SUMMARY
We currently lack a comprehensive understanding of the mechanisms underlying neural tube formation and their contributions to neural tube defects (NTDs). Developing a model to study such a complex morphogenetic process is critical, especially one that models human-specific aspects. Three-dimensional, hESCs-derived neural rosettes (NRs) provide a powerful resource for in vitro modeling of human neural tube formation. Epigenomic maps reveal enhancer elements unique to NRs relative to 2D systems. A master regulatory network illustrates that key NR properties are related to their epigenomic landscapes. We found that folate-associated DNA methylation changes were enriched within NR regulatory elements near genes involved in neural tube formation and metabolism. Our comprehensive regulatory maps offer insights into the mechanisms by which folate may prevent NTDs. Lastly, our distal regulatory maps provide a better understanding of the potential role of neurological disorder-associated SNPs.
In Brief
Neural rosettes offer a 3D model of neural tube development. Valensisi et al. show that these cells have distinct epigenomic landscapes compared to 2D neural stem cells. Cis-regulatory elements identified in NRs harbor folate-associated CpGs, which may be important during neural tube development, and neurodevelopmental disorder associated variants.
INTRODUCTION
Advances in 3D culture systems have opened the door to unprecedented opportunities for recapitulating the in vivo features of human CNS development (Kelava and Lancaster, 2016). Neural rosettes (NRs) derived from human embryonic stem cells (hESCs) can recapitulate the molecular and morphogenetic sequence of events from gastrulation to neural tube (NT) formation (Deglincerti et al., 2016; Kelava and Lancaster, 2016; Zhang et al., 2001). These 3D tubular structures emerge from embryoid bodies (EBs) with a differentiation timing comparable to that seen in vivo (Pankratz et al., 2007). NRs display apical-basal polarity typical of neuroepithelium and radial organization similar to that of radial glial cells, a primary neural progenitor cell population present during development and in the adult brain (Conti and Cattaneo, 2010). Previous studies have explored molecular markers and signaling pathways establishing the identity and differentiation potential of NRs and showed that NRs better recapitulate the in vivo properties of bona fide neural stem cells (NSCs) (Elkabetz et al., 2008; Koch et al., 2009). Recently, 3D culture systems were used to investigate molecular mechanisms and signaling pathways governing self-organization of NT formation and NT closure both in mouse and non-human primates (Meinhardt et al., 2014; Zhu et al., 2015). These studies serve as proof of concept of the power of 3D culture systems for modeling NT formation and its disruption.
Disruption in NT formation and closure leads to neural tube defects (NTDs) (Copp and Greene, 2012), the second most common birth defects and a global public health burden (Zaganjor et al., 2015). An important player in NTD etiology is folate (Nazki et al., 2014), a critical component in the one-carbon metabolism pathway providing methyl groups for a range of biochemical reactions, including methylation of DNA and histones. How folate affects NT formation is still poorly understood, especially for human-specific aspects (Wallingford et al., 2013), but major evidence points to changes in DNA methylation and gene expression (Copp and Greene, 2012).
Epigenomic remodeling plays a major role in development and cell fate determination and alterations to DNA methylation and histone modifications, here referred to as epigenetics, have been associated with disease (Romanoski et al., 2015). To date, we are still missing a comprehensive understanding of epigenetic mechanisms for neural tube formation and NTDs. Recently, epigenomic dynamics of neural differentiation from hESCs were investigated using monolayer culture systems, namely 2D NSCs, (Xie et al., 2013; Ziller et al., 2015). However, 2D culture systems lack the complexity to recapitulate morphogenetic properties of the neural tube, a key aspect to understand how its disruption leads to NTDs. Here, we leverage the 3D nature of hESC-derived NRs to explore the epigenomic regulation of NT formation and possible applications for studying NTDs and other neurological disorders connected with early development.
RESULTS
Distinct neural rosette transcriptional profiles compared to 2D neural stem cells
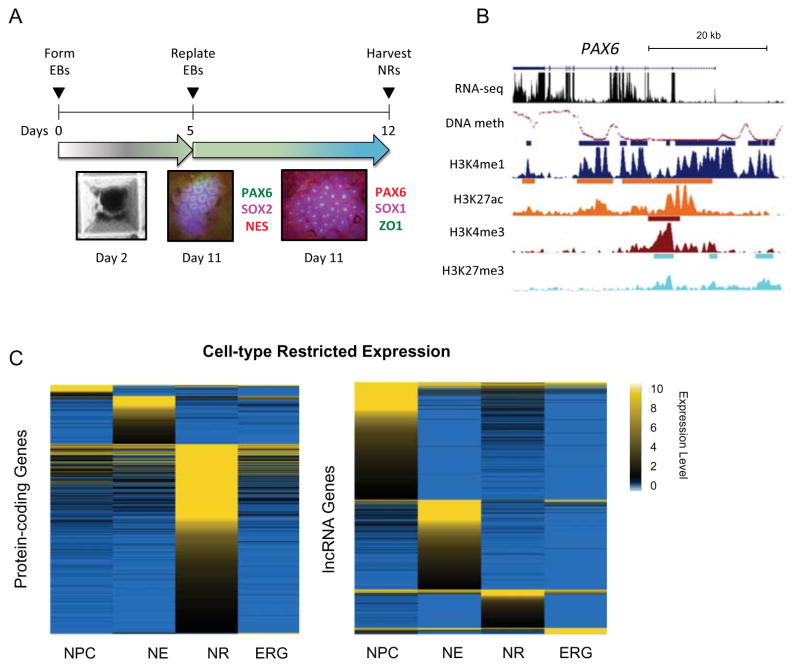
Neural rosettes (NRs) were generated from hESCs via formation of embryonic bodies (EBs) (Figure 1A, Supplemental Experimental Procedures). To gain insights into the role of epigenomic regulation in defining NR identity and neural tube formation, we generated comprehensive maps of gene expression by strand-specific RNA-sequencing (RNA-seq); histone H3 modifications via ChIP-seq that are commonly used to predict promoter and enhancer elements and their functional status - lysine 4 trimethylation (H3K4me3) and monomethylation (H3K4me1), lysine 27 acetylation (H3K27ac) and trimethylation (H3K27me3); and genome-wide DNA methylation by whole genome bisulfite sequencing (WGBS) (Figure 1B). To explore molecular features that set apart NRs from 2D NSCs, we compared the NR transcriptome and epigenome to publicly available data for three 2D NSCs that match the developmental window of NRs, namely neural progenitor cells (NPCs) (Xie et al., 2013), neuroepithelia (NE) and early radial glia (ERG) (Ziller et al., 2015).
Figure 1. Distinct neural rosette transcriptional profiles compared to 2D neural stem cells.
