiation stage, during which genes for morphogenesis and phototransduction are expressed; and 3) late differentiation stage, which includes the axonal growth, synapse formation and outer segment (OS) biogenesis (Swaroop et al. 2010). Top2b functions in both cell-autonomous and non-cell-autonomous manners in regulating late stage photoreceptor development by controlling key genes in the photoreceptor transcriptional network.
INTRODUCTION
Rod and cone photoreceptors constitute the largest cell population in the mammalian retina (Brzezinski and Reh 2015; Carter-Dawson and LaVail 1979; Cepko 2015). A recent study using single-cell RNA-seq profiling has determined that the percentages of rods and cones in the mouse are 65.6% and 4.2%, respectively (Macosko et al. 2015). Lineage studies showed that both cell types are derived from multipotent retinal progenitor cells (Reviewed by: Brzezinski and Reh 2015; Cepko 2015; Wang and Cepko 2016). Birth-dating studies have established that the genesis of retinal cell types is a highly conserved process. In the mouse, the production of cone cells starts around embryonic day 12 (E12), peaks at E14–15, and the majority of cone cells are differentiated by birth. Meanwhile the production of rod cells starts around E14, peaks at birth and the majority of rods are generated by postnatal day 8–12 (P8–12) (Rapaport et al. 2004; Wang et al. 2014; Young 1985b). Three major stages are well defined in photoreceptor cell development: 1) cell proliferation and cell fate determination, during which the multipotent retinal progenitors proliferate and their competence being restricted as photoreceptor cell precursors; 2) early differentiation stage, during which genes for morphogenesis and phototransduction are expressed; and 3) late differentiation stage, which includes the axonal growth, synapse formation and outer segment (OS) biogenesis (Swaroop et al. 2010). The development of the OS is well documented with electron microscopic studies (Nilsson 1964; Steinberg et al. 1980; Tokuyasu and Yamada 1959). However, the molecular mechanism underlying the photoreceptor cell differentiation is not completely understood. Studies have shown that photoreceptor development involves a complex gene regulatory network including several key factors, e.g., Otx2, Crx, Nr2e3, Nrl, Rorb, Opn1sw/mw/lw, etc. (Brzezinski and Reh 2015; Chen et al. 1997; Peng et al. 2005; Swaroop et al. 2010; Wang and Cepko 2016). Mutations in one or more of these critical genes cause loss of the OS and further complete loss of photoreceptors that leads to several retinopathies, e.g., retinitis pigmentosa, and loss of vision (Coppieters et al. 2007; Freund et al. 1997; Swain et al. 1997; Weitz et al. 1992).
Topoisomerase II beta (Top2b) is an enzyme that controls and alters the topologic states of DNA during transcription (Wang 2002). The onset of Top2b expression is observed in progenitor cells that just finished the final division and is actively involved in neural development (Lyu and Wang 2003; Tiwari et al. 2012; Tsutsui et al. 1993; Tsutsui et al. 2001). Many studies have established a multifaceted role of Top2b in 1) resolving the topological constraints in early-stage neuronal gene expression (Madabhushi et al. 2015); 2) neurite guidance during late stage neuronal development (Nevin et al. 2011; Yang et al. 2000); 3) cerebral stratification (Lyu and Wang 2003); and 4) control of developmentally regulated genes in the brain and neural retina (Lyu et al. 2006). We have shown severe defects in differentiation and survival of retinal neurons and Muller glia in Dkk3-Cre mediated Top2b conditional conditional knockout animals (cKO) (Li et al. 2014). However, due to the pleiotropic effect of Top2b knockout on gene expression, the mechanism underlying Top2b functions, e.g., cell-autonomous (primary) or non-cell-autonomous (secondary), during retinal development is unclear.
In this study, we show that Top2b plays an important role in photoreceptor development in both cell-autonomous and non-cell-autonomous manner. Transcriptome analysis by RNA-seq reveals that Top2b controls key genes in photoreceptor regulatory network and genes linked to retinopathies.
MATERIALS AND METHODS
Sample collection
All studies performed were conducted under the strict guidelines and consent of the Institutional Animal Care and Use Committee (IACUC) of Rutgers, The State University of New Jersey. Top2b cKO animals were generated by breeding a strain of Dkk3-Cre mice (Sato et al. 2007) with the Top2b-floxed mice on 129SvEv background. Three to five animals from each genotype were used for each stage. No animal was excluded. Gender of the animals was not reported since no difference between different sexes was observed. Animals were euthanized with CO2 inhalation. For immunohistochemistry, retinae were dissected from the animals and fixed with 4% (w/v) PFA for 1 hr. Following fixation, samples were washed three times with 1× PBS for 10 min each, soaked in 30% (w/v) sucrose for 2~3 days, embedded in cryo-preserving media (Tissue Tek® OCT compound, SAKURA FINETEK USA INC, Torrance, CA, US) and stored at −80°C. Cryo-sectioning was performed with a cryostat (Thermo Scientific, Waltham, MA, US). Sections of samples were air-dried for 10 min and stored at −80°C. For RNA-seq analysis, P0 and P6 retinae (n≥4 for each genotype on each stage) were dissected rapidly free of other ocular tissue and frozen with liquid nitrogen. For total RNA isolation, retinal samples were transferred into 500 μl of TRIzol reagent (Invitrogen), and isolation was performed according to the manufacturer's instructions.
Immunohistochemistry
Immunofluorescence staining was performed on mouse eye sections as described previously (Li et al. 2014). The primary antibodies used in this study are listed in Table 1. Images were captured using a Zeiss Axio Imager M1 fluorescence microscope and analyzed using AxioVision 4.8 (Zeiss, Germany). For proper comparison between control and cKO samples, all images were captured from dorsal-medial region of the retina. Cell counting and measurements were performed in the central region of the retina and the regions were randomly selected. For each data points, samples from at least three animals were calculated for an average and standard deviation. Two tailed heteroscedastic Student’s T-test was employed to calculate p-values.
Table 1.
