Abstract
After a century of incremental research, technological advances, coupled with a need for sustainable crop yield increases, have reinvigorated the study of beneficial plant-microbe interactions with attention focused on how microbiomes alter plant phenotypes. We review recent advances in plant microbiome research, and describe potential applications for increasing crop productivity. The phylogenetic diversity of plant microbiomes is increasingly well characterized, and their functional diversity is becoming more accessible. Large culture collections are available for controlled experimentation, with more to come. Genetic resources are being brought to bear on questions of microbiome function. We expect that microbial amendments of varying complexities will expose rules governing beneficial plant-microbe interactions contributing to plant growth promotion and disease resistance, enabling more sustainable agriculture.
Introduction
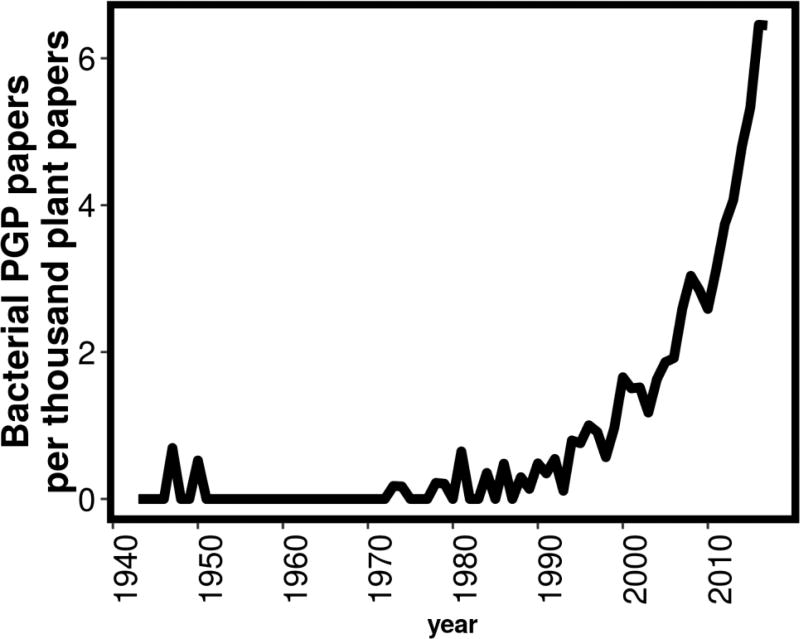
The manipulation of soil microbiomes to optimize crop productivity is an ancient practice; records can be traced to ~300 BC [1]. It is interesting to note that although soil microbiomes are now touted as a cornerstone of the next green revolution [2], the first commercial bioinoculant, “nitrogin”, was patented in 1896 [3], during the golden age of microbiology and preceding the Haber-Bosch process by 15 years. Currently, the Organic Materials Review Institute (OMRI) lists 174 products under the category of “microbial inoculants” and 274 products under the category “microbial products”, either as crop fertilizers or as crop management tools. The number of publications associated with plant growth promoting (PGP) microbes, has been growing exponentially since the ‘90s (Figure 1). Few, if any, of these are associated with mechanistic studies or modes of action; exceptions being biological nitrogen fixation by rhizobia on legumes [4], and auxin [5] or ACC-deaminase [6] –mediated phytostimulation. However, genetic limitations to function in planta and variable field effects have sharply limited their widespread deployment. We therefore need to forge a deeper understanding of (a) the mechanisms governing microbial invasion and persistence into standing heterogeneous communities in diverse locations, soils and hosts; and (b) the genetics, in both partners, that drives colonization and delivery of plant phenotypes by microbes. The advent of culture-independent microbial ecology, powered by development of high-throughput analytic technologies, has enabled increasingly systematic study of the plant-associated ecological context in which microbial inoculants could be applied; and of mechanisms of plant control over colonization by beneficial microbes. Novel approaches also offer opportunities to bridge current gaps between plant-productivity phenotypes and understanding of the underlying mechanisms [7–9].
Figure 1.
The number of articles about bacterial plant growth promotion per year per thousand plant-related papers, found in the PubMed database, using the search term (("plant development"[MeSH Terms] OR ("plant"[All Fields] AND "development"[All Fields]) OR "plant development"[All Fields] OR ("plant"[All Fields] AND "growth"[All Fields]) OR "plant growth"[All Fields]) AND promoting[All Fields] AND ("microbiology"[Subheading] OR "microbiology"[All Fields] OR "bacteria"[All Fields] OR "bacteria"[MeSH Terms])) OR (("plant development"[MeSH Terms] OR ("plant"[All Fields] AND "development"[All Fields]) OR "plant development"[All Fields] OR ("plant"[All Fields] AND "growth"[All Fields]) OR "plant growth"[All Fields]) AND promoting[All Fields] AND rhizobacteria[All Fields]).
Screening of large isolate collections
The limited taxonomy of plant-associated microbes, compared with the vast diversity of soil microorganisms [9–11], suggests that plants are a highly selective microbial niche and thus that general rules may be inferred for plant colonization by microbes. Shotgun metagenomics to compare plant-associated microbiome functions can be used to search for plant colonization markers [12,13]; this can be complemented by read-binning and assembly of bacterial genomes from plant-associated environments [14]. However, metagenomic datasets from different rhizospheres exhibit little overlap in plant-enriched functions [13,15]. On the other hand, plant-associated microbiomes contain a relatively high cultivable fraction of microbes, particularly bacteria [16–19]. It is therefore feasible in plant microbiome research to mitigate the limitations of culture independent methods by generating and studying taxonomically and functionally representative culture collections from plant-associated habitat. Returning to culture-dependent microbial surveys allows the construction of increasingly complex experimental ecology systems for understanding plant-microbe interactions, while providing material for the discovery of potential PGP inoculants. Large-scale isolation, genome sequencing and functional screening efforts are underway in both academic and industrial settings (http://news.monsanto.com/press-release/corporate/novozymes-and-monsanto-complete-closing-bioag-alliance). The definition of large scale, in fact, has changed rapidly from hundreds [16] to tens of thousands [20] of strains. Recent plant-associated bacterial and fungal isolate collections (summarized in Table 1) are derived from sugarcane [20]; grapevine [21–23]; potato [24]; tomato [25]; eucalyptus [25]; rice [26,27]; ancient wheat ancestors [19]; lettuce [28]; Arabidopsis [16, 29]; poplar [29]; and from plants growing in an arsenic-contaminated soils [30]. The increasing volume of isolate collections will tax existing repositories; yet the genomic diversity contained in the bacterial isolates that are being obtained is not nearing saturation [16]. Mechanisms to curate, share and standardize metadata for strains from these collections are needed.
Table 1.