(A) Top: schematic of the neural rosette differentiation protocol. Bottom: Immunostaining for neural markers in NRs at day 11 of differentiation. (B) UCSC Genome Browser snapshot of PAX6 locus (chr11:31,800,000–31,850,000) showing normalized signals of DNA methylation level (mCG/CG), RNA-seq (read count expressed as TPM), and ChIP-seq reads (RPKM, input normalized). ChIP-seq peak calls are shown as bars above each track. (C) Heatmap representation of expression levels (TPM) of entropy-based cell-type specific protein-coding (left) and lncRNA (right) genes. See related Supplemental Experimental Procedures, Figure S1 and Table S1.
We first focused on the NR transcriptome. Given that all cells will express key genes for neural stem cells and neurodevelopment, we used an entropy-based method (Schug et al., 2005) to define cell-type specific gene expression from our global transcriptional analysis. We identified 415 protein-coding genes and 122 long noncoding RNAs (lncRNAs) with expression restricted to NRs (Figure 1C and Table S1A). To validate our findings, we also assessed differential expression in a pairwise comparison fashion using Rank Product (Figure S1A and Table S1B), and observed high concordance with the entropy-based analysis, with 96% NR-restricted genes recovered in the top 10% for Rank Product for NR vs all 2D NSCs (Figure S1B and Table S1C). Among NR-restricted protein-coding genes, we found several genes whose function during neurulation can potentially influence NT formation. One such gene was T-box 3 (TBX3), known for its role in maintaining pluripotency in mouse ESCs (Niwa et al., 2009). Interestingly, recent evidence showed that TBX3 knockdown in human ESCs decreases neural rosette formation (Esmailpour and Huang, 2012), suggesting that it may also be important during neurulation. The expression of betaine-homocysteine methyltransferase (BHMT), a member of the one-carbon metabolism pathway, was also found to be restricted to NRs. Genetic variants in BHMT have been reported to influence the risk for spina bifida (Boyles et al., 2006; Morin et al., 2003). Another such gene was ephrin receptor A5 (EPHA5), which is involved in regulation of cell adhesion during cranial development. EphA5−/− mice display anencephaly, likely due to the cranial neural folds failing to fuse in the dorsal midline (Holmberg et al., 2000). This collection of genes is a strong indication of a neural tube phenotype.
Epigenomic landscapes at neural rosette promoters
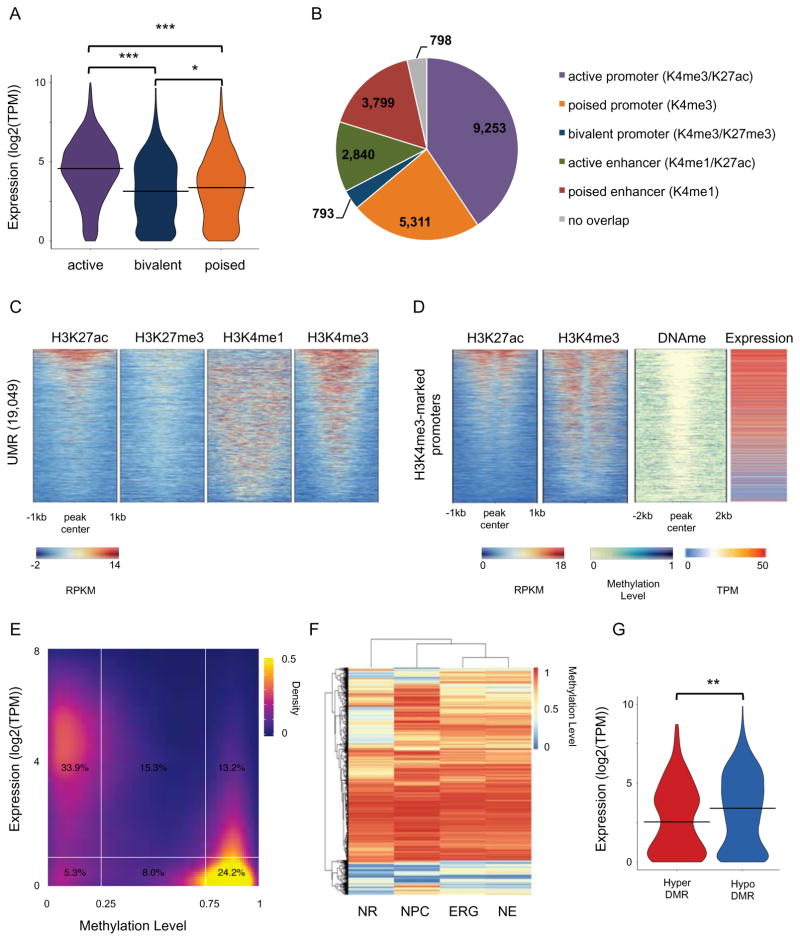
A large fraction of chromatin-enriched regions were unique to NRs with respect to the other cell populations (Figure S2A and Table S2), with the exception of H3K4me3. H3K4me3 marked 13,880 transcription start sites (TSS; gene-level annotations), compared to nearly 20,000 TSSs marked by H3K4me3 in 2D NSCs (Figure S2B). However, NR promoters were not devoid of chromatin dynamics. H3K27ac showed between 39% and 68% of peaks to be NR-unique in pairwise comparisons (Figure S2A and Table S2), of which 2,780 were found at promoters (Figure S2C). GO term analysis of these putative NR-specific active promoters suggested these genes are involved in general developmental functions (Figure S2D). H3K27me3 showed between 65% and 67% of peaks to be NR-unique (Figure S2A and Table S2). We identified 830 bivalent promoters (H3K4me3 and H3K27me3) in NRs (6% of all H3K4me3 marked regions), a signature that is a key process during lineage and cell type specification (Voigt et al., 2013), and observed that the NR bivalent signature was highly cell-type specific (Figure S2E). GO term analysis of NR bivalent genes showed enrichment for processes such as glycoprotein metabolism, metal ion binding, and voltage-gated channel activity (Figure S2F). The poised nature of the promoter chromatin state suggests a delayed developmental expression of these genes, potentially at a time when distinct neurons are specified. Next, we showed how different classes of H3K4me3-marked promoters, namely active (co-enriched for H3K27ac), bivalent (co-enriched for H3K27me3) and poised (no co-enrichment for H3K27ac or H3K27me3) correlated with the expression level of the genes (Figure 2A).
Figure 2. Epigenomic landscapes at neural rosette promoters.
(A) Violin plot of distribution of the expression values (log2(TPM)) of genes for active (H3K4me3/H3K27ac), poised (H3K4me3 only) and bivalent (H3K4me3/H3K27me3) promoters. *** T-test p-value < 0.0001. * T-test p-value ≤ 0.01. (B) Pie chart showing overlap of UMRs with chromatin-marked promoters and enhancers. (C) Heatmap representation of ChIP-seq signal intensity (RPKM, input normalized) for H3K4me1, H3K4me3, H3K27me3 and H3K27ac over 2kb windows centered at UMRs. (D) Heatmap representation of ChIP-seq signal intensity (RPKM, input normalized) for H3K4me3 and H3K27ac over 1kb windows centered at TSS, DNA methylation levels over 2kb windows centered at TSS, and gene expression (TMP) for H3K4me3-marked promoters. (E) Density plot showing gene expression (log2(TPM)) and DNA methylation levels for all promoters (defined as +/− 1kb around TSS; Gencode 19). (F) Heatmap and clustering visualization of aggregate mC levels (mCG/CG) in DMRs (delta-mC > 0.2) from all comparisons for positions with > 5X coverage in all cell types. 31,395 DMRs are shown in the heatmap. (G) Violin plot of distribution of the expression values (log2(TPM)) of genes which promoters harbour a DMR. ** T-test p-value < 0.001. See related Figure S2 and Table S2.