Antibodies
| Antibody name | Immunogen | Manufacturer and catalog #, RRID, species, mono- or poly-clonal |
Dilution (with PBS) |
|---|---|---|---|
| Cleaved-Casp3 | Endogenous levels of the large fragment (17/19 kDa) of activated caspase-3 resulting from cleavage adjacent to Asp175. |
Cell Signaling, 9661, AB_2341188, rabbit, polyclonal |
1:1600 |
| Crx | Amino acids 166 –285 mapping near the C-terminus of CRX of human origin. |
Santa Cruz Biotechnology, sc- 30150, AB_2276566, rabbit, polyclonal), |
1:100 |
| Nr2e3 | VPS36 fusion protein ag5549 | Proteintech, 14262-1-AP, rabbit, polyclonal |
1:100 |
| Opn1mw/lw | Extracellular doman of human OPN1LW; mouse OPN1MW |
Santa Cruz Biotechnology, sc- 22117, AB_2300956, goat, polyclonal |
1:100 |
| Opn1sw | N-terminus of the opsin protein encoded by OPN1SW |
Santa Cruz Biotechnology, sc- 14363, AB_2158332, goat, polyclonal |
1:100 |
| Rhodopsin 4D2 | N-terminal region of rhodopsin | Gift from Dr. Robert S. Molday from University of British Columbia, Canada, AB_2315273, mouse, monoclonal |
1:100 |
| Synaptophysin | A synaptosome prepared from rat retina. |
Sigma-Aldrich, s5768, AB_477523, mouse, monoclonal |
1:400 |
| Top2b (Topo IIbeta) |
Amino acids 1341–1626 of Topo IIβ of human origin. |
Santa Cruz Biotechnology, sc- 25330, AB_628384, mouse, monoclonal |
1:300 |
| Tuj-1 | microtubules derived from rat brain | Abcam, ab14545, mouse, monoclonal |
1:1000 |
| vGlut1 | Strep-Tag® fusion protein of rat VGLUT 1 (aa 456 – 560) |
Synaptic Systems, 135302, AB_887877, rabbit, polyclonal |
1:300 |
Helium ion microscopic imaging
Retinae from control and Top2b cKO mice were collected, fixed and sectioned as described previously (Li et al. 2014). Each slide contains 8~10 sections, which are 10~12 μm thick. For chemical drying, the slides were subjected to sequential dehydration in graded ethanol (Fisher Scientific, Waltham, MA, USA) (50, 70, 80, 90, 100%) baths at twenty minute intervals. After the final twenty minutes of ethanol dehydration, solutions of different ratios of ethanol and hexamethyldisilazane (HMDS, UltraPure Solutions, Inc., Castroville, CA, USA) (25, 50, 75, 100%) were added sequentially to replace the inner-tissue solution from ethanol to HMDS. The tissue samples were removed from the 100% HMDS bath, placed in a fume hood with cover for 12 hours, and stored in a newly charged desiccation chamber under vacuum. For helium ion microscopy (HIM, ORION Plus, Carl Zeiss, Peabody, MA, USA), sample was mounted onto an HIM stub with carbon tape and images was taken with different fields of view using ORION PLUS software.
Cre-mediated Top2b knockout and shRNA-mediated Top2b knockdown
For Top2b gene knockout in postnatal mouse retinae, a CAG-Cre-GFP construct (Addgene plasmid #13776) was injected into the sub-retinal space of P0 pups of the Top2bf/f mice (Lyu and Wang 2003). For Top2b knockdown, an effective Top2b shRNA (shTop2b) and a control plasmid construct using LentiLox 3.7 vector (Azarova et al. 2010) were used. Plasmid DNA injection and electroporation were performed following a published protocol (Matsuda and Cepko 2004). Briefly, the plasmids were mixed with FastGreen (0.025%) and ~ 0.5 μl of DNA was injected into the subretinal space between the retina and choroid of P0 wild-type mouse pups. After injection, tweezer-type electrodes (model 520, 7 mm diameter, BTX, San Diego) briefly soaked in PBS were placed to softly hold the heads of the pups, and five 80V square pulses of 50ms duration with 950ms intervals were applied using a pulse generator ECM830 (BTX, San Diego, CA, USA). Usually DNA was transfected only into right eyes. Transfected retinae were harvested at various stages and sectioned for immunostaining as described (Li et al. 2014).
RNA-seq analysis
Whole transcriptome RNA libraries were sequenced using the SOLiD System (Applied Biosystems). Data analysis was performed according to published protocols (Trapnell et al., 2012) with minor modifications. Briefly, colorspace data of raw 50 bp reads was aligned to the mouse genome (mm9 or GRCm37) using Bowtie (Langmead et al., 2009). Gene expression levels were analyzed with Cufflinks, with differentially expressed genes determined by Cuffdiff (Trapnell et al., 2010). Full raw and processed data have been deposit to NCBI with GEO accession number GSE86187. Coverage plots were generated using BEDTools (http://code.google.com/p/bedtools/) with BEDGraph and BigWig files, which were then visualized in the UCSC Genome Browser (http://genome.ucsc.edu).
Ingenuity Pathway Analysis
Top2b-dependent genes (TDGs) (fold change ≥ 1.5, p-value ≤ 0.05) were determined using Cuffdiff (Trapnell et al. 2012) and further analyzed using Ingenuity Pathway Analysis tools (www.qiagen.com/ingenuity). Significance of the three output panels (i.e., Biological process, Pathway, Disease) was calculated using right tailed Fisher’s Exact Test.
Analysis of Top2b-dependent genes with Gene Ontology database and disease databases
TDGs related to neuronal cell death, neurite outgrowth and visual system development were analyzed with open-access databases including the Gene Ontology (http://www.geneontology.org); multiple retinal disease databases (RetNet, https://sph.uth.tmc.edu/Retnet/; Disease Atlas, NextBio, http://www.nextbio.com); and mitochondrial disease related gene lists (Yu-Wai-Man et al. 2011) for their functions and disease-related features.
RESULTS
Top2b deficiency causes developmental defects in photoreceptor outer segment formation
In our previous study, we showed that Top2b conditional knockout (cKO; Top2bf/f;DKK3-Cre) in early retinal progenitor cells leads to defects in differentiation and survival of all retinal cell types (Li et al. 2014). In this study, we attempt to define the molecular mechanism of Top2b function using the photoreceptor cell as a model. First, retinal samples from the cKO (Top2b−/−) and control (Top2b+/+ and Top2b+/−) mice at various stages were analyzed by immunostaining with rod marker Rhodopsin (Fig. 1A), s-cone marker Opn1sw (Fig. 1B) and m/l-cone marker Opn1mw/lw (Fig. 1C). Rhodopsin expression was detected only in a few cells in the outer nuclear layer (ONL) of both the control and cKO samples starting at E19.5, and became extensive at P7 and P14 (Fig. 1A). At P7 and P14, noticeably thinner ONLs were observed in the cKO retina (Fig. 1A, D); and there was no or little Rhodopsin accumulation near the outer limiting membrane (OLM) (purple arrows in Fig. 1A). By P14, Rhodopsin signal was not detected or significantly reduced in the outer segment (OS) with strong signal in the cell bodies (Fig. 1A, E). In addition, Opn1sw labeled s-cones showed dramatic degeneration at P0, P7, and P14 (Fig. 1B). Opn1sw+ cells were observed with shorter processes at P0 (arrowheads in Fig. 1B). No neural process was detected at P7 and P14 (Fig. 1B). The OS was only observed in control samples (purple arrows in Fig. 1B) but not in cKO samples. By P14, the vast majority of Opn1sw+ s-cones were degenerated in the cKO retinae (Fig. 1B, F). Moreover, examination of the m/l-cones by anti-Opn1mw/lw antibody staining showed that in the cKO opsin was accumulated in small regions but no OS can be identified (purple arrows in Fig. 1C). Strong signal of Opn1mw/lw was found in cytoplasm (yellow arrows in Fig. 1C). It is also noted that the Rhodopsin and Opn1mw/lw labeled ONL cells intruded into the inner nuclear layer (INL) in P14 cKO samples (Fig. 1A, C).
Figure 1. Top2b deletion leads to malformation of outer segment structure in both rod and cone cells.