Summary of recent microbial culture collections from plant-associated environments.
| Specimen | Plant compartment | No. of isolates |
Domain | Analyses performed | Reference |
|---|---|---|---|---|---|
| Arabidopsis thaliana | Shoot, root and soil | 7976 | Bacteria | 16S rRNA sequenced 432 genomes sequenced. | [16] |
| Arabidopsis thaliana | Root | 196 | Bacteria | Genomes sequenced | [29] |
| Populus deltoides | Root | 203 | Bacteria | Genomes sequenced | [29] |
| Eucalyptus | Rhizosphere and rhizoplane | 298 | Bacteria | Control of bacterial wilt in Eucalyptus and Solanum lycocersicum caused by Ralstonia solanacearum | [25] |
| Oryza sativa | Phyllosphere | 86 | Bacteria | Indole acetic acid (IAA) production, nitrogen fixation | [26] |
| Oryza sativa | Root, stem and leaves | 1318 | Bacteria | Automated Ribosomal Intergenic Spacer Analysis 762 members 16S rRNA sequenced 689 members identified at the genus level The 228 members of the working collection were analyzed for in vitro PGP and plant immunity traits | [27] |
| Saccharum officinarum | Rhizosphere, endophytic root and endophytic stalk compartments | 5137 colony communities | Bacteria | 16S rRNA sequenced | [20] |
| Solanum tuberosum | Phylloplane and interiors, soil and tubers surface | 243 | - | Interaction assays with plant pathogens | [24] |
| Soil | Soil within the rhizosphere of different autochthonous plants | 80 | Bacteria | Arsenic resistance Arsenate reductase activity | [30] |
| Triticum dicoccoides Aegilops sharonensis Triticum aeastivum | Stems and seeds | 686 | Fungi | 514 members ITS sequenced Categorization into cOTUs at 97% sequence similarity | [19] |
| Vitis vinifera Lepidium draba L. | Rhizosphere and root | 500 | Bacteria | A range of bacterial features known to contribute to PGP, stress tolerance or bio-control | [22] |
| Vitis vinifera | Root, shoot and leaves | 381 | Bacteria | 377 members evaluated for features known to contribute to PGP. Evaluation of their effects on root development in Arabidopsis thaliana. | [21,98] |
| Vitis vinifera | Rhizosphere and endosphere | 510 | Bacteria | 8 strains examined for an array of PGP abilities in vitro, focusing both on conventional and drought-related PGP traits | [23] |
While ongoing attempts exist to screen isolates in the field (see supplementary table 1 for a summary of recent PGP experiments in laboratory and field settings), the most common approach is to utilize a pre-screening strategy to select candidate strains for further analysis. Pre-screening strategies include in vitro screening for known PGP-related activities such as 1-aminocyclopropane-1-carboxylate (ACC) deaminase [31], phosphate solubilization [32], nitrogen fixation [33], or enhancement of plant immune system function [34–36]. Of the 1151 bacterial strains screened in [21,22,27,29], 332 strains solubilized phosphate, 229 strains produced auxin; ACC deaminase activity was found in 85 of 729 strains and bacterial nitrogen fixation was measured in 54 of 229 strains. These screening methods, however, are more likely to confirm the known, rather than to discover novel mechanism of PGP. Furthermore, none of these traits are actually correlated with the magnitude of PGP. Thus the suite of PGP traits that are commonly tested does not predict plant-associated phenotypes, and suggests that untapped mechanisms await discovery.
Genome sequencing of strain collections (Figure 2) [16,37] might provide a richer screening tool for sets of PGP traits that could be readily detected in genomes [38]. For example, the presence of minimal Nif and full Phn gene cassettes and genes required for indole acetic acid (IAA) production corresponded to the respective phenotypes in Paenibacillus polymixa genomes, albeit at variable levels [39]. This indicates that identification of appropriate genomic markers and screening of genome collections might provide a faster and less labor-intensive alternative to physiological screening, while also providing the opportunity for the discovery of correlated and novel PGP-associated genes.
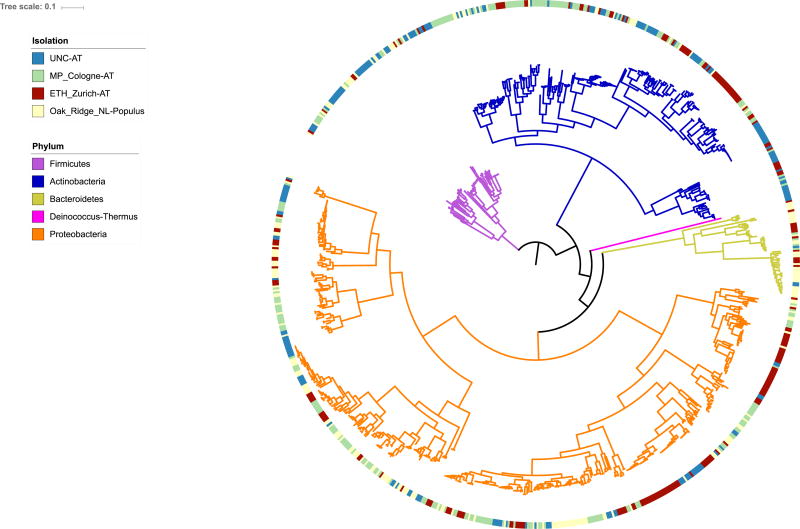
Figure 2. Diversity of genome-sequenced plant associated bacterial isolates from Arabidopsis and Populus currently available on IMG/JGI.
An approximately-maximum-likelihood phylogenetic tree of 831 plant-associated bacteria isolated from Arabidopsis thaliana roots (blue and green bars) and shoots (red bars); and Populus deltoides roots (yellow bars). Tree branches are colored by Phylum. Purple: Firmicutes, Blue: Actinobacteria, Yellow: Bacteroidetes, Pink: Deinococcus-thermus and Orange: Proteobacteria. The tree was constructed using a concatenated filtered alignment of 31 single copy genes [97]. The genome assemblies of each of the three different sequencing projects can be accessed via the IMG/JGI portal by using the following project IDs:
“Genome sequencing of Arabidopsis leaf and root microbiota representing the majority of bacterial species in their natural communities.” A. thaliana. Max Planck Cologne and ETH Zurich “Plant associated metagenomes-Microbial community diversity and host control of community assembly across model and emerging plant ecological genomics systems.” A. thaliana. University of North Carolina “Populus root and rhizosphere microbial communities from Tennessee, USA” – P. deltoides The tree can be visualized and downloaded on iTOL [98] using the following link: http://itol.embl.de/tree/1522317731415721485965060 or via the user Understanding_the_plant_microbiome_COPB.
Ecological considerations for plant beneficial function of microbes in the field
Ultimately, beneficial phenotypes will need to be operative in the field. A successful microbial inoculant has to invade and persist in the context of indigenous microbes and local abiotic conditions in variable settings, and to establish a compatible interaction with the host that includes molecular détente with the plant immune system. Studies of successional dynamics of plant microbiota suggest that upon emergence, initial seed microbiomes rapidly give way to different, soil-derived communities that are still changing days following emergence [40]. Throughout the growing season, this soil-derived community undergoes continuous succession in both above-ground [41,42] and below-ground [43] fractions of the plant. Thus, even if PGP inoculants colonize the plant initially, their persistence over time is not guaranteed. Measuring persistence of bacterial inoculants in soil poses technical difficulties, as the inoculant needs to be identified from within a complex community. Heterologous bacterial inoculants can persist in soil for up to seven weeks [23,27,44–46], but whether they are at levels necessary to continuously provide PGP activity is not clear. Methods to detect persistence include culture-based enumeration using re-isolation of antibiotic resistant inoculants [27,44], or culture-independent measurement of relative abundance of the inoculant’s 16S rRNA gene in the soil, via DGGE [23,44]; amplicon sequencing [45] or by metagenomic sequencing [46].