DNA methylation remodeling is an essential component of epigenetic regulation during development (Smith and Meissner, 2013). Recent studies showed that unmethylated and low-methylated regions (UMR/LMRs) can be used to predict regulatory elements (Burger et al., 2013; Stadler et al., 2011). UMRs are defined as regions with average methylation lower than 10%, are mostly found at promoters, and have a high CG density such as CpG islands (CGIs). We identified 19,049 UMRs with an average size of 2kb (Figure S2G) and confirmed their expected genomic locations (Figure S2H). Consistent with their genomic location, a larger fraction of UMRs colocalized with H3K4me3-marked promoters and encompassed chromatin-marked promoters in different states (Figure 2B). Contrasting UMRs with the chromatin maps, we observed that almost all UMRs were located at chromatin-marker regulatory region (Figure 2C). As for the UMRs that overlapped H3K4me1-marked regions, those are predominantly promoter-proximal enhancers. Consistently with their chromatin signature, H3K4me3-marked promoters showed to be largely demethylated while their genes appeared to be expressed (Figure 2D). This was consistent when we considered methylation at promoters genome-wide, where genes with demethylated promoters were expressed at a high level, while methylation in promoters (> 75%) corresponded to genes being undetected or expressed at a low level (TPM <1) (Figure 2E).
To compare NRs to 2D NSC methylomes, we defined differentially methylated regions (DMRs) for each pairwise comparison (Figure S2I). When considering regions that were differentially methylated in at least one of the three pairwise comparisons between NRs and 2D NSCs, we found 28,102 DMRs, whereas only 15,769 DMRs were found in pairwise comparisons among 2D NSCs, suggesting that DNA methylation signature in NRs is more different with respect to any 2D NSCs than 2D NSCs are among each other. Furthermore, NRs tend to harbor hypomethylated DMRs when compared to any 2D NSC (Figure 2F and Figure S2L) - 25,195 hypomethylated DMRs out of 28,102 DMRs. Of the 1,143 NR-specific promoter DMRs, 459 were hypomethylated and their genes showed significantly lower expression than genes of hypermethylated promoters (Figure 2G).
NR regulatory enhancer networks and dynamics during nervous system development
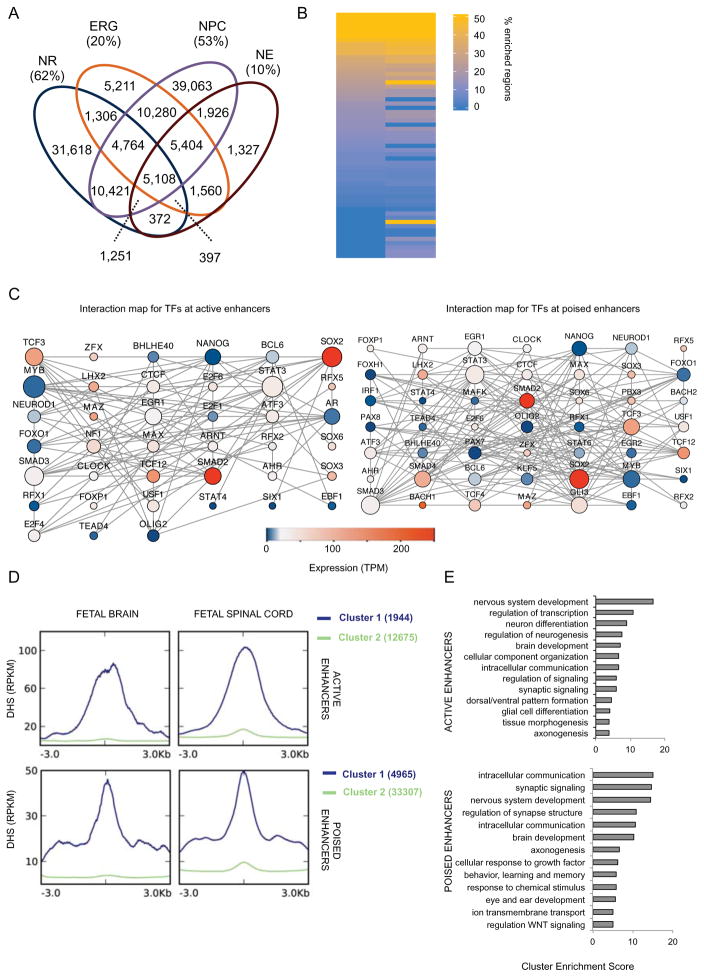
Enhancers play a major role in cell-type specific transcriptional regulation (Shlyueva et al., 2014). The H3K4me1 enhancer signature (Heintzman et al., 2009) was the most cell-type specific epigenomic modification when compared against all 2D NSC population, with 62% (38,181) peaks unique to NRs (Figure 3A). Co-enrichment of the activating H3K27ac modification with H3K4me1 reveals enhancers in an active state, while elements marked by H3K4me1 alone tend to be in a poised state (Creyghton et al., 2010; Hawkins et al., 2011; Rada-Iglesias et al., 2011). Using these criteria, we identified 22,475 active and 41,797 poised enhancers in NRs. Collectively, these results show that NRs have a quite distinct transcriptional and epigenomic landscape compared to 2D neural stem cells, and highlight a role for distal regulatory elements in defining NR epigenomic signature.
Figure 3. NR regulatory enhancer networks and dynamics during nervous system development.
(A) Venn diagram shows the overlap for all H3K4me1 peaks among NR, NPC, NE and ERG. Fraction of the cell-type unique peaks is also given as percentage of the total number of peaks. (B) Heatmap representation of number (expressed as percentage of the total) of enhancer regions were a motif was significantly enriched (q-value < 0.001). Only motifs whose TF was expressed (TPM>1) in NR are shown. See Table S3A for complete list. (C) In silico protein-protein interaction maps for TFs whose motifs are enriched at active (left) and poised (right) enhancers. Size of node is proportional to the number of interactions; color represents expression value (TPM) of the gene as measured by RNA-seq. (D) Distribution of the average enrichment of DHS signals (RPKM) over 6kb windows centered at active (top) and poised (bottom) enhancers. K-mean clustering (K=2) was used to identify enhancers enriched for DHS (cluster 1, in blue). Cluster 2, in green, represents enhancers lacking enrichment for DHS. (E) Bar plots show the most significantly enriched clusters of GO terms associated with NNGs in Cluster 1 of D. See Table S3C-D for NNGs of active and poised enhancers, respectively. See related Figure S3 and Table S3.