OS structure in retinal sections were examined by immunostaining with rod marker Rhodopsin (A), s-cone marker Opn1sw (B), m/l-cone marker Opn1mw/lw (C), and helium ion microscopy (HIM) (G). In Top2b cKO, there was a significant reduction of OS thickness in all photoreceptor cells. OS structure was mostly missing or barely identifiable in HIM. Purple arrows indicate the OS, red arrowheads indicate the processes of photoreceptor cells, and the yellow arrows indicate marker expression inside cell bodies. Images in boxed areas are shown in a higher magnification. The thickness of the ONL and OSL of rod cells were measured as indicated by the bar lines and plotted (D and E). The number of s-cone cells was counted and plotted (F). OLM, outer limiting membrane; ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer; OS, outer segment; OSL, outer segment layer; RPE, retinal pigment epithelium. Scale bars A-C: 50μm/10μm (enlarged); scale bar in G = 5μm. Data in plots are represented as median ± max/min (n ≥ 3). p-values were determined by Student’s T-test.
To further determine the defects in the OS at a micrometer ultrastructural level, we examined retinal sections using a helium ion microscopy (HIM, Zeiss, Germany). In both P14 and P21 control retinae, the OS of both rod and cone cells were found between the retinal pigment epithelial cells (RPE) layer and the ONL (arrows in Fig. 1G). In contrast, there was no distinguishable OS structure in cKO retinae at these two stages (Fig. 1G). In addition, the retina in cKO samples was largely detached from the RPE.
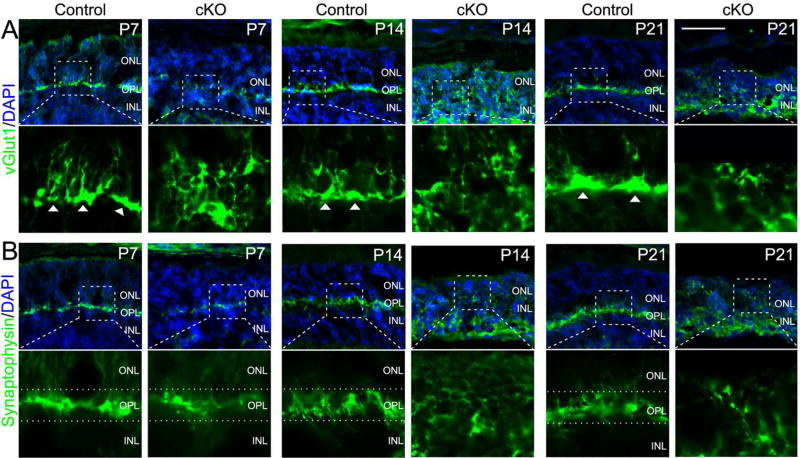
Aberrant synapse formation in Top2b cKO retinae
Although we reported missing outer plexiform layer (OPL) in our previous study (Li et al. 2014), it was not clear if it was due to the absence of horizontal cells or interruption of synaptic connections between photoreceptor cells and bipolar cells. The observed phenotypes in the OS lead us to speculate that axonal/dendritic growth, and synapse formation of photoreceptor cells may be affected in the cKO mouse retinae. Therefore, retinal sections of the control and cKO animals were stained with antibodies against vGlut1 (a vesicular glutamate transporter) (Fig. 2A) and Synaptophysin (a synaptic vesicle protein) (Fig. 2B). vGlut1 is required for synaptic vesicles with glutamate at the synaptic terminals (Johnson et al. 2003) and is usually found in the pre-synapses (Sherry et al. 2003). Expression of vGlut1 in control retinae suggested the extension of photoreceptor cell synapse into OPL at P7 and formation of bulb-shaped structure at P14 and P21 (arrowheads in Fig. 2A). In cKO retinae, however, vGlut1 was expanded in the cytoplasm and no bulb-shaped structure can be found (Fig. 2A). In addition, Synaptophysin is expressed in all vesicular synaptic terminals in both the outer plexiform layer (OPL) and inner plexiform layer (IPL) of the retina (Brandstatter et al. 1996; Spiwoks-Becker et al. 2001). Synatophysin signals were prominent in the synapses within the OPL (dotted lines in Fig. 2B) at P7, 14 and 21. In the cKO, Synaptophysin signals at P14 and P21 became more prominent in the cytoplasm of the ONL cells and the OPL structure is disrupted (Fig. 2B).
Figure 2. Defective synapse formation in Top2b cKO photoreceptor cells.
Retinal sections from the control and Top2b cKO mice were stained with the synaptic vesicular glutamate transporter marker vGlut1 (A) and synaptic vesicle marker Synaptophysin (B). Arrowheads in (A) indicate the synaptic ends of photoreceptor cells. Dotted lines in (B) indicate the OPL. Boxed areas are shown in a higher magnification. ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer. Scale bar = 50μm.
Increased photoreceptor cell death in the ONL of Top2b cKO
Defects in axonal/dendritic growth and the formation of synapse and OS usually lead to photoreceptor degeneration (Swaroop et al. 2010). Thus, we analyzed photoreceptor cell death by immunostaining with apoptosis marker activated caspase 3 (Casp3; Fig. 3A). Compared with the control, a significantly increased number of Casp3+ cells were observed in the ONL of cKO retinae at P7, P14 and P21 (Fig. 3B).
Figure 3. Increased cell death in Top2b cKO photoreceptor cells.
Retinal sections from the control and Top2b cKO mice were stained with cell death marker activated Casp3. Arrowheads indicate the Casp3+ cells in the ONL (A). The number of Casp3+ cells in the ONL was counted and plotted in (B). ONL, outer nuclear layer; INL, inner nuclear layer. Scale bar = 50μm. Data in plots are represented as median ± max/min (n ≥ 3). p-values were determined by Student’s T-test.
Cell-autonomous role of Top2b in rod OS formation by mosaic cell analysis
Since the cKO animals showed defects in almost all retina cell types (Li et al. 2014), it is unclear whether the observed defects in photoreceptors were due to cell-autonomous or non-cell-autonomous effects. To address this issue, we performed a mosaic analysis by targeted Top2b knockout in postnatal rod photoreceptor cells (rods) by injection and electroporation of plasmid construct CAG-Cre-GFP into the eye in P0 Top2bf/f animals (Lyu and Wang 2003). Animals were harvested and analyzed at P7. We found that almost all of GFP+ cells (Top2b knockout) were rods and located in the INL, while the majority of DsRed+ rods (control) were in the ONL (Fig. S1), a phenotype of delayed cell differentiation. In addition, no significant cell death was detected in GFP+ cells (Fig. S1C). These results support a cell-autonomous role of Top2b in rod cell differentiation. To further confirm this cell-autonomous function of Top2b, shRNA-mediated gene silencing analysis was performed. Plasmid DNA shTop2b-GFP was injected and electroporated into P0 retina of the wildtype mice. Transfected retinae were examined at P14 and P21 after P0 electroporation (Fig. 4A). Transfection led to strong reporter GFP expression in rods in the ONL (Fig. 4B). The number of GFP+ cells was counted and the percentage of GFP+ cells in the total number of ONL cells was determined (Fig. 4C). The efficiency of Top2b knockdown was evaluated by immunostaining analysis with anti-Top2b antibody, which showed a reduction or elimination of Top2b expression in the transfected cells (arrows in Fig. 4B). Over 97% of GFP+ cells at P7 and almost 100% at both P14 and P21 were negative for Top2b staining (Fig. 4C), indicating a successful gene knockdown. Comparing shTop2b knockdown samples with the controls (scrambled shRNA), there was a significant reduction of OS length (Fig. 4F, bars and chart) and disorganized OS (Fig. 4F, white arrows) in transfected GFP+ cells at P21. In contrast to the dramatic phenotypes observed in cKO animals, there was no significant difference of ONL thickness, ONL cell number (Fig. 4E), or synapses (yellow arrows, Fig. 4F), indicating Top2b knockdown did not cause a dramatic cell loss. Thus, these results further support a cell-autonomous role of Top2b in rod cell differentiation.