Diversity of the inoculum
The diversity of a microbial inoculum can widen available plant-associated niches and enhance productivity [47–50]. However, interpreting the effect of microbial (and plant) diversity on microbiome function must be done in a nuanced and context-dependent manner [51]. Complex inocula can provide plants with stronger disease resistance [52–57] and growth promotion [58,59] than single strains. Use of a complex inoculum, rather than single strains, improved arsenic sequestration efficiency of the hyper-accumulator fern Pteris vittata [30], tripling phytoremediation efficiency. In other cases, however, consortia were equal to, or worse than, some individual strains tested, as evidenced by the growth of grapevines under drought stress [23]. Consortia can consist of either closely related strains used to expand the niche breadth of a certain trait [54,55], or of distantly related strains providing PGP via different mechanisms, thus contributing to an overall additive effect [59]. However, increasing strain richness, for example within the biocontrol species Pseudomonas fluorescens, can cause community collapse and subsequent loss of plant protection [60]. Multispecies inocula can be used to exploit positive microbe-microbe interactions. For instance, bacteria can enhance germ tube elongation and hyphal branching in arbuscular mycorrhizal fungi (AMF), promoting the symbiotic development of the AMF on potato plants [61].
The prospect of inoculating crops with consortia rather than single strains exponentially increases the complexity of experimental screening systems. This demands solid design and an analytic framework that is experimentally tractable. Synthetic bacterial communities consisting of up to hundreds of strains have been shown to colonize plants in a reproducible pattern under gnotobiotic conditions [16,37,57], providing a powerful research tool to study microbial PGP and microbe-microbe interactions.
Plant-microbe interactions in soil
Any plant microbiome is a direct function of the microbial meta-community found in the soil around it [10,11,43], a community that in turn can be deeply impacted by agricultural practices [62,63]. Continuously growing crops in agricultural soils can result in pathogen buildup [56] but also in the emergence of disease-suppressive soils: soils that convey resistance to plant pathogens and can contain biocontrol agents within their resident bacterial community [36]. The observation of disease suppressive soils has been linked to shifts in microbial community composition and activity [56,64–66]. Microbial communities in the soil can induce other phenotypes in plants. Artificial selection experiments [66] have shown that iteratively selecting soil slurries can alter plant biomass [68]. Biotic plant-soil feedback was recently shown to be dependent on the type of mycorrhizal fungi or biologically nitrogen fixing (BNF) bacteria that associate with plants [69,70]. Plants associating with AMF or with BNF exhibited conspecific growth inhibition, a long standing observation that can be linked to density dependent predation or disease [71]. In contrast, plants associating with ectomycorrhizal fungi (EF) exhibited the opposite trend: local conspecifics facilitate growth linked to stronger protection from pathogens provided by EF. EF fungi preferentially link to, and transfer carbon between, kindred trees in ectomycorrhizal networks [72], potentially explaining conspecific facilitation. As the spatial density of monoculture, agricultural crops resembles that of EF-associated plants, the mechanisms that evolved to protect from pathogens in EF-associated species may be relevant to how biological disease suppression is applied to crops. Ultimately, both sides of this ancient interaction need to be understood if we are to harness them for agricultural productivity.
Genetic control of beneficial plant-microbe interactions
Plants assemble distinct root [10,11,73] and shoot [18,74,75] microbiomes from the surrounding soil and air. Significant differences in microbe community composition were detected between plant species [13,73,76–79], and between natural accessions of the same species, though to date the intraspecific genetic contribution to microbiome assembly is quite low [10,11,15,18,74,80,81]. Nevertheless, these findings demonstrate that host genetics contributes to plant microbiome assembly; whether the observed heritability will be actionable with respect to plant breeding using genetic approaches based on natural variation to identify causal host genes remains to be demonstrated.
Plants detect microbes via pattern-recognition receptors that bind microbe-associated molecular patterns (MAMPs), triggering a basal defense sufficient to halt the growth of most pathogenic microbes [82,83]. Most non-pathogenic bacteria and fungi associated with plants are sure to produce their own MAMPs, which prompts the question of how beneficial microbes and plants manage to avoid elimination of the microbes via an immune response. Plants can presumably discriminate pathogens from non-pathogens and respond by either resisting microbial growth, ignoring it, or actively supporting it on or within plant tissues. The transcriptional response of Arabidopsis leaves differs when inoculated with different non-pathogenic members of its natural microbiota [35]. While Methylobacterium extorquens induces almost no transcriptional response, Sphingomonas melonis activates the expression of defense related genes that partly overlap with those triggered by the pathogen Pseudomonas syringae DC3000 [34]. This may represent a mechanism of plant defense priming [84] driven by the plant microbiome. The response pattern to non-pathogenic bacteria can differ both across plant species [13] and across accessions within a single species [45]. While some Arabidopsis accessions are colonized by, and establish a beneficial relationship with Pseudomonas fluorescens, other accessions actively inhibit growth of the same strains in their roots.
Given the critical function of defense phytohormones in the plant immune system, it is not surprising that plant microbiome composition is influenced by defense phytohormone signaling. Experiments using a set of mutants with altered defense phytohormone synthesis and/or perception demonstrated that salicylic acid and/or salicylic acid-mediated events influence the root microbiome composition at multiple taxonomic levels [37]. These data suggest that microbial inoculants will not act as “one size fits all”, and may need to be specifically tested down to the host genotype level [74].
The plant microbiome structure changes upon infection [12,53]. This could represent a general response to the plant defense mechanisms, but may also reflect changes made to the habitat by the pathogen [85,86]. Antifungal traits are enriched in barley following infection with Fusarium graminearum, potentially via changes in exudate composition [87]. A study of tomato plants challenged with the pathogen Ralstonia solanacearum revealed that the root exudation profile changed upon pathogen infection, increasing the secretion of phenolic compounds. Exudates of infected plants were correlated with changes in soil microbial abundances, and this response could be emulated in vitro using caffeic acid, one of the phenolic compounds secreted [88]. Similar patterns of pest-inhibition were shown in response to insect herbivores. Inoculation of Arabidopsis with Pseudomonas fluorescens WCS417r altered the plant volatile emission profile in response to an herbivorous caterpillar; this correlated with recruitment of an insecticidal parasitoid wasp [89].
Plant defense mechanisms also impact other drivers of plant-microbe interactions, like plant nutrition [90]. Mutant analysis in Lotus japonicus demonstrated that the root nodulation pathway plays a role not only in nitrogen-fixing symbiosis, but also in the establishment of taxonomically diverse root microbiome [79]. Modern molecular approaches are also being applied to understanding nitrogen-fixing symbioses in non-nodulating plants. Use of dual host-microbe transcriptomics demonstrated that the capacity of a nitrogen-fixing Burkholderia strain to form microaerobic biofilms on sugarcane roots is preceded by reduced motility and immunogenicity, followed by metabolic adaptation to the sugar-rich plant environment. The plant does not activate an immune response, but does change its root morphology, and supplies the bacterium with photosynthates [91], a response pattern that is analogous to the process of infection by BNF in legumes [92].