To predict the regulatory network shaping the enhancer landscape in NRs, we performed transcription factor binding site (TFBS) motif enrichment analysis on both active and poised enhancer regions (Figure 3B, Figure S3A and Table S3A-B) and constructed in silico protein-protein interaction maps for TFs with detectable transcripts in NRs (Figure 3C and Figure S3B). Our analysis suggests that SOX2 plays a central role in TF networks anchored at either active or poised enhancers. SOX2 is expressed throughout development and is involved in maintaining pluripotency as well as differentiation of several lineages (Sarkar and Hochedlinger, 2013). In NSCs, SOX2 is critical for maintaining neural progenitor identity by inhibiting premature differentiation toward the neuronal lineage (Graham et al., 2003; Lee et al., 2014). Another major hub in both networks is TCF3. TCF3 is a key player in controlling the balance between differentiation and maintenance of neural progenitors (Kuwahara et al., 2014) by antagonizing Wnt/β-Catenin signaling pathway (Atlasi et al., 2013). TCF3 is also necessary for establishing the anterior-posterior axis (Merrill et al., 2004). Among TF motifs uniquely enriched at active enhancers, we observed E2F4, a member of a family of TFs involved in regulating cell-cycle. E2f4−/− mice have brain malformations and abnormal fetal expression of key neural marker such as Pax6, Nkx2.1, and Dlx (Swiss and Casaccia, 2010). Among TF motifs uniquely enriched at poised enhancers is the transcriptional repressor GLI3, a mediator of the Shh signaling pathway. Gli3 mediates the graded Shh signal (Stamataki et al., 2005) and is essential for establishing dorsal-ventral patterning in the neural tube (Persson et al., 2002). Given GLI3 is expressed in NRs, its enrichment at poised enhancers may shed light on the dynamic state of these elements
Next, we asked how the NR enhancer landscape might change during nervous system development. To address this question we obtained DNase I hypersensitivity sites (DHS) data for human fetal spinal cord (15 weeks of gestation) and brain (18 weeks of gestation) from the Roadmap Epigenome Project (Thurman et al., 2012). We found that approximately 13% of both active and poised enhancers are hypersensitive at these later stages in neurodevelopment, suggesting they remain or become active (Figure 3D – Clusters 1). We assigned hypersensitive enhancers to the nearest neighboring genes (NNGs) (Table S3C-D) and performed functional annotation clustering analysis of associated GO terms (Figure 3E). For those active NR enhancers that are hypersensitive during brain and spinal cord development, top enriched clusters suggested these genes may play a role in more general neurodevelopmental processes, for instance transcription factor activity and neuron differentiation. Top GO term clusters for poised NR enhancers, that potentially become active as the brain and spinal cord form, were instead enriched for terms linked to synapse signaling and axonogenesis. These latter functions, associated with more mature neuronal activities, are consistent with the poised for activation state of the enhancers. To validate these findings, we also used an alternative method for assigning regulatory elements to putative gene targets, GREAT (McLean et al., 2010), and observed more than 72% concordance with NNGs (Figure S3C); GO term analysis of GREAT assigned putative targets showed high similarity as well (Figure S3D).
Broad distal regulatory domains reveal master regulators of neural tube formation
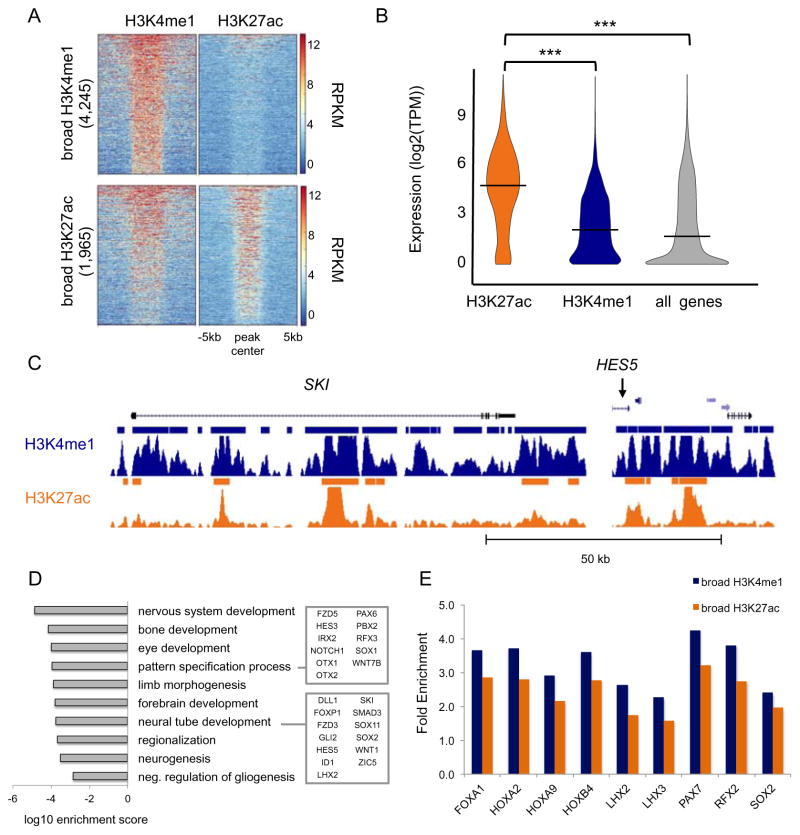
Recently a great deal of interest has emerged around broad distal regulatory domains – comprising sites stitched together, namely super-enhancers (Hnisz et al., 2013; Whyte et al., 2013) or stretch enhancers – defined largely by broad acetylation peaks (Parker et al., 2013). Besides sharing all the properties of average sized enhancers (commonly considered less than 1kb), super-enhancers are often located in close proximity to known master regulators, e.g. pluripotent genes and oncogenes, and are deemed to act as switches to determine cell fate. To identify broad enhancers, and their putative target genes based on genomic proximity, we first defined broad H3K4me1 and H3K27ac domains as peaks longer than 3kb and identified 4,245 (7%) and 1,964 (6%) peaks, respectively (Figure S4A). While most broad H3K27ac peaks were co-enriched for H3K4me1, indicating enhancers in an active state, a large fraction of broad H3K4me1 domains appeared to exist in a poised state (Figure 4A). Consistent with their functional state, active broad enhancers were largely found near highly expressed genes, while expression values of nearest genes to poised broad enhancers were significantly lower (Figure 4B). All expressed genes were compared as a control.
Figure 4. Broad distal regulatory domains reveal master regulators of neural tube formation.