Figure 4. Targeted Top2b knock-down in photoreceptors reduces OS thickness.
DNA mixture containing shTop2b-GFP or scrambled shRNA-GFP constructs was injected and electroporated into sub-retinal space of P0 mouse to transfect photoreceptor progenitors. (A) Schematic shows the timeline of electroporation and sample harvest and analysis at P7, P14 and P21. The resulting transfected rod photoreceptors were stained with anti-Top2b antibody (B). Arrows indicate transfected cells that lack of Top2b staining. (C) Plots of the number of GFP+ cells and percentage of GFP+ cells in the ONL per 100μm region. (D) Plot of the percentage of shTop2b-GFP+/Top2b− cells. (E) Plots of the number of cells and thickness of the ONL in the transfected regions. (F) Reduced thickness in the OSL/ISL of shTop2b transfected retinae at P21. White arrows indicate transfected OS/IS (GFP+). The length of OS was measured as indicated by the vertical bars. Synapses of the transfected cells are similar in the control or shTop2b transfected samples (yellow arrows). Boxed areas are shown in a higher magnification on the side or in the bottom right corner of each image. IS, inner segment; ISL, inner segment layer; OS, outer segment; OSL, outer segment layer; ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. Scale bars = 50μm. Data in plots are represented as median ± max/min (n≥3). p-values were determined by Student’s T-test.
Top2b deficiency affects the expression of key genes in a transcriptional network for photoreceptor differentiation and maintenance
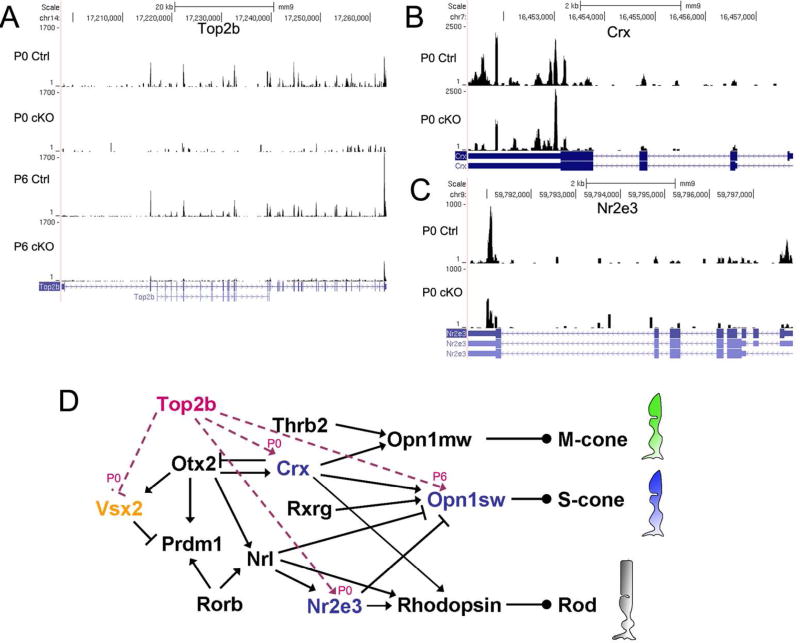
Top2b is known to regulate gene transcription (Wang 2002). To determine genes controlled by Top2b, we performed transcriptome analysis using RNA-seq data from control and cKO retinae at P0 and P6. The two time points, i.e., P0 and P6, were selected because: 1) P0 is at the peak of rod photoreceptor genesis and cone maturation; 2) P6 is at the middle-stage of rod differentiation; and 3) rods comprise the vast majority (94%) of the photoreceptor cell population (Brzezinski and Reh 2015; Young 1985a). Thus sequencing result for these two stages largely represent the genes required for photoreceptor cell differentiation and maturation. Four libraries were constructed: P0 control, P0 cKO, P6 control, and P6 cKO (Table S1). Differentially expressed genes contrasting the control and Top2b cKO samples were determined using Cufflinks and Cuffdiff (Trapnell et al. 2012; Trapnell et al. 2010). We identified 699 genes at P0 and 180 genes at P6 (Fig. 5A, B; Table S2, 3) that were significantly changed in expression level (|fold change| ≥ 1.5 (|log2(fold change)| ≥ 0.585), p-value ≤ 0.05), including Top2b itself (Fig. 6A). These differentially expressed genes are thus referred to as the Top2b-dependent genes (TDG). No significant change was detected in the expression of 20 known housekeeping genes (Table S4), confirming that the sequencing and calculations of transcript abundances was of good quality.
Figure 5. RNA-seq analysis identifies Top2b-dependent genes.
RNA-seq analysis identified 699 Top2b-dependent genes (TDGs) at P0 (A) and 180 TDGs at P6 (B) plotted against the log2(fold change) of their expression levels between WT and cKO. Up/down-regulated TDGs are represented by orange/blue dots and their names are labeled in green/pink, respectively. TDGs are also categorized according to their gene ontology (GO) and shown in the pie diagrams.
Figure 6. Top2b regulates key genes in photoreceptor transcriptional network.
Several key genes in the transcriptional network for photoreceptor development were recognized as Top2b-dependent genes (TDGs) by RNA-seq analysis. Coverage plots of Top2b (A), Crx (B), and Nr2e3 (C) show differential expression between control and Top2b cKO samples. The number of sequence reads mapped to their genes is plotted on their genomic locations. The maps of alternative spliced transcripts of these genes are also shown underneath the plot. (D) a schematic drawing of photoreceptor transcriptional network with Top2b as key modulator. Names of the down-regulated genes are shown in blue; up-regulated gene Vsx2 is in orange. Magenta lines indicate the effect of Top2b on those key genes.
To explore the role of Top2b in photoreceptor development, TDGs were analyzed by Ingenuity Pathway Analysis (IPA, www.ingenuity.com, Redwood City, CA, USA), a web-based tool for statistical analysis and biological interpretation of gene expression, in parallel with analysis via literature review and functional classification from the Gene Ontology Consortium. Top2b dependent interactions, pathways, and disease related gene pools were obtained to predict its regulatory role. Interestingly, the analysis reveals six TDGs, i.e., Crx, Nr2e3, Vsx2, Opn1sw, Glo1 and Smim13 (Table 2) that are critical for rod and cone photoreceptor cell fate determination, differentiation and maturation (Cepko 2015; Furukawa et al. 1997; Hennig et al. 2008; Swaroop et al. 2010). The altered expression level of Crx and Nr2e3 was shown in coverage plots (Fig. 6B–C). It is likely that Top2b modulates photoreceptor cell differentiation by regulating the transcription of these pivotal genes in the regulatory network (Fig. 6D).
Table 2.