Regulatory overlap exists between the plant immune system and nutritional stress, as recently demonstrated in the context of the plant phosphate starvation response (PSR) [93]. Signaling dependent on the phytohormone jasmonic acid (JA) and JA accumulation is induced during PSR in Arabidopsis. The Arabidopsis pho1 mutant, deficient in shoot phosphate accumulation, exhibited an activation of JA signaling leading to increased resistance to insect herbivory [94]. PSR signaling and concomitant dampening of the immune system can also regulate beneficial fungal colonization [95,96]. Fungus-mediated PGP also requires PEN2-dependent indole glucosinolate metabolism, a component of plant defense [95]. Experiments using both wild soil and a synthetic bacterial community demonstrated that Arabidopsis PSR mutants assemble a different root microbiome than wild type plants, even when grown in phosphate-sufficient soil. Further, the master transcriptional regulator of PSR directly integrates this nutritional stress response and immune system outputs [97]. These early examples suggest that the coordination of defense and nutrition is critical to driving microbiome function.
Conclusions
The study of plant microbiomes has benefited from ecological studies on one-hand and reductionist mechanistic discoveries on the other. Both schools of thought are yielding increasingly profound insight into the ecological processes that govern plant-microbe interactions as well as the molecular mechanisms that facilitate them. The generation of large isolate collections and the study of synthetic microbial communities in combination with plant genetic resources, will allow us to bridge this gap and to conduct reductionist, hypothesis driven studies in increasingly complex ecological contexts up to field tests. These advances have the potential to transform our understanding of plant-microbe interactions in nature and in agriculture, and will contribute significantly to the next green revolution.
Supplementary Material
Highlights.
Soil microbiomes induce reproducible plant phenotypes
Large collections of plant-associated microbes are available for research
Plant growth promoting microbial inoculants can persist in soil for weeks
Co-regulation of immune system function and nutritional stress responses exists
Deployment of consortia may enable more resilient plant phenotypes than single strains
Acknowledgments
Supported by NSF INSPIRE grant IOS-1343020 and DOE-USDA Feedstock Award DE-SC001043 to J.L.D. S.H.P was supported by NIH Training Grant T32 GM067553 and was a Howard Hughes Medical Institute International Student Research Fellow. J.L.D is an Investigator of the Howard Hughes Medical Institute, supported by the HHMI and the Gordon and Betty Moore Foundation (GBMF3030). O.M.F is supported by NIH NRSA Fellowships F32-GM117758.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vessey JK. Plant growth promoting rhizobacteria as biofertilizers. Plant Soil. 2003;255:571–586. [Google Scholar]
- 2•.Parnell JJ, Berka R, Young HA, Sturino JM, Kang Y, Barnhart DM, DiLeo MV. From the Lab to the Farm: An Industrial Perspective of Plant Beneficial Microorganisms. Front. Plant Sci. 2016;7:1110. doi: 10.3389/fpls.2016.01110. This review discusses some practical considerations for the developemnt of commercial microbial inoculatns, and includes field data for some recent commercial products. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sahoo RK, Bhardwaj D, Tuteja N. Plant Acclimation to Environmental Stress. Springer New York; 2013. Biofertilizers: A Sustainable Eco-Friendly Agricultural Approach to Crop Improvement; pp. 403–432. [Google Scholar]
- 4.Desbrosses GJ, Stougaard J. Root Nodulation: A Paradigm for How Plant-Microbe Symbiosis Influences Host Developmental Pathways. Cell Host Microbe. 2011;10:348–358. doi: 10.1016/j.chom.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 5.Dobbelaere S, Croonenborghs A, Thys A, Vande Broek A, Vanderleyden J. Phytostimulatory effect of Azospirillum brasilense wild type and mutant strains altered in IAA production on wheat. Plant Soil. 1999;212:153–162. [Google Scholar]
- 6.Li J, Ovakim DH, Charles TC, Glick BR. An ACC deaminase minus mutant of Enterobacter cloacae UW4 no longer promotes root elongation. Curr. Microbiol. 2000;41:101–105. doi: 10.1007/s002840010101. [DOI] [PubMed] [Google Scholar]
- 7.Herrera Paredes S, Lebeis SL. Giving back to the community: microbial mechanisms of plant-soil interactions. Funct. Ecol. 2016;30:1043–1052. [Google Scholar]
- 8.Vorholt JA. Microbial life in the phyllosphere. Nat. Rev. Microbiol. 2012;10:828–840. doi: 10.1038/nrmicro2910. [DOI] [PubMed] [Google Scholar]
- 9.Bulgarelli D, Schlaeppi K, Spaepen S, van Themaat EVL, Schulze-Lefert P. Structure and Functions of the Bacterial Microbiota of Plants. Annu. Rev. Plant Biol. 2013;64:807–838. doi: 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- 10.Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, Tremblay J, Engelbrektson A, Kunin V, del Rio TG, et al. Defining the core Arabidopsis thaliana root microbiome. Nature. 2012;488:86–90. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bulgarelli D, Rott M, Schlaeppi K, Ver Loren van Themaat E, Ahmadinejad N, Assenza F, Rauf P, Huettel B, Reinhardt R, Schmelzer E, et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature. 2012;488:91–95. doi: 10.1038/nature11336. [DOI] [PubMed] [Google Scholar]
- 12.Chapelle E, Mendes R, Bakker PaH, Raaijmakers JM. Fungal invasion of the rhizosphere microbiome. ISME J. 2015;10:1–4. doi: 10.1038/ismej.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ofek-Lalzar M, Sela N, Goldman-Voronov M, Green SJ, Hadar Y, Minz D. Niche and host-associated functional signatures of the root surface microbiome. Nat. Commun. 2014;5:4950. doi: 10.1038/ncomms5950. [DOI] [PubMed] [Google Scholar]
- 14.Finkel OM, Delmont TO, Post AF, Belkin S. Metagenomic Signatures of Bacterial Adaptation to Life in the Phyllosphere of a Salt-Secreting Desert Tree. Appl. Environ. Microbiol. 2016;82:2854–61. doi: 10.1128/AEM.00483-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bulgarelli D, Garrido-Oter R, Münch PC, Weiman A, Dröge J, Pan Y, McHardy AC, Schulze-Lefert P. Structure and Function of the Bacterial Root Microbiota in Wild and Domesticated Barley. Cell Host Microbe. 2015;17:392–403. doi: 10.1016/j.chom.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16•.Bai Y, Müller DB, Srinivas G, Garrido-Oter R, Potthoff E, Rott M, Dombrowski N, Münch PC, Spaepen S, Remus-Emsermann M, et al. Functional overlap of the Arabidopsis leaf and root microbiota. Nature. 2015;528:364–369. doi: 10.1038/nature16192. This article reports on the generation of large scale isolate collections from the roots and shoots of A. thaliana and genome sequences of a large subset of these collections. It further provides a proof of concept for the establishment of synthetic communities consisting of hundreds of strains in gnotobiotic plants, offering a new experimental platform for studying the ecology of the plant microbiome. [DOI] [PubMed] [Google Scholar]