(A) Heatmap representation of the ChIP-seq signal intensity (RPKM, input normalized) over a 10kb window centered at H3K4me1 and H3K27ac peaks across broad H3K27ac peaks (top) and broad H3K4me1 peaks (bottom). (B) Violin plot of distribution of the expression values (log2(TPM)) of the NNGs for broad H3K4me1 (blue) and H3K27ac (orange) peaks. Expression of all genes detected by RNA-seq in NRs is shown as a control. *** ANOVA p-value < 0.0001. (C) UCSC Genome Browser snapshots of SKI and HES5 loci. ChIP-seq signals (RPKM, input normalized) for H3K4me1 (blue) and H3K27ac (orange) are displayed. Solid bars indicate peaks called by MACSv1.4. (D) Bar plot shows the most significantly enriched GO terms associated with NNGs of broad enhancers. (E) Bar plot of TFBS motifs significantly enriched (q-value < 0.001) at broad enhancers versus standard enhancers. Enrichment at broad H3K4me1 (blue) and H3K27ac (orange) regions is expressed as ratio between fraction of broad regions and fraction of standard regions enriched for the given TFBS motif. See related Figure S4 and Table S4.
Among putative targets of active broad enhancers were SKI and HES5 (Figure 4C; see Table S4A-B for complete list). HES5 is a primary effector of the Notch signaling pathway and plays a critical role in neural tube development (Louvi and Artavanis-Tsakonas, 2006; Yoon and Gaiano, 2005). In neural rosettes, HES5 prevents the switch from neural to neuronal identity (Abranches et al., 2009). Similarly, SKI functions as a repressor of TGF-beta signaling and may play a role in neurulation and patterning of the neural tube (Berk et al., 1997). Overall, GO term analysis showed that putative targets are primarily involved in processes specific to neural tube formation, such as establishment of rostral-caudal axis and regionalization of the nervous system (Figure 4D). Using GREAT to assign broad enhancers to putative gene targets, we observed over 75% concordance with NNG method (Figure S4B).
Next, we explored which TFs were more likely to be found at broad enhancers than at standard enhancers. We performed TFBS motif enrichment analysis of both active and poised broad enhancers using standard peaks as background (Figure 4E and Table S4C-D). We found that LHX2 and HOXA9 motifs are enriched at both broad enhancer classes. Both factors are known to co-localized with Sox2 during early neurodevelopment in mice and are critical determinants of regional-specific Sox2 occupancy. For example, Lhx2 is more enriched in rostral regions (anterior fate) and HoxA9 is more enriched in caudal regions (posterior fate) (Hagey et al., 2016). While SOX2 and LHX2 genes are highly expressed in NRs, HOXA9 was not detected in NRs. The co-presence of motifs for both anterior and posterior factors, may contribute to a key feature of NRs, which is an anterior identity while possessing the capability of responding to posterior patterning cues (Conti et al., 2005; Elkabetz et al., 2008; Pankratz et al., 2007). Taken together, these results depict the use of broad enhancers near master regulators that underlie key properties of NRs, such as dependency on Notch signaling for maintaining neural stem cell identity and unrestricted differentiative potentials. At the same time, NRs, like the neural tube, may be primed to respond to imminent regionalization signaling by the likely accessibility of key TFs, such as HOXA9 at broad enhancers.
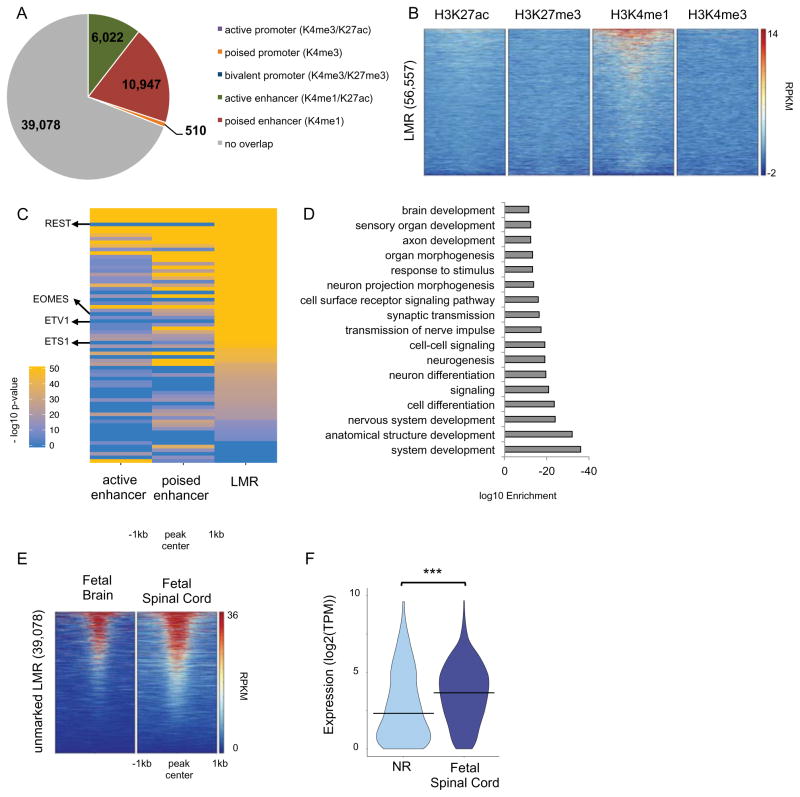
LMRs predict distal regulatory elements and their developmental dynamics
LMRs have an average methylation ranging from 10% to 50%, are low CG dense regions, tend to be enriched for H3K4me1, DHS as well as for p300/CBP, and are primarily located distal to promoters in intergenic or intronic regions (Burger et al., 2013; Stadler et al., 2011). We identified 56,558 LMRs (500bp average size) in the NR methylome (Figure S2G) and determined their genomic annotations (Figure S5A). We contrasted LMRs with the NR-specific hypomethylated DMRs. We found that NR hypomethylated DMRs largely occur at LMRs (48% of 2,995 hypo-DMRs common across all three pairwise comparisons against 2D NSCs; Figure S5B), whereas only a small fraction (6%) were found to be at UMRs. This was consistent with the observation that NR hypomethylated DMRs largely occur in intergenic and intronic regions (Figure S5C) and are enriched for H3K4me1 and to a lesser extent for H3K27ac, but not for H3K4me3 (Figure S5D). We then integrated all LMRs with our chromatin maps (Figure 5A–B), and as expected based on their genomic localization, LMRs were depleted of promoter mark H3K4me3, while they were enriched for the enhancer mark H3K4me1.
Figure 5. LMR predictions of distal regulatory domains and their developmental dynamics.
(A) Pie chart showing the overlap of LMRs with chromatin-marked promoters and enhancers. (B) Heatmap representation of the ChIP-seq signal intensity (RPKM, input normalized) for H3K4me1, H3K4me3, H3K27me3 and H3K27ac over 2kb windows centered at LMRs. (C) Heatmap representation of negative log10(p-value) for significantly (q-value < 0.01) enriched TF binding site motifs at active enhancers, poised enhancers and unmarked LMRs. Only motifs whose TF was expressed (TPM>1) in NR are shown. See Table S5B. (D) Bar plot of the most significantly enriched GO terms associated with NNGs of unmarked LMRs. See also Table S5C. (E) Heatmap representation of the DHS signal (RPKM) for human fetal brain and spinal cord (18 and 15 weeks of gestation, respectively) over 2kb windows centered at unmarked LMRs. (F) Violin plot of expression (log2(TPM)) of NNGs in NRs and fetal spinal cord (15 weeks of gestation) for unmarked LMRs where REST binding site motif was found to be significantly enriched. *** T-test p-value < 0.0001. See related Figure S5 and Table S5.