List of Top2b dependent genes critical for photoreceptor cell development identified by RNA-seq.
| Gene symbol |
WT FPKM | cKO FPKM | Log2(fold- change) |
p_value | |
|---|---|---|---|---|---|
| P0 | Crx | 61.41 | 26.66 | −1.21 | 3.26E-06 |
| Nr2e3 | 21.04 | 5.95 | −1.82 | 2.57E-06 | |
| Glo1 | 80.36 | 34.34 | −1.23 | 1.84E-06 | |
| Smim13 | 8.56 | 3.66 | −1.22 | 0.00187 | |
| Vsx2 | 11.49 | 26.76 | 1.22 | 4.86E-04 | |
| P6 | Opn1sw | 29.47 | 11.45 | −1.36 | 3.16E-12 |
| Glo1 | 194.61 | 107.153 | −0.86 | 3.27E-07 |
FPKM: Fragments Per Kilobase of transcript per Million mapped reads.
Top2b associated genes are linked with photoreceptor-related retinopathy
The degeneration of photoreceptor cells causes many retinal diseases that ultimately lead to blindness (Clarke et al. 2000). To determine the association of Top2b with these retinal diseases, TDGs were searched against the databases of genes related to retinopathy (RetNet; NextBio Disease Atlas; (Montana and Corbo 2008)). Arl6, Atxn7, Bbs7, Crx, Dmd, Nr2e3, Pde6b, Pxmp3, Rbp4, Trpm1, Abca4, Iqcb1, Opn1sw and Pgk1 were found to be linked to various retinal diseases, e.g., retinitis pigmentosa, S-cone syndrome, congenital stationary night blindness and syndromes including retinopathies such as Bardet-Biedl syndrome (Table 3).
Table 3.
Differential expression of retinopathy related genes in Top2b cKO retinae.
| Stage | gene | log2 (fold change) |
p_value | q_value | disease | reference |
|---|---|---|---|---|---|---|
| P0 | Arl6 | −2 | 0.00161861 | 0.030496 | recessive Bardet-Biedl syndrome |
(Chiang et al. 2004; Fan et al. 2004) |
| Atxn7 | −2 | 0.00299047 | 0.048919 | dominant spinocerebellar ataxia w/ macular dystrophy or retinal degeneration |
(Aleman et al. 2002) |
|
| Bbs7 | −4.1 | 0.000536956 | 0.012973 | recessive Bardet Biedl syndrome |
(Badano et al. 2003) |
|
| Crx | −1.2 | 3.25587E-06 | 0.000281 | dominant cone-rod dystrophy; recessive, dominant and de novo Leber congenital amaurosis; dominant retinitis pigmentosa. |
(Furukawa et al. 1999; Hanein et al. 2004; Menotti- Raymond et al. 2010) |
|
| Dmd | −1.5 | 0.000626471 | 0.014728 | oregon eye disease (probably) |
(D'Souza et al. 1995) |
|
| Nr2e3 | −1.8 | 2.56724E-06 | 0.000241 | recessive enhanced S- cone syndrome; recessive retinitis pigmentosa in Portuguese Crypto Jews; Goldmann-Favre syndrome; dominant retinitis pigmentosa; combined dominant and recessive retinopathy |
(Coppieters et al. 2007; Escher et al. 2009; Sharon et al. 2003) |
|
| Pxmp3 | −1.4 | 0.000157789 | 0.005179 | recessive Refsum disease, infantile form |
(Gartner et al. 1992) |
|
| Trpm1 | −4.3 | 8.81098E-06 | 0.000589 | recessive congenital stationary night blindness, complete |
(Audo et al. 2009) | |
| Pde6b | 1.93 | 0.00273468 | 0.045837 | recessive retinitis pigmentosa; dominant congenital stationary night blindness |
(Riess et al. 1992) | |
| Rbp4 | 5.5 | 0.000100318 | 0.003672 | recessive RPE degeneration |
(Seeliger et al. 1999) |
|
|
P6 |
Iqcb1 | <−1024 | 3.03261E-11 | 2.15792E-08 | recessive Senior-Loken syndrome; recessive Leber congenital amaurosis |
(Estrada-Cuzcano et al. 2011; Otto et al. 2005) |
| Opn1sw | −1.4 | 3.15836E-12 | 2.5511E-09 | dominant tritanopia | (Weitz et al. 1992) | |
| Pgk1 | −1.1 | 2.70792E-08 | 1.09363E-05 | retinitis pigmentosa with myopathy |
(Tonin et al. 1993) | |
| Abca4 | 1.71 | 1.77636E-15 | 1.90E-12 | recessive Stargardt disease, juvenile and late onset; recessive macular dystrophy; recessive retinitis pigmentosa; recessive fundus flavimaculatus; recessive cone-rod dystrophy; |
(Gerber et al. 1995; Maugeri et al. 2000; Molday et al. 2000) |
Genes involve in photoreceptor cell-related retinal diseases are shown in bold.
DISCUSSION
Photoreceptors constitute the majority of the retinal cell population, and their development expands from embryonic to neonatal stages (Brzezinski and Reh 2015; Rapaport et al. 2004; Wang and Cepko 2016; Young 1985a; Young 1985b). In this study, we report a combinatorial cell-autonomous and non-cell-autonomous role of Top2b during late-stage photoreceptor differentiation and maturation.
Top2b functions in the late-stage differentiation and maintenance of photoreceptor cells
Dkk3-Cre mediated cKO resulted in Top2b deletion in retinal progenitor cells and all the progeny in all retinal cell types (Li et al. 2014), making it a valuable approach to study Top2b function in the development of all retinal cell lineages including photoreceptors. In Top2b cKOs, both cones and rods were generated but failed to fully differentiate. Deficits exist in: i) the OS maturation and maintenance (Fig. 1); ii) accumulated Opsin in the cell body (Fig. 1A–C); iii) synapse formation (Fig. 2); and iv) cell degeneration and cell death in both rods and cones (Fig. 3). The defects in the OS with traditional fluorescence microscopy were confirmed by helium ion microscopy (HIM). HIM provides a new approach to investigate nanometer-scale surface features of biological samples (Joens et al. 2013). With this technique, detailed differences on surface ultrastructure of photoreceptors between the control and cKO mice were revealed clearly (Fig. 1G). Similar defects were also observed in late-stage development in the animals lacking Crx (Furukawa et al. 1999), Nrl (Daniele et al. 2005), or Vsx2 (Rutherford et al. 2004), except that in these animals there was an increase in the number of cone cells. In addition, the lack of OS and the degeneration of photoreceptor cells are usually observed in the opsin-deficient animals (Daniele et al. 2011; Lem et al. 1999). It is believed that mis-localized opsin is frequently associated with retinal degeneration (den Hollander et al. 2008; Deretic 2006) and has been indicated as a major cause of photoreceptor cell death in the absence of heterotrimeric kinesin-2 function (Louie et al. 2010). The enriched Rhodopsin and Opn1sw/mw/lw in photoreceptor cell bodies of cKO retinae (Fig. 1A–C) may explain the dramatic cell loss found in the ONL (Fig. 3). Since cone cells represent 3–5% of all photoreceptor cells, the dramatic cell loss observed in the ONL is most likely due to the rod cell phenotype.