- 17.Burch AY, Do PT, Sbodio A, Suslow TV, Lindow SE. High-Level Culturability of Epiphytic Bacteria and Frequency of Biosurfactant Producers on Leaves. Appl. Environ. Microbiol. 2016;82:5997–6009. doi: 10.1128/AEM.01751-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bodenhausen N, Horton MW, Bergelson J. Bacterial Communities Associated with the Leaves and the Roots of Arabidopsis thaliana. PLoS One. 2013;8:e56329. doi: 10.1371/journal.pone.0056329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ofek-Lalzar M, Gur Y, Ben-Moshe S, Sharon O, Kosman E, Mochli E, Sharon A. Diversity of fungal endophytes in recent and ancient wheat ancestors Triticum dicoccoides and Aegilops sharonensis. FEMS Microbiol. Ecol. 2016;92:fiw152. doi: 10.1093/femsec/fiw152. [DOI] [PubMed] [Google Scholar]
- 20.Armanhi JSL, de Souza RSC, de Araújo LM, Okura VK, Mieczkowski P, Imperial J, Arruda P. Multiplex amplicon sequencing for microbe identification in community-based culture collections. Sci. Rep. 2016;6:29543. doi: 10.1038/srep29543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baldan E, Nigris S, Romualdi C, D’Alessandro S, Clocchiatti A, Zottini M, Stevanato P, Squartini A, Baldan B. Beneficial Bacteria Isolated from Grapevine Inner Tissues Shape Arabidopsis thaliana Roots. PLoS One. 2015;10:e0140252. doi: 10.1371/journal.pone.0140252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Samad A, Trognitz F, Compant S, Antonielli L, Sessitsch A. Shared and host-specific microbiome diversity and functioning of grapevine and accompanying weed plants. Environ. Microbiol. 2016 doi: 10.1111/1462-2920.13618. [DOI] [PubMed] [Google Scholar]
- 23.Rolli E, Marasco R, Vigani G, Ettoumi B, Mapelli F, Deangelis ML, Gandolfi C, Casati E, Previtali F, Gerbino R, et al. Improved plant resistance to drought is promoted by the root-associated microbiome as a water stress-dependent trait. Environ. Microbiol. 2015;17:316–331. doi: 10.1111/1462-2920.12439. [DOI] [PubMed] [Google Scholar]
- 24.Cray JA, Connor MC, Stevenson A, Houghton JDR, Rangel DEN, Cooke LR, Hallsworth JE. Biocontrol agents promote growth of potato pathogens, depending on environmental conditions. Microb. Biotechnol. 2016;9:330–354. doi: 10.1111/1751-7915.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santiago TR, Grabowski C, Rossato M, Romeiro RS, Mizubuti ESG. Biological control of eucalyptus bacterial wilt with rhizobacteria. Biol. Control. 2015;80:14–22. [Google Scholar]
- 26.Venkatachalam S, Ranjan K, Prasanna R, Ramakrishnan B, Thapa S, Kanchan A. Diversity and functional traits of culturable microbiome members, including cyanobacteria in the rice phyllosphere. Plant Biol. 2016;18:627–637. doi: 10.1111/plb.12441. [DOI] [PubMed] [Google Scholar]
- 27.Bertani I, Abbruscato P, Piffanelli P, Subramoni S, Venturi V. Rice bacterial endophytes: Isolation of a collection, identification of beneficial strains and microbiome analysis. Environ. Microbiol. Rep. 2016;8:388–398. doi: 10.1111/1758-2229.12403. [DOI] [PubMed] [Google Scholar]
- 28.Cipriano MAP, Lupatini M, Lopes-Santos L, da Silva MJ, Roesch LFW, Destéfano SAL, Freitas SS, Kuramae EE. Lettuce and rhizosphere microbiome responses to growth promoting Pseudomonas species under field conditions. FEMS Microbiol. Ecol. 2016:92. doi: 10.1093/femsec/fiw197. [DOI] [PubMed] [Google Scholar]
- 29.Levy A, Clingenpeel S, Salas González I, Herrera Paredes A, Stillman K, Monteiro F, Alvarez BR, Lundberg DS, Lu T-Y, Lebeis S, et al. Genomic determinants of bacterial adaptation to plants. submitted. [Google Scholar]
- 30•.Lampis S, Santi C, Ciurli A, Andreolli M, Vallini G. Promotion of arsenic phytoextraction efficiency in the fern Pteris vittata by the inoculation of As-resistant bacteria: a soil bioremediation perspective. Front. Plant Sci. 2015;6:80. doi: 10.3389/fpls.2015.00080. This article shows that bacterial isolates can be mined and used as innoculants not only for crop improvement but also to enhance phytoremediation. Hyperaccumulator plants that were innoculated with bacteria isolated from arsenic contaminated soil increased arsenic sequestration by up to 3-fold. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glick BR, Penrose DM, Li J. A Model For the Lowering of Plant Ethylene Concentrations by Plant Growth-promoting Bacteria. J. Theor. Biol. 1998;190:63–68. doi: 10.1006/jtbi.1997.0532. [DOI] [PubMed] [Google Scholar]
- 32.Richardson AE. Prospects for using soil microorganisms to improve the acquisition of phosphorus by plants. Funct. Plant Biol. 2001;28:897. [Google Scholar]
- 33.Lin L, Li Z, Hu C, Zhang X, Chang S, Yang L, Li Y, An Q. Plant Growth-Promoting Nitrogen-Fixing Enterobacteria Are in Association with Sugarcane Plants Growing in Guangxi, China. Microbes Environ. 2012;27:391–398. doi: 10.1264/jsme2.ME11275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pieterse CMJ, Zamioudis C, Berendsen RL, Weller DM, Van Wees SCM, Bakker PAHM. Induced Systemic Resistance by Beneficial Microbes. Annu. Rev. Phytopathol. 2014;52:347–375. doi: 10.1146/annurev-phyto-082712-102340. [DOI] [PubMed] [Google Scholar]
- 35•.Vogel C, Bodenhausen N, Gruissem W, Vorholt JA. The Arabidopsis leaf transcriptome reveals distinct but also overlapping responses to colonization by phyllosphere commensals and pathogen infection with impact on plant health. New Phytol. 2016;212:192–207. doi: 10.1111/nph.14036. This paper demonstrates via RNA-seq that plants responds differently to members of its natural phyllosphere microbiome, and that while some bacteria induce plant defense genes, others have only a marginal effect on plant gene expression. [DOI] [PubMed] [Google Scholar]
- 36.Raaijmakers JM, Mazzola M. Soil immune responses. Science. 2016;352:1392–1393. doi: 10.1126/science.aaf3252. [DOI] [PubMed] [Google Scholar]
- 37•.Lebeis SL, Herrera Paredes S, Lundberg DS, Breakfield N, Gehring J, McDonald M, Malfatti S, Glavina del Rio T, Jones CD, Tringe SG, et al. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science. 2015;349:860–864. doi: 10.1126/science.aaa8764. This paper reveals the role of the plant hormone salicylic acid in selecting root microbiota of A. thaliana. [DOI] [PubMed] [Google Scholar]