The larger fraction of LMRs was devoid of chromatin marks. It has been suggested that TF binding may be both necessary and sufficient to induce local demethylation and establish LMRs (Stadler et al., 2011). We asked whether unmarked LMRs might represent distal regulatory regions that are poised to become active at a later stage in development, but have yet to acquire indicative chromatin marks. To explore this hypothesis, we first sought to explore whether these elements might be able to bind TFs relevant for neural differentiation. Thus, we performed TFBS motif enrichment analysis on the unmarked LMRs (Table S5A) and compared to active and poised enhancers (Table S3A-B). We found that unmarked LMRs were enriched for a similar set of motifs to those found at enhancers (Figure 5C and Table S5B), with 73% of motifs shared with enhancers (Figure S5E), suggesting that these regions have the ability to function as distal regulatory elements.
Among the top enriched motifs in unmarked LMRs, but not found in H3K4me1-marked enhancers, was the motif for the repressor REST, known for its role in maintaining neural stem cells and preventing premature neuronal differentiation (Huang et al., 2011) The presence of repressive TF motifs at LMRs may prevent these putative poised enhancers from activating, and also explain why some LMRs lack an enhancer chromatin signature.
Other motifs enriched only in unmarked LMRs were for EOMES/TBR2 and members of the ETS family. EOMES/TBR2 is well characterized for its role in controlling differentiation in radial glia towards neurons during early brain development (Arnold et al., 2008; Sessa et al., 2008). Eomes/Tbr2 also mediates Pax6 control of the balance of NSC self-renewal and neurogenesis (Sansom et al., 2009). The ETS family of TFs are known for their role in regulating a spectrum of developmental processes. For example, Etv1 is involved in differentiation of dopaminergic neurons (Flames and Hobert, 2009) and cerebellum granule cells (Abe et al., 2011), while Ets1 plays a role in neural crest differentiation (Wang et al., 2015).
Next, we assigned unmarked LMRs to NNGs (Table S5C). As validation, 85% of putative target genes identified by GREAT were also identified by NNG approach (Figure S5F). Putative targets of unmarked LMRs were highly involved in development of the nervous system (Figure 5D; Figure S5G). Interestingly, the GO terms associated with these genes (e.g. transmission of the nerve impulse and axonogenesis) are indicative of a function at a later stage of development relative to neural tube formation, such as when neurons are formed, which would be consistent with a poised regulatory element.
Furthermore, we contrasted unmarked LMRs with fetal brain and spinal cord DHS and found that over half (23,448 of 39,078) are hypersensitive at later stages in development (Figure 5E). To further supporting the poised for activation state of unmarked LMRs, we compared the expression of putative target genes of unmarked LMRs containing the REST motif between NRs and fetal spinal cord and found that the expression of these genes significantly increased at a later stage in development (Figure 5F). Upregulation of these genes suggest the elements switch to an active state.
Collectively, these results support our hypothesis that unmarked LMRs may predict distal regulatory elements prior to acquiring chromatin features more commonly used to identify enhancers. Given that NRs and other 2D NSCs express a large repertoire of neural TFs that are present throughout development (see above and Ziller et al., 2015), these factors may establish poised enhancers (both H3K4me1-only and LMR-only elements), which activate upon localization of additional cell-specific factors.
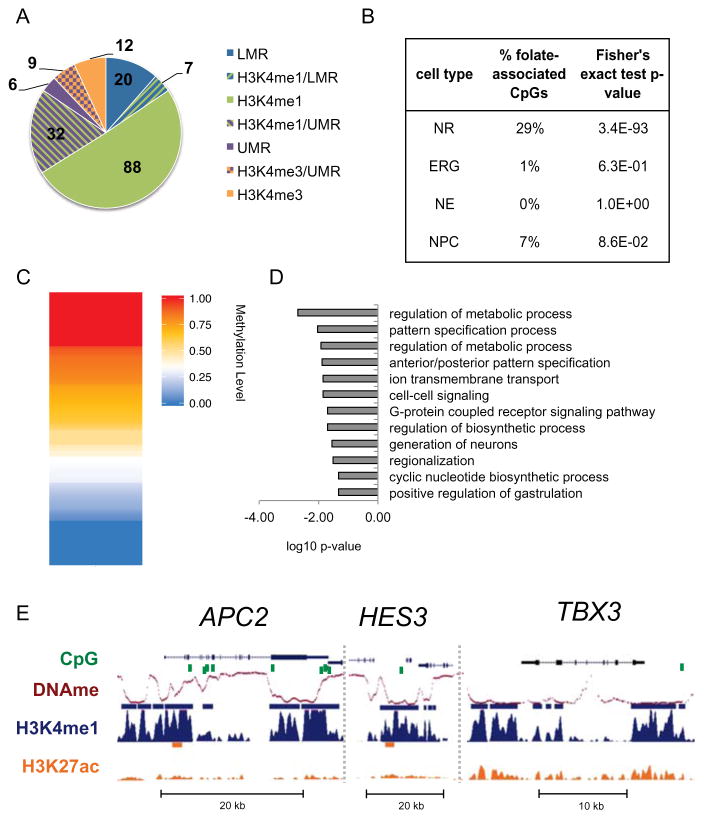
Folate-associated CpGs overlap NR regulatory elements
High levels of folate during the periconceptional period are the most effective means to reduce the risk of neural tube defects (NTDs) (Zaganjor et al., 2015). Recently, a large study correlated a wide range of maternal folate levels during early stages of pregnancy with DNA methylation in cord blood at birth in 1,988 healthy mother-newborn pairs (Joubert et al., 2016). Methylation levels at 443 individual CpGs were correlated with folate levels. We sought to explore how our model of NT formation could unravel the regulatory mechanism whereby folate-associated methylation influences neural tube formation. To do so, we assayed the genomic context for the 443 folate-associated CpGs and found that most were located in intergenic or intronic regions, where enhancers are more likely to be located (Figure S6A). About 40% of the folate-associated CpGs are at a regulatory element, with 3/4 of those overlapping H3K4me1-marked enhancers (Figure 6A). By contrast, overlapping folate-associated CpGs with 2D NSC H3K4me1-marked enhancers showed little to no overlap (Figure 6B), supporting a potential functional relevance of these CpGs in neural tube formation. Analysis of the folate-associate CpGs at NR regulatory elements showed that about half of the folate-associate CpGs are indeed demethylated (methylation level < 50%) in the NR methylome (Figure 6C).
Figure 6. Folate-associated CpGs overlap NR regulatory elements.