Cell-autonomous and non-cell-autonomous role of Top2b in photoreceptor differentiation
Dkk3-Cre mediated Top2b cKO provides valuable insight into retinal development. However, it has limitations for addressing cell-autonomous vs. non-cell-autonomous and primary vs. secondary effects of Top2b on photoreceptor differentiation. Thus, we performed mosaic analysis using shRNA-mediated Top2b knockdown in postnatal developing photoreceptor cells. Shorter OS was observed in the sample transfected with shTop2b (Fig. 4F) without dramatic cell loss (Fig. 4D), suggesting that a cell-autonomous function of Top2b is primarily affecting OS maturation and maintanence. Moreover, in contrast to a dramatic synapse degeneration (Fig. 2), cell degeneration and cell death (Fig. 3) observed in the cKO, Top2b knockdown did not cause a noticeable difference in the thickness of the ONL or morphology of synapses (Fig. 4E, F). Cell-autonomous effects alone cannot account for the severe phenotypes observed in the retinae of cKO mice. Therefore, the multi-level defects in photoreceptor differentiation in cKO animals are likely due to both cell-autonomous (primary) and non-cell-autonomous (secondary) effects of Top2b. As photoreceptors directly form synaptic connections with the horizontal and bipolar cells, alterations in these two cell types usually lead to defects in photoreceptor cells. A study has shown that the loss of horizontal cells leads to partial rod photoreceptor cell degeneration, retraction of axons from the OPL and partial cell death (Sonntag et al. 2012). Since defects of the horizontal cells but not the bipolar cells were observed in Top2b cKO animals (Li et al. 2014), it is likely that horizontal cell degeneration exacerbates the photoreceptor phenotype. However, in the same study (Sonntag et al. 2012) the s-cones remain largely unchanged, which is different from our observation of a dramatic s-cone cell loss (Fig. 1B). It will be intriging to find out what is responsible for the s-cone dystrophy.
Surprisingly, synapse formation was not affected in the mosaic retinae with shTop2b knockdown (Fig. 4F), suggesting that Top2b may not play a cell-autonomous role in this process. Top2b is known to be important for axon guidance and neurite growth of motor neurons (Yang et al. 2000). We also found reduced optic nerve and disrupted IPL, OPL in cKO retina (Li et al. 2014). These observations suggest a possibility that Top2b plays a different role among various types of neurons.
Top2b controls key genes in photoreceptor gene-regulatory network
Rods constitute the majority of the cells in the mouse retina (Brzezinski and Reh 2015; Carter-Dawson and LaVail 1979; Cepko 2015). TDGs, e.g., Vsx2, Crx, Nr2e3, Opn1sw, Glo1 and Smim13, identified by RNA-seq analysis (Figs 5, 6) are key genes in regulatory network that dictates photoreceptor differentiation and homeostasis. Thus, Top2b functions as a key modulator at multiple levels in this hierarchical transcriptional regulatory network (Fig. 6E). Among the six photorecepor-related TDGs, Vsx2 mutation leads to delayed Crx expression and rod development (Rutherford et al. 2004). Crx and Nr2e3 are known to be essential genes in controlling the rod/cone cell fate determination (Peng et al. 2005; Peng and Chen 2005) and development of the OS (Furukawa et al. 1997). These studies are consistent with the observed deficits in photoreceptor cells in Top2b cKO animals, except that mutation of Nr2e3 can led to excess s-cones (Peng et al. 2005). It is possible that Top2b deletion affects Crx and Nr2e3 expression and consequently affects Opn1sw and Rhodopsin expression at a later stage during photoreceptor differentiation. Alternatively, Top2b may directly regulate Opn1sw expression by binding to a distant regulatory region ~200kb upstream Opn1sw transcription starting site (data from GSM1516578 in (Madabhushi et al. 2015)). As Opn1sw encodes the blue cone pigment (Nathans et al. 1986), the reduction in Opn1sw expression may also be a contributor to the dramatic loss of the OS and the number of s-cones in the cKO.
Several TDGs have been known to function in neurite growth and retinal cell maintenance including Igf1, Creb1, Hras, Mark1, Grin1, Cdkn1a, Serpine2, Ptprz1, Gnao1, Drd2, Pdia3 and Dpysl3. Although neurite degeneration and neural cell death can be separate events (Ikegami and Koike 2003), our observation favors the idea that the impaired neurite outgrowth accelerates retinal cell death.
Finally, mice lacking Top2b present a transcriptome profile that is prone to several photoreceptor-related retinal diseases, e.g., retinitis pigmentosa, S-cone syndrome, and Bardet-Biedl syndrome (Table 3). The identification of retinopathy-related TDGs suggests that Top2b function is required for not only normal development but also the prevention of retinal diseases.
Supplementary Material
Significance Statement.
Photoreceptors sense light signals with their outer segments and transduce the signals to other cells in the neural retina, enabling animals to see. Defects in photoreceptors cause their own death, eventually leading to severe eye diseases and often blindness. Extensive studies have revealed that many genes are involved in the differentiation of photoreceptors from retinal progenitor cells. In this study, we show that Topoisomerase II beta (Top2b) contributes to the late-stage differentiation/maturation of photoreceptors, especially in the formation of outer segments and synapses by affecting the expression of key genes in the photoreceptor transcriptional network and genes linked to retinopathies.
Acknowledgments
We thank Dr. Robert S. Molday for a monoclonal antibody to Rhosopsin 4D2, Dr. Gabriella D'Arcangelo for an antibody to Synaptophysin, and Drs. Torgny Gustafsson and Slava Manichev for helium ion microscope imaging. We also thank the members in Cai lab for proof reading the manuscript and providing helpful suggestions.
Support: This work was supported in part by the grants from the National Institute of Health [EY018738 to L.C., R21NS0855691 to K. -B. L.]; the New Jersey Commission on Spinal Cord Research [12FEL001 to Y.L.]; and the University City Science Center’s QED Award [K.-B. L.].
Footnotes
CONFLICT OF INTERESTS
The authors declare no conflict of interests in this work.
AUTHORS’ CONTRIBUTIONS
Conceptualization and Methodology, L. C. and Y. L.; Investigation, Y. L. and Y. L. L.; Animal resource, Y. L. L and L. C.; shRNA-mediated gene knockdown study, H. H. and Y. L.; RNA library construction M. R. S. and R. P. H.; RNA-seq analysis, R. P. H., L. C. and Y. L.; HIM imaging, Y. L., H. Y. C. and K. -B. L.; Writing – Original Draft, Y. L. and L. C.; Writing – Editing, L. C., Y. L., R. P. H., and K. -B. L.
Data Accessibility
RNA-seq data reported in this study have been deposited at NCBI; the GEO accession number is GSE86187.