- 38.Bruto M, Prigent-Combaret C, Muller D, Moënne-Loccoz Y. Analysis of genes contributing to plant-beneficial functions in plant growth-promoting rhizobacteria and related Proteobacteria. Sci. Rep. 2014;4:6261. doi: 10.1038/srep06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie J, Shi H, Du Z, Wang T, Liu X, Chen S. Comparative genomic and functional analysis reveal conservation of plant growth promoting traits in Paenibacillus polymyxa and its closely related species. Sci. Rep. 2016;6:21329. doi: 10.1038/srep21329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barret M, Briand M, Bonneau S, Préveaux A, Valière S, Bouchez O, Hunault G, Simoneau P, Jacques M-A. Emergence Shapes the Structure of the Seed Microbiota. Appl. Environ. Microbiol. 2015;81:1257–1266. doi: 10.1128/AEM.03722-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Copeland JK, Yuan L, Layeghifard M, Wang PW, Guttman DS. Seasonal Community Succession of the Phyllosphere Microbiome. Mol. Plant-Microbe Interact. 2015;28:274–285. doi: 10.1094/MPMI-10-14-0331-FI. [DOI] [PubMed] [Google Scholar]
- 42.Shade A, Mcmanus PS, Handelsman J. Unexpected Diversity during Community Succession in the Apple Flower Microbiome. MBio. 2013;4:e00602–12. doi: 10.1128/mBio.00602-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43•.Edwards J, Johnson C, Santos-medellín C, Lurie E, Kumar N. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA. 2015 doi: 10.1073/pnas.1414592112. This research article provides a comprehensive picture of the community composition of the rice root microbiome, including an illuminating view of the successional patterns that shape it. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schreiter S, Sandmann M, Smalla K, Grosch R. Soil Type Dependent Rhizosphere Competence and Biocontrol of Two Bacterial Inoculant Strains and Their Effects on the Rhizosphere Microbial Community of Field-Grown Lettuce. PLoS One. 2014;9:e103726. doi: 10.1371/journal.pone.0103726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45••.Haney CH, Samuel BS, Bush J, Ausubel FM. Associations with rhizosphere bacteria can confer an adaptive advantage to plants. Nat. Plants. 2015;1:15051. doi: 10.1038/nplants.2015.51. This paper shows, using a suite of different methods, that different accessions of A. thaliana differ in their recruitment of P. fuorescens, providing a mechanistic understanding of fine taxonomic shifts observed in surveys. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krober M, Wibberg D, Grosch R, Eikmeyer F, Verwaaijen B, Chowdhury SP, Hartmann A, Pühler A, Schlüter A. Effect of the strain Bacillus amyloliquefaciens FZB42 on the microbial community in the rhizosphere of lettuce under field conditions analyzed by whole metagenome sequencing. Front. Microbiol. 2014;5:252. doi: 10.3389/fmicb.2014.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crutsinger GM, Collins MD, Fordyce JA, Gompert Z, Nice CC, Sanders NJ. Plant Genotypic Diversity Predicts Community Structure and Governs an Ecosystem Process. Science. 2006;313:966–968. doi: 10.1126/science.1128326. [DOI] [PubMed] [Google Scholar]
- 48.Cordero OX, Polz MF. Explaining microbial genomic diversity in light of evolutionary ecology. Nat. Rev. Microbiol. 2014;12:263–273. doi: 10.1038/nrmicro3218. [DOI] [PubMed] [Google Scholar]
- 49.Weidner S, Koller R, Latz E, Kowalchuk G, Bonkowski M, Scheu S, Jousset A. Bacterial diversity amplifies nutrient-based plant-soil feedbacks. Funct. Ecol. 2015 doi: 10.1111/1365-2435.12445. [DOI] [Google Scholar]
- 50.Gudelj I, Weitz JS, Ferenci T, Claire Horner-Devine M, Marx CJ, Meyer JR, Forde SE. An integrative approach to understanding microbial diversity: from intracellular mechanisms to community structure. Ecol. Lett. 2010;13:1073–1084. doi: 10.1111/j.1461-0248.2010.01507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shade A. Diversity is the question, not the answer. ISME J. 2017;11:1–6. doi: 10.1038/ismej.2016.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yasmin S, Zaka A, Imran A, Zahid MA, Yousaf S, Rasul G, Arif M, Mirza MS. Plant growth promotion and suppression of bacterial leaf blight in rice by inoculated bacteria. PLoS One. 2016;11:1–19. doi: 10.1371/journal.pone.0160688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53•.Agler MT, Ruhe J, Kroll S, Morhenn C, Kim S-T, Weigel D, Kemen EM. Microbial Hub Taxa Link Host and Abiotic Factors to Plant Microbiome Variation. PLOS Biol. 2016;14:e1002352. doi: 10.1371/journal.pbio.1002352. This paper uses co-occurrence networks derived from a large scale survey of Arabidopsis-associated microbial communities to identify “hub” taxa, which strongly interact with other community members. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu J, Wei Z, Friman VP, Gu SH, Wang XF, Eisenhauer N, Yang TJ, Ma J, Shen QR, Xu YC, et al. Probiotic Diversity Enhances Rhizosphere Microbiome Function and Plant Disease Suppression. MBio. 2016;7:e01790–16. doi: 10.1128/mBio.01790-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wei Z, Yang T, Friman V-P, Xu Y, Shen Q, Jousset A. Trophic network architecture of root-associated bacterial communities determines pathogen invasion and plant health. Nat. Commun. 2015;6:8413. doi: 10.1038/ncomms9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Santhanam R, Luu VT, Weinhold A, Goldberg J, Oh Y, Baldwin IT. Native root-associated bacteria rescue a plant from a sudden-wilt disease that emerged during continuous cropping. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E5013–20. doi: 10.1073/pnas.1505765112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Niu B, Paulson JN, Zheng X, Kolter R. Simplified and representative bacterial community of maize roots. Proc. Natl. Adac. Sci. U.S. A. 2015;114:E2450–E2459. doi: 10.1073/pnas.1616148114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baas P, Bell C, Mancini LM, Lee MN, Conant RT, Wallenstein MD. Phosphorus mobilizing consortium Mammoth P™ enhances plant growth. PeerJ. 2016;4:e2121. doi: 10.7717/peerj.2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Timm CM, Pelletier DA, Jawdy SS, Gunter LE, Henning JA, Engle N, Aufrecht J, Gee E, Nookaew I, Yang Z, et al. Two Poplar-Associated Bacterial Isolates Induce Additive Favorable Responses in a Constructed Plant-Microbiome System. Front. Plant Sci. 2016:7. doi: 10.3389/fpls.2016.00497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Becker J, Eisenhauer N, Scheu S, Jousset A. Increasing antagonistic interactions cause bacterial communities to collapse at high diversity. Ecol. Lett. 2012;15:468–474. doi: 10.1111/j.1461-0248.2012.01759.x. [DOI] [PubMed] [Google Scholar]
- 61.Loján P, Demortier M, Velivelli SL, Pfeiffer S, Suárez JP, de Vos P, Doyle Prestwich B, Sessitsch A, Declerck S. Impact of plant growth-promoting rhizobacteria on root colonization potential and life cycle of Rhizophagus irregularis following co-entrapment into alginate beads. J. Appl. Microbiol. 2016 doi: 10.1111/jam.13355. [DOI] [PubMed] [Google Scholar]
- 62.Hartmann M, Frey B, Mayer J, Mäder P, Widmer F. Distinct soil microbial diversity under long-term organic and conventional farming. ISME J. 2015;9:1177–1194. doi: 10.1038/ismej.2014.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fierer N, Ladau J, Clemente JC, Leff JW, Owens SM, Pollard KS, Knight R, Gilbert JA, McCulley RL. Reconstructing the Microbial Diversity and Function of Pre-Agricultural Tallgrass Prairie Soils in the United States. Science. 2013;342:621–624. doi: 10.1126/science.1243768. [DOI] [PubMed] [Google Scholar]