(A) Pie chart showing overlap of 174 folate-associated CpGs from Joubert et al., 2016 and NR regulatory elements. Fisher’s exact test log10 p-value < -5 for all overlaps shown. (B) Percentages of the 443 folateassociated CpGs identified by Joubert et al., 2014 overlapping enhancers from NRs and 2D NSCs. Statistical significance of the overlap was calculated using Fisher’s exact test. (C) Heatmap representation of methylation level at the 174 folate-associated CpGs that overlap NR regulatory elements. (D) Bar plot of the most significantly enriched GO terms associated with NNGs of regulatory elements that overlap folate-associated CpGs. (E) UCSC genome browser snapshots of ChIP-seq signals (RPKM, input normalized) for H3K4me1, H3K4me3, H3K27me3 and H3K27ac at some loci that contain folate-associated CpGs. Peaks calls are shown as bars above each track. See related Figure S6 and Table S6.
In line with these results, GO term analysis (Table S6A) of NNGs of the folate-associated CpGs at NR regulatory elements showed enrichment for neurodevelopment and metabolic processes (Figure 6D; Table S6B-C). 73% of NNGs were validated using GREAT and GO term analysis of GREAT putative targets showed high concordance (Figure S6B–C). We found that over half of the putative target genes for folate-associated CpGs at NR regulatory elements are expressed in NRs (TPM > 1; Figure S6D). Among putative target genes (Figure 6E), we found TBX3, a gene with expression restricted to NRs. TBX3 is associated with several GO terms related to neural tube patterning and axis establishment. Another related gene was FOXA2, a transcriptional activator and effector of SHH signaling. Several genetic models showed that this pathway is important for proper neural tube closure and disruption of Shh signaling leads to NTDs (Murdoch and Copp, 2010). In mice, Foxa2 mediates Shh signaling in the floor plate, a critical organizing center for neural tube patterning (Cho et al., 2014; Mansour et al., 2011). Given the poised state of putative FOXA2 enhancers and that the gene is not expressed in NRs (TPM < 1), it may be possible that this gene is activated at a later stage of NT development. Notably, the APC2 locus harbors nine individual folate-associate CpGs, eight of which are within NR enhancers. APC2 is involved in the regulation of WNT signaling and is expressed in both the fetal and adult human brain. A study in mice showed an association between folate and DNA methylation in its homologous gene, APC (Sie et al., 2013).
It is worth noting that several NNGs of enhancers that contain folate-associate CpGs are implicated in signaling pathways that regulate neural tube axis establishment, that is dorsal-ventral and rostral-caudal axes. This process likely plays an important role in NT closure and NTDs. NT closure is a continuous process along the tube and key signaling pathways, such as SHH and Notch pathways, drive this process and play a role in regulating timing of closure along the NT (Greene and Copp, 2009). Interestingly, we observed enhancers that overlapped folate-associate CpGs are highly enriched for broad enhancers (28 out of 127; Fisher’s test p-value = 3.8e-32), over three times more than expected, given that approximately 6–7% of all NR enhancers are broad. Among the putative target genes of broad enhancers that contain a folate-associate CpG is HES3, a regulator of the Notch signaling pathway, required to maintain neural stem cell population by inhibiting premature differentiation into neurons (Imayoshi et al., 2010) (Figure 6E). Mouse studies showed that Hes3 mutants develop NTDs (Hirata et al., 2001), likely as results of premature neural differentiation before neural tube closure is complete (Copp and Greene, 2012).
These results reveal insights into mechanisms by which folate may influence neurodevelopment by altering methylation levels at distal regulatory elements such as enhancers. Furthermore, it demonstrates the functional relevance of our in vitro 3D model of human neural tube formation, which failed to be captured when analyzing regulatory maps in 2D NSCs (Figure 6B).
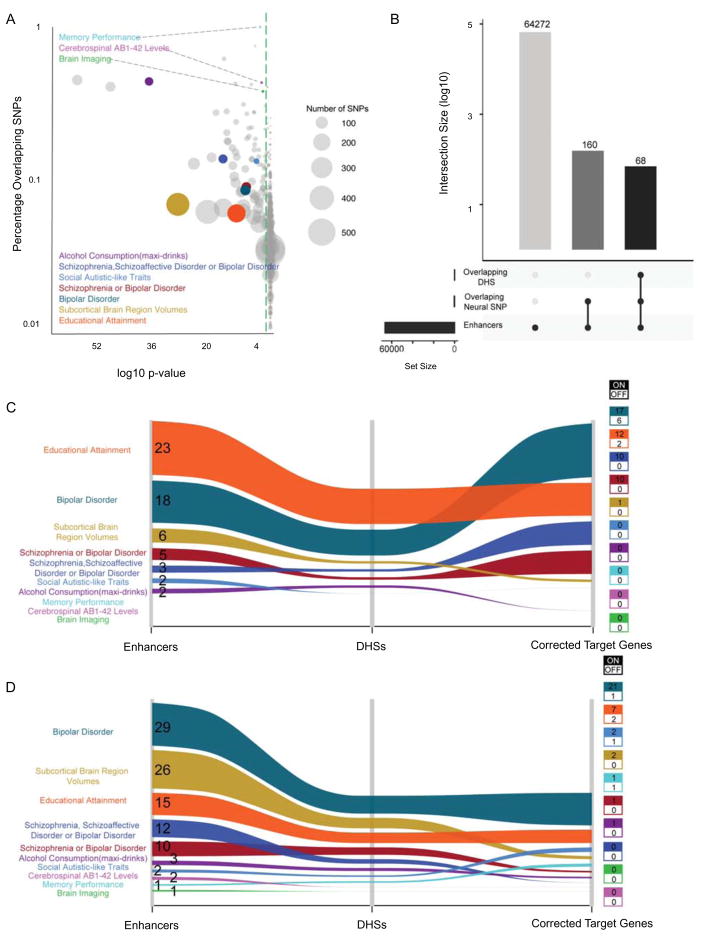
NR enhancers are enriched for genetic variants associated with neurological disorders
Recent studies have shown that non-coding single nucleotide polymorphisms (SNPs) associated with risk for disease may be enriched at enhancer elements (Corradin and Scacheri, 2014; Hawkins et al., 2010b; 2013). Given the limited genetic studies for NTDs that have resulted in very few candidates for this condition, a large collection of NTD-associated SNPs is not available for analysis at NR enhancers. Nonetheless, we asked whether we could observe enrichment for other neurological disorders or conditions potentially founded in neurodevelopment. To do so, we collected SNPs associated with a variety of neurological traits and other conditions from public data (573 traits from the NHGRI-EGI GWAS Catalog: https://www.ebi.ac.uk/gwas/) and examined their overlap within NR enhancers (Table S7A-B). We screened both lead SNPs and SNPs in linkage disequilibrium (r2 ≥ 0.8) for localization at NR enhancers. Our analysis showed that for approximately 20% of these traits (123 out of 573) a significant enrichment (q-value < 0.05) of the associated SNPs was found at NR enhancers, of which 10 were neurological disorders and traits, including Schizophrenia and Autism (Figure 7A). For both these neurological disorders, an increasing body of evidence is emerging supporting a neurodevelopmental etiology (Donovan and Basson, 2016; Stachowiak et al., 2013).