REFERENCES
- Azarova AM, Lin RK, Tsai YC, Liu LF, Lin CP, Lyu YL. Genistein induces topoisomerase IIbeta- and proteasome-mediated DNA sequence rearrangements: Implications in infant leukemia. Biochem Biophys Res Commun. 2010;399(1):66–71. doi: 10.1016/j.bbrc.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandstatter JH, Lohrke S, Morgans CW, Wassle H. Distributions of two homologous synaptic vesicle proteins, synaptoporin and synaptophysin, in the mammalian retina. J Comp Neurol. 1996;370(1):1–10. doi: 10.1002/(SICI)1096-9861(19960617)370:1<1::AID-CNE1>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Brzezinski JA, Reh TA. Photoreceptor cell fate specification in vertebrates. Development. 2015;142(19):3263–3273. doi: 10.1242/dev.127043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter-Dawson LD, LaVail MM. Rods and cones in the mouse retina. I. Structural analysis using light and electron microscopy. J Comp Neurol. 1979;188(2):245–262. doi: 10.1002/cne.901880204. [DOI] [PubMed] [Google Scholar]
- Cepko CL. The Determination of Rod and Cone Photoreceptor Fate. Annual Review of Vision Science. 2015;1(1):211–234. doi: 10.1146/annurev-vision-090814-121657. [DOI] [PubMed] [Google Scholar]
- Chen S, Wang QL, Nie Z, Sun H, Lennon G, Copeland NG, Gilbert DJ, Jenkins NA, Zack DJ. Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron. 1997;19(5):1017–1030. doi: 10.1016/s0896-6273(00)80394-3. [DOI] [PubMed] [Google Scholar]
- Clarke G, Heon E, McInnes RR. Recent advances in the molecular basis of inherited photoreceptor degeneration. Clin Genet. 2000;57(5):313–329. doi: 10.1034/j.1399-0004.2000.570501.x. [DOI] [PubMed] [Google Scholar]
- Coppieters F, Leroy BP, Beysen D, Hellemans J, De Bosscher K, Haegeman G, Robberecht K, Wuyts W, Coucke PJ, De Baere E. Recurrent mutation in the first zinc finger of the orphan nuclear receptor NR2E3 causes autosomal dominant retinitis pigmentosa. Am J Hum Genet. 2007;81(1):147–157. doi: 10.1086/518426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniele LL, Insinna C, Chance R, Wang J, Nikonov SS, Pugh EN., Jr A mouse M-opsin monochromat: retinal cone photoreceptors have increased M-opsin expression when S-opsin is knocked out. Vision Res. 2011;51(4):447–458. doi: 10.1016/j.visres.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniele LL, Lillo C, Lyubarsky AL, Nikonov SS, Philp N, Mears AJ, Swaroop A, Williams DS, Pugh EN., Jr Cone-like morphological, molecular, and electrophysiological features of the photoreceptors of the Nrl knockout mouse. Invest Ophthalmol Vis Sci. 2005;46(6):2156–2167. doi: 10.1167/iovs.04-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hollander AI, Roepman R, Koenekoop RK, Cremers FP. Leber congenital amaurosis: genes, proteins and disease mechanisms. Prog Retin Eye Res. 2008;27(4):391–419. doi: 10.1016/j.preteyeres.2008.05.003. [DOI] [PubMed] [Google Scholar]
- Deretic D. A role for rhodopsin in a signal transduction cascade that regulates membrane trafficking and photoreceptor polarity. Vision Res. 2006;46(27):4427–4433. doi: 10.1016/j.visres.2006.07.028. [DOI] [PubMed] [Google Scholar]
- Freund CL, Gregory-Evans CY, Furukawa T, Papaioannou M, Looser J, Ploder L, Bellingham J, Ng D, Herbrick JA, Duncan A, Scherer SW, Tsui LC, Loutradis-Anagnostou A, Jacobson SG, Cepko CL, Bhattacharya SS, McInnes RR. Cone-rod dystrophy due to mutations in a novel photoreceptor-specific homeobox gene (CRX) essential for maintenance of the photoreceptor. Cell. 1997;91(4):543–553. doi: 10.1016/s0092-8674(00)80440-7. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Morrow EM, Cepko CL. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell. 1997;91(4):531–541. doi: 10.1016/s0092-8674(00)80439-0. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Morrow EM, Li T, Davis FC, Cepko CL. Retinopathy and attenuated circadian entrainment in Crx-deficient mice. Nature genetics. 1999;23(4):466–470. doi: 10.1038/70591. [DOI] [PubMed] [Google Scholar]
- Hennig AK, Peng GH, Chen S. Regulation of photoreceptor gene expression by Crx-associated transcription factor network. Brain research. 2008;1192:114–133. doi: 10.1016/j.brainres.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami K, Koike T. Non-apoptotic neurite degeneration in apoptotic neuronal death: pivotal role of mitochondrial function in neurites. Neuroscience. 2003;122(3):617–626. doi: 10.1016/j.neuroscience.2003.08.057. [DOI] [PubMed] [Google Scholar]
- Joens MS, Huynh C, Kasuboski JM, Ferranti D, Sigal YJ, Zeitvogel F, Obst M, Burkhardt CJ, Curran KP, Chalasani SH, Stern LA, Goetze B, Fitzpatrick JA. Helium Ion Microscopy (HIM) for the imaging of biological samples at sub-nanometer resolution. Sci Rep. 2013;3:3514. doi: 10.1038/srep03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J, Tian N, Caywood MS, Reimer RJ, Edwards RH, Copenhagen DR. Vesicular neurotransmitter transporter expression in developing postnatal rodent retina: GABA and glycine precede glutamate. J Neurosci. 2003;23(2):518–529. doi: 10.1523/JNEUROSCI.23-02-00518.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lem J, Krasnoperova NV, Calvert PD, Kosaras B, Cameron DA, Nicolo M, Makino CL, Sidman RL. Morphological, physiological, and biochemical changes in rhodopsin knockout mice. Proc Natl Acad Sci U S A. 1999;96(2):736–741. doi: 10.1073/pnas.96.2.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Hao H, Tzatzalos E, Lin RK, Doh S, Liu LF, Lyu YL, Cai L. Topoisomerase IIbeta is required for proper retinal development and survival of postmitotic cells. Biol Open. 2014;3(2):172–184. doi: 10.1242/bio.20146767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie CM, Caridi G, Lopes VS, Brancati F, Kispert A, Lancaster MA, Schlossman AM, Otto EA, Leitges M, Grone HJ, Lopez I, Gudiseva HV, O'Toole JF, Vallespin E, Ayyagari R, Ayuso C, Cremers FP, den Hollander AI, Koenekoop RK, Dallapiccola B, Ghiggeri GM, Hildebrandt F, Valente EM, Williams DS, Gleeson JG. AHI1 is required for photoreceptor outer segment development and is a modifier for retinal degeneration in nephronophthisis. Nat Genet. 2010;42(2):175–180. doi: 10.1038/ng.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu YL, Lin CP, Azarova AM, Cai L, Wang JC, Liu LF. Role of topoisomerase IIbeta in the expression of developmentally regulated genes. Mol Cell Biol. 2006;26(21):7929–7941. doi: 10.1128/MCB.00617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu YL, Wang JC. Aberrant lamination in the cerebral cortex of mouse embryos lacking DNA topoisomerase IIbeta. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(12):7123–7128. doi: 10.1073/pnas.1232376100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161(5):1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madabhushi R, Gao F, Pfenning AR, Pan L, Yamakawa S, Seo J, Rueda R, Phan TX, Yamakawa H, Pao PC, Stott RT, Gjoneska E, Nott A, Cho S, Kellis M, Tsai LH. Activity-Induced DNA Breaks Govern the Expression of Neuronal Early-Response Genes. Cell. 2015;161(7):1592–1605. doi: 10.1016/j.cell.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Cepko CL. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc Natl Acad Sci U S A. 2004;101(1):16–22. doi: 10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montana CL, Corbo JC. Inherited diseases of photoreceptors and prospects for gene therapy. Pharmacogenomics. 2008;9(3):335–347. doi: 10.2217/14622416.9.3.335. [DOI] [PubMed] [Google Scholar]