- 64.Cha JY, Han S, Hong HJ, Cho H, Kim D, Kwon Y, Kwon SK, Crüsemann M, Bok Lee Y, Kim JF, et al. Microbial and biochemical basis of a Fusarium wilt-suppressive soil. ISME J. 2016;10:119–129. doi: 10.1038/ismej.2015.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shen Z, Ruan Y, Xue C, Zhong S, Li R, Shen Q. Soils naturally suppressive to banana Fusarium wilt disease harbor unique bacterial communities. Plant Soil. 2015;393:21–33. [Google Scholar]
- 66.Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, Schneider JHM, Piceno YM, DeSantis TZ, Andersen GL, Bakker PA, et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. 2011;332:1097–100. doi: 10.1126/science.1203980. [DOI] [PubMed] [Google Scholar]
- 67.Mueller UG, Sachs JL. Engineering Microbiomes to Improve Plant and Animal Health. Trends Microbiol. 2015;23:606–617. doi: 10.1016/j.tim.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 68.Swenson W, Wilson DS, Elias R. Artificial ecosystem selection. Proc. Natl. Acad. Sci. 2000;97:9110–9114. doi: 10.1073/pnas.150237597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bennett JA, Maherali H, Reinhart KO, Lekberg Y, Hart MM, Klironomos J. Plant-soil feedbacks and mycorrhizal type influence temperate forest population dynamics. Science. 2017;355:181–184. doi: 10.1126/science.aai8212. [DOI] [PubMed] [Google Scholar]
- 70••.Teste FP, Kardol P, Turner BL, Wardle DA, Zemunik G, Renton M, Laliberté E. Plant-soil feedback and the maintenance of diversity in Mediterranean-climate shrublands. Science. 2017;355:173–176. doi: 10.1126/science.aai8291. These two research articles present large-scale surveys reveiling a strong correlation between density dependent spacial distributions of plants and the type of mycorriza with which they associate. [DOI] [PubMed] [Google Scholar]
- 71.Janzen DH. Herbivores and the number of tree species in tropical forests. Am. Nat. 1970;104:501–528. [Google Scholar]
- 72.Pickles BJ, Wilhelm R, Asay AK, Hahn AS, Simard SW, Mohn WW. Transfer of 13 C between paired Douglas-fir seedlings reveals plant kinship effects and uptake of exudates by ectomycorrhizas. New Phytol. 2016 doi: 10.1111/nph.14325. [DOI] [PubMed] [Google Scholar]
- 73.Schlaeppi K, Dombrowski N, Oter RG, Ver Loren van Themaat E, Schulze-Lefert P. Quantitative divergence of the bacterial root microbiota in Arabidopsis thaliana relatives. Proc. Natl. Acad. Sci. U. S. A. 2014;111:585–92. doi: 10.1073/pnas.1321597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Horton MW, Bodenhausen N, Beilsmith K, Meng D, Muegge BD, Subramanian S, Vetter MM, Vilhjálmsson BJ, Nordborg M, Gordon JI, et al. Genome-wide association study of Arabidopsis thaliana leaf microbial community. Nat. Commun. 2014;5:5320. doi: 10.1038/ncomms6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Maignien L, DeForce EA, Chafee ME, Eren AM, Simmons SL. Ecological succession and stochastic variation in the assembly of Arabidopsis thaliana phyllosphere communities. MBio. 2014;5:e00682–13. doi: 10.1128/mBio.00682-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bouffaud M-L, Poirier M-A, Muller D, Moënne-Loccoz Y. Root microbiome relates to plant host evolution in maize and other Poaceae. Environ. Microbiol. 2014;16:2804–2814. doi: 10.1111/1462-2920.12442. [DOI] [PubMed] [Google Scholar]
- 77.Laforest-Lapointe I, Messier C, Kembel SW. Host species identity, site and time drive temperate tree phyllosphere bacterial community structure. Microbiome. 2016;4:27. doi: 10.1186/s40168-016-0174-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Redford AJ, Bowers RM, Knight R, Linhart Y, Fierer N. The ecology of the phyllosphere: geographic and phylogenetic variability in the distribution of bacteria on tree leaves. Environ. Microbiol. 2010;12:2885–93. doi: 10.1111/j.1462-2920.2010.02258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zgadzaj R, Garrido-Oter R, Jensen DB, Koprivova A, Schulze-Lefert P, Radutoiu S. Root nodule symbiosis in Lotus japonicus drives the establishment of distinctive rhizosphere, root, and nodule bacterial communities. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E7996–E8005. doi: 10.1073/pnas.1616564113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Peiffer JA, Spor A, Koren O, Jin Z, Tringe SG, Dangl JL, Buckler ES, Ley RE. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. 2013;110:6548–6553. doi: 10.1073/pnas.1302837110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81••.Wagner MR, Lundberg DS, del Rio TG, Tringe SG, Dangl JL, Mitchell-Olds T. Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat. Commun. 2016;7:12151. doi: 10.1038/ncomms12151. This is a report of a large-scale field experiment aiming to disentangle the effects of genotype, environment, age and year of harvest on plant microbiomes, illustrating the importance of genotype-by-environment interactions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 83.Böhm H, Albert I, Fan L, Reinhard A, Nürnberger T. Immune receptor complexes at the plant cell surface. Curr. Opin. Plant Biol. 2014;20:47–54. doi: 10.1016/j.pbi.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 84.Martinez-Medina A, Flors V, Heil M, Mauch-Mani B, Pieterse CM, Pozo MJ, Ton J, van Dam NM, Conrath U. Recognizing Plant Defense Priming. Trends Plant Sci. 2016;21:818–822. doi: 10.1016/j.tplants.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 85•.Xin XF, Nomura K, Aung K, Velásquez AC, Yao J, Boutrot F, Chang JH, Zipfel C, He SY. Bacteria establish an aqueous living space in plants crucial for virulence. Nature. 2016;539:524–529. doi: 10.1038/nature20166. This paper demonstrates facilitation of pathogenic bacteira by increasing the humidity in the apoplast. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86••.Yamada K, Saijo Y, Nakagami H, Takano Y. Regulation of sugar transporter activity for antibacterial defense in Arabidopsis. Science. 2016 doi: 10.1126/science.aah5692. paah5692. This paper demonstrates a mechanism by which plants deprive pathogenic bacteria of extracellular sugar as a defense strategy. [DOI] [PubMed] [Google Scholar]
- 87.Dudenhöffer JH, Scheu S, Jousset A. Systemic enrichment of antifungal traits in the rhizosphere microbiome after pathogen attack. J. Ecol. 2016;104:1566–1575. [Google Scholar]
- 88.Gu Y, Wei Z, Wang X, Friman V-P, Huang J, Wang X, Mei X, Xu Y, Shen Q, Jousset A. Pathogen invasion indirectly changes the composition of soil microbiome via shifts in root exudation profile. Biol. Fertil. Soils. 2016;52:997–1005. [Google Scholar]