Figure 7. NR enhancers are enriched for genetic variants associated with neurological disorders.
(A) Bubble chart showing traits and conditions from public data (https://www.ebi.ac.uk/gwas/) for which both lead SNPs and SNPs in linkage disequilibrium (r2 ≥ 0.8) were found inside NR enhancers. Circle size represents the number of SNPs overlapped by enhancers for each trait. Colored circles represent traits associated with neurological disorders. Grey circles represent traits not associated with neurological disorders. Green dashed line indicates p-value = 0.05 (hypergeometric test with Bonferroni correction). (B) Bar chart shows number of enhancers overlapping neural related SNPs and DHS (with associated target genes) as represented by bar height. Overlapping conditions are shown by linked dots below bar. (C and D) Alluvial plots showing the number of active (C) and poised (D) enhancers overlapped by neural condition SNPs, overlapping DHS with gene targets and the count of target genes. Colored boxes show the number of target genes expressed (on = TPM > 1) or off.
From the 10 enriched neurological disorders and traits, we found their overlapping SNPs to reside in 120 NR enhancers (Figure 7B), 59 of which were active (Figure 7C) and 101 were in a poised state (Figure 7D). Next, we sought to identify the putative target genes for the 120 NR enhancers. We leveraged a recent study that provides prediction of target genes for over 500,000 putative regulatory elements using correlations between DHS signal and gene expression levels across 72 human cell types (Sheffield et al., 2013). This allowed us to predict gene targets for 68 NR enhancers (Figure 7B). Among both active and poised enhancers, we observed that vast majority of putative target genes were expressed in NRs (Table S7B; Figure 7C–D). For example, among predicted gene targets expressed in NRs was autism susceptibility candidate 2 gene (AUTS2), a gene implicated in neurodevelopment and neurological disorders, including autism spectrum disorders, intellectual disability, and developmental delay (Oksenberg and Ahituv, 2013). Notably, SNPs associated with subcortical brain region volumes are more abundant at poised enhancers and their target genes are largely off in NRs. Altered activation of these enhancers and target genes may provide insight into encephalopathies.
Overall, our results suggested that our model can be useful for evaluating the impact of genetic risk factors for neurological disorders associated with dysfunction of early development. For variants at active enhancers, our results likely reveal insights into the developmental origins of these disorders. Given that just over half (63%) of the enhancer SNPs are within poised enhancers, the impact of these variants is likely realized later in development.
DISCUSSION
We generated an initial set of transcriptomic and epigenomic maps in a 3D human NSC culture system to build a comprehensive regulatory framework of hESC-derived neural rosettes, which largely recapitulate neural tube development unlike 2D systems. A key feature of NRs is their unrestricted differentiative potential within the neural lineage (Elkabetz et al., 2008; Koch et al., 2009). Maintaining neural stemness and inhibiting premature progression toward neuronal fate is critical. Our results show that these key properties may be mediated by epigenomic features. Examination of in silico TF-TF interaction maps derived from putative enhancer binders revealed two important nodes, which were occupied by SOX2 and TCF3. Both of which are known to play a role in maintaining neural stem cell identity (Atlasi et al., 2013; Graham et al., 2003; Kuwahara et al., 2014). Similarly, repression of TGF-beta signaling and activation of Notch signaling (Louvi and Artavanis-Tsakonas, 2006; Yoon and Gaiano, 2005) are two fundamental events in early neurodevelopment and establishing NR identity. Our study suggests that a repressor of TGF-beta signaling, SKI, and an effector of Notch signaling, HES5, may be transcriptionally regulated via broad enhancers, which resemble super/stretch enhancers typically enriched near master regulators.
Another key feature of NRs is the anterior identity while possessing the capability of responding to patterning cues (Conti et al., 2005; Elkabetz et al., 2008; Pankratz et al., 2007). This developmental potential and ability to respond to signaling cues may be mediated by the largely poised enhancer landscape. We found that broad enhancers, predominantly poised, were highly enriched in proximity to genes involved in neural tube regionalization. Furthermore, we identified motifs for TFs associated with the anterior and posterior phenotype at enhancers. The regulatory networks anchored in chromatin-marked regulatory elements provides a better understanding of the epigenetic mechanisms guiding gene regulation, whereby patterning of the neural tube is established and orchestrated.
Little is known about the molecular mechanisms that link maternal folate levels to mechanisms of neurodevelopment and malformations such as NTDs during the early stages of human embryogenesis (Nazki et al., 2014). Our study demonstrates the power of human in vitro modeling to fill this gap. This study highlights several major points. Our findings clearly demonstrate that NRs are a more relevant model for studying epigenetic mechanisms of folate, as we found little to no overlap between folate-associated CpGs and 2D NSC enhancer maps. This provides a framework to explain how folate-associated effects on DNA methylation may in turn influence NT formation and NTD. Moreover, enhancers are likely a critical mediator of mechanisms that link folate to NT formation. Not only are the majority of reported folate-associated CpGs enriched outside of promoters and genic regions, but, more specifically, our findings show that three quarters of those overlapping NR regulatory map are at enhancers, as defined by histone modifications or a DNA methylation signature. This is in line with the ever-increasing understanding of the central role that enhancers play in development and predisposition to disease. Future studies that make using of genome-wide DNA methylation maps are likely to reveal more extensive folate-associated CpGs, as currently studies have utilized array technologies with minimal CpG representation.
Our study also provides insights into other neurodevelopment related disorders and traits. Enhancer variants offer an unprecedented opportunity to unravel how genetic variants in the non-coding genome can influence risk for many diseases and traits. Cell-type specific contexts are key for assessment of the effects of enhancer variants. Representing an early stage of neurodevelopment, NRs provide a useful tool for investigating neurological disorders and traits that may have a developmental origin, like Schizophrenia and Autism. Furthermore, we were able to identify their putative target genes and found among them genes involved in autism spectrum disorders, intellectual disability, and developmental delay (Oksenberg and Ahituv, 2013). In conclusion, this study provides comprehensive epigenomic maps of regulatory elements in neural rosettes and supports the use of this neural stem cells population as 3D human model for studying neural tube formation and NTDs.
EXPERIMENTAL PROCEDURES
For detailed descriptions see Supplemental Experimental Procedures.
NR differentiation
hES cells (WA09, WiCell) were cultured on Matrigel in mTeRS1 (Stemcell Technologies). NRs were differentiated for 5 days in STEMdiff™ Neural Induction Medium according to the manufacturer’s guides and AggreWell™ 800 plates (Stemcell Technologies) and then transferred to poly-ornithine and laminin coated plates and cultured in STEMdiff™ Neural Induction Medium for 7 days. NR 3D tubular-structures were harvested at day 12 of differentiation.
Genome-wide assays, Library Construction and Sequencing
ChIP-seq was performed as previously described (Hawkins et al., 2010a) using 2 million cells per ChIP