- Nathans J, Thomas D, Hogness DS. Molecular genetics of human color vision: the genes encoding blue, green, and red pigments. Science. 1986;232(4747):193–202. doi: 10.1126/science.2937147. [DOI] [PubMed] [Google Scholar]
- Nevin LM, Xiao T, Staub W, Baier H. Topoisomerase II{beta} is required for lamina-specific targeting of retinal ganglion cell axons and dendrites. Development. 2011;138(12):2457–2465. doi: 10.1242/dev.060335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson SE. Receptor Cell Outer Segment Development and Ultrastructure of the Disk Membranes in the Retina of the Tadpole (Rana Pipiens) J Ultrastruct Res. 1964;11:581–602. doi: 10.1016/s0022-5320(64)80084-8. [DOI] [PubMed] [Google Scholar]
- Peng GH, Ahmad O, Ahmad F, Liu J, Chen S. The photoreceptor-specific nuclear receptor Nr2e3 interacts with Crx and exerts opposing effects on the transcription of rod versus cone genes. Hum Mol Genet. 2005;14(6):747–764. doi: 10.1093/hmg/ddi070. [DOI] [PubMed] [Google Scholar]
- Peng GH, Chen S. Chromatin immunoprecipitation identifies photoreceptor transcription factor targets in mouse models of retinal degeneration: new findings and challenges. Vis Neurosci. 2005;22(5):575–586. doi: 10.1017/S0952523805225063. [DOI] [PubMed] [Google Scholar]
- Rapaport DH, Wong LL, Wood ED, Yasumura D, LaVail MM. Timing and topography of cell genesis in the rat retina. J Comp Neurol. 2004;474(2):304–324. doi: 10.1002/cne.20134. [DOI] [PubMed] [Google Scholar]
- Rutherford AD, Dhomen N, Smith HK, Sowden JC. Delayed expression of the Crx gene and photoreceptor development in the Chx10-deficient retina. Invest Ophthalmol Vis Sci. 2004;45(2):375–384. doi: 10.1167/iovs.03-0332. [DOI] [PubMed] [Google Scholar]
- Sato S, Inoue T, Terada K, Matsuo I, Aizawa S, Tano Y, Fujikado T, Furukawa T. Dkk3-Cre BAC transgenic mouse line: a tool for highly efficient gene deletion in retinal progenitor cells. genesis. 2007;45(8):502–507. doi: 10.1002/dvg.20318. [DOI] [PubMed] [Google Scholar]
- Sherry DM, Wang MM, Bates J, Frishman LJ. Expression of vesicular glutamate transporter 1 in the mouse retina reveals temporal ordering in development of rod vs. cone and ON vs. OFF circuits. J Comp Neurol. 2003;465(4):480–498. doi: 10.1002/cne.10838. [DOI] [PubMed] [Google Scholar]
- Sonntag S, Dedek K, Dorgau B, Schultz K, Schmidt KF, Cimiotti K, Weiler R, Lowel S, Willecke K, Janssen-Bienhold U. Ablation of retinal horizontal cells from adult mice leads to rod degeneration and remodeling in the outer retina. J Neurosci. 2012;32(31):10713–10724. doi: 10.1523/JNEUROSCI.0442-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiwoks-Becker I, Vollrath L, Seeliger MW, Jaissle G, Eshkind LG, Leube RE. Synaptic vesicle alterations in rod photoreceptors of synaptophysin-deficient mice. Neuroscience. 2001;107(1):127–142. doi: 10.1016/s0306-4522(01)00345-1. [DOI] [PubMed] [Google Scholar]
- Steinberg RH, Fisher SK, Anderson DH. Disc morphogenesis in vertebrate photoreceptors. J Comp Neurol. 1980;190(3):501–508. doi: 10.1002/cne.901900307. [DOI] [PubMed] [Google Scholar]
- Swain PK, Chen S, Wang QL, Affatigato LM, Coats CL, Brady KD, Fishman GA, Jacobson SG, Swaroop A, Stone E, Sieving PA, Zack DJ. Mutations in the cone-rod homeobox gene are associated with the cone-rod dystrophy photoreceptor degeneration. Neuron. 1997;19(6):1329–1336. doi: 10.1016/s0896-6273(00)80423-7. [DOI] [PubMed] [Google Scholar]
- Swaroop A, Kim D, Forrest D. Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nat Rev Neurosci. 2010;11(8):563–576. doi: 10.1038/nrn2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari VK, Burger L, Nikoletopoulou V, Deogracias R, Thakurela S, Wirbelauer C, Kaut J, Terranova R, Hoerner L, Mielke C, Boege F, Murr R, Peters AH, Barde YA, Schubeler D. Target genes of Topoisomerase IIbeta regulate neuronal survival and are defined by their chromatin state. Proc Natl Acad Sci U S A. 2012;109(16):E934–E943. doi: 10.1073/pnas.1119798109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuyasu K, Yamada E. The fine structure of the retina studied with the electron microscope. IV. Morphogenesis of outer segments of retinal rods. J Biophys Biochem Cytol. 1959;6:225–230. doi: 10.1083/jcb.6.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui K, Okada S, Watanabe M, Shohmori T, Seki S, Inoue Y. Molecular cloning of partial cDNAs for rat DNA topoisomerase II isoforms and their differential expression in brain development. Journal of Biological Chemistry. 1993;268(25):19076–19083. [PubMed] [Google Scholar]
- Tsutsui K, Sano K, Kikuchi A, Tokunaga A. Involvement of DNA topoisomerase IIbeta in neuronal differentiation. The Journal of biological chemistry. 2001;276(8):5769–5778. doi: 10.1074/jbc.M008517200. [DOI] [PubMed] [Google Scholar]
- Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3(6):430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- Wang S, Cepko CL. Photoreceptor Fate Determination in the Vertebrate Retina. Invest Ophthalmol Vis Sci. 2016;57(5):ORSFe1–ORSFe6. doi: 10.1167/iovs.15-17672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Sengel C, Emerson MM, Cepko CL. A gene regulatory network controls the binary fate decision of rod and bipolar cells in the vertebrate retina. Dev Cell. 2014;30(5):513–527. doi: 10.1016/j.devcel.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitz CJ, Miyake Y, Shinzato K, Montag E, Zrenner E, Went LN, Nathans J. Human tritanopia associated with two amino acid substitutions in the blue-sensitive opsin. Am J Hum Genet. 1992;50(3):498–507. [PMC free article] [PubMed] [Google Scholar]
- Yang X, Li W, Prescott ED, Burden SJ, Wang JC. DNA topoisomerase IIbeta and neural development. Science. 2000;287(5450):131–134. doi: 10.1126/science.287.5450.131. [DOI] [PubMed] [Google Scholar]
- Young RW. Cell differentiation in the retina of the mouse. Anat Rec. 1985a;212(2):199–205. doi: 10.1002/ar.1092120215. [DOI] [PubMed] [Google Scholar]
- Young RW. Cell proliferation during postnatal development of the retina in the mouse. Brain research. 1985b;353(2):229–239. doi: 10.1016/0165-3806(85)90211-1. [DOI] [PubMed] [Google Scholar]
- Yu-Wai-Man P, Griffiths PG, Chinnery PF. Mitochondrial optic neuropathies - disease mechanisms and therapeutic strategies. Progress in retinal and eye research. 2011;30(2):81–114. doi: 10.1016/j.preteyeres.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.