- 89.Pangesti N, Weldegergis BT, Langendorf B, van Loon JJA, Dicke M, Pineda A. Rhizobacterial colonization of roots modulates plant volatile emission and enhances the attraction of a parasitoid wasp to host-infested plants. Oecologia. 2015;178:1169–1180. doi: 10.1007/s00442-015-3277-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hacquard S, Garrido-Oter R, González A, Spaepen S, Ackermann G, Lebeis S, McHardy AC, Dangl JL, Knight R, Ley R, et al. Microbiota and Host Nutrition across Plant and Animal Kingdoms. Cell Host Microbe. 2015;17:603–616. doi: 10.1016/j.chom.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 91.Paungfoo-Lonhienne C, Lonhienne TGA, Yeoh YK, Donose BC, Webb RI, Parsons J, Liao W, Sagulenko E, Lakshmanan P, Hugenholtz P, et al. Crosstalk between sugarcane and a plant-growth promoting Burkholderia species. Sci. Rep. 2016;6:37389. doi: 10.1038/srep37389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cao Y, Halane MK, Gassmann W, Stacey G. The Role of Plant Innate Immunity in the Legume-Rhizobium Symbiosis. Annu. Rev. Plant Biol. 2017;68:12.1–12.27. doi: 10.1146/annurev-arplant-042916-041030. [DOI] [PubMed] [Google Scholar]
- 93.López-Arredondo DL, Leyva-González MA, González-Morales SI, López-Bucio J, Herrera-Estrella L. Phosphate nutrition: improving low-phosphate tolerance in crops. Annu. Rev. Plant Biol. 2014;65:95–123. doi: 10.1146/annurev-arplant-050213-035949. [DOI] [PubMed] [Google Scholar]
- 94.Khan GA, Vogiatzaki E, Glauser G, Poirier Y. Phosphate Deficiency Induces the Jasmonate Pathway and Enhances Resistance to Insect Herbivory. Plant Physiol. 2016;171:632–44. doi: 10.1104/pp.16.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95••.Hiruma K, Gerlach N, Sacristán S, Nakano RT, Hacquard S, Kracher B, Neumann U, Ramírez D, Bucher M, O’Connell RJ, et al. Root Endophyte Colletotrichum tofieldiae Confers Plant Fitness Benefits that Are Phosphate Status Dependent. Cell. 2016;165:464–474. doi: 10.1016/j.cell.2016.02.028. This paper demonstrates that, under phosphate starvation conditions, the endophytic fungus, Colletotrichum tofieldiae, provides PGP to Arabidopsis, but the plant must restrain fungal growth to avoid pathogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hacquard S, Kracher B, Hiruma K, Münch PC, Garrido-Oter R, Thon MR, Weimann A, Damm U, Dallery J-F, Hainaut M, et al. Survival trade-offs in plant roots during colonization by closely related beneficial and pathogenic fungi. Nat. Commun. 2016;7:11362. doi: 10.1038/ncomms11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97••.Castrillo G, Teixeira PJPL, Herrera Paerdes S, Law TF, de Lorenzo L, Feltcher ME, Finkel OM, Breakfield N, Mieczkowski P, Jones CD, et al. Direct integration of phosphate starvation and immunity in response to a root microbiome. Nature. 2017;543:513–518. doi: 10.1038/nature21417. This paper demonstrates that the master transcriptional regulators of phosphate stress response, PHR1, also directly repress defense in response to a root microbiome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Baldan E, Nigris S, Populin F, Zottini M, Squartini A, Baldan B. Identification of culturable bacterial endophyte community isolated from tissues of Vitis vinifera “Glera.”. Plant Biosyst. -An Int. J. Deal. with all Asp. Plant Biol. 2014;148:508–516. [Google Scholar]
- 99.Wu M, Eisen JA. A simple, fast, and accurate method of phylogenomic inference. Genome Biol. 2008;9:R151. doi: 10.1186/gb-2008-9-10-r151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Letunic I, Bork P. Interactive tree of life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2006;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 101.Mengual C, Schoebitz M, Azcón R, Roldán A. Microbial inoculants and organic amendment improves plant establishment and soil rehabilitation under semiarid conditions. J. Environ. Manage. 2014;134:1–7. doi: 10.1016/j.jenvman.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 102.Latz E, Eisenhauer N, Scheu S, Jousset A. Plant identity drives the expression of biocontrol factors in a rhizosphere bacterium across a plant diversity gradient. Funct. Ecol. 2015;29:1225–1234. [Google Scholar]
- 103.Yuan Z, Druzhinina IS, Labbé J, Redman R, Qin Y, Rodriguez R, Zhang C, Tuskan GA, Lin F. Specialized Microbiome of a Halophyte and its Role in Helping Non-Host Plants to Withstand Salinity. Sci. Rep. 2016;6:32467. doi: 10.1038/srep32467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Oteino N, Lally RD, Kiwanuka S, Lloyd A, Ryan D, Germaine KJ, Dowling DN. Plant growth promotion induced by phosphate solubilizing endophytic Pseudomonas isolates. Front. Microbiol. 2015;6:745. doi: 10.3389/fmicb.2015.00745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Henning JA, Weston DJ, Pelletier DA, Timm CM, Jawdy SS, Classen AT. Root bacterial endophytes alter plant phenotype, but not physiology. PeerJ. 2016;4:e2606. doi: 10.7717/peerj.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Vargas L, Santa Brígida AB, Mota Filho JP, de Carvalho TG, Rojas CA, Vaneechoutte D, Van Bel M, Farrinelli L, Ferreira PCG, Vandepoele K, et al. Drought Tolerance Conferred to Sugarcane by Association with Gluconacetobacter diazotrophicus: A Transcriptomic View of Hormone Pathways. PLoS One. 2014;9:e114744. doi: 10.1371/journal.pone.0114744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Buysens C, César V, Ferrais F, Dupré de Boulois H, Declerck S. Inoculation of Medicago sativa cover crop with Rhizophagus irregularis and Trichoderma harzianum increases the yield of subsequently-grown potato under low nutrient conditions. Appl. Soil Ecol. 2016;105:137–143. [Google Scholar]
- 108.Keyser CA, Jensen B, Meyling NV. Dual effects of Metarhizium spp. and Clonostachys rosea against an insect and a seed-borne pathogen in wheat. Pest Manag. Sci. 2016;72:517–526. doi: 10.1002/ps.4015. [DOI] [PubMed] [Google Scholar]
- 109.Silveira APD, da Sala VMR, Cardoso EJBN, Labanca EG, Cipriano MAP. Nitrogen metabolism and growth of wheat plant under diazotrophic endophytic bacteria inoculation. Appl. Soil Ecol. 2016;107:313–319. [Google Scholar]
- 110.Colla G, Rouphael Y, Bonini P, Cardarelli M. Coating seeds with endophytic fungi enhances growth, nutrient uptake, yield and grain quality of winter wheat. Int. J. Plant Prod. 2015;9:171–190. [Google Scholar]
- 111.Leggett M, Newlands NK, Greenshields D, West L, Inman S, Koivunen ME. Maize yield response to a phosphorus-solubilizing microbial inoculant in field trials. J. Agric. Sci. 2015;153:1464–1478. doi: 10.1017/S0021859614001166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ibañez F, Arroyo ME, Angelini J, Tonelli ML, Muñoz V, Ludueña L, Valetti L, Fabra A. Non-rhizobial peanut nodule bacteria promote maize (Zea mays L.) and peanut (Arachis hypogaea L.) growth in a simulated crop rotation system. Appl. Soil Ecol. 2014;84:208–212. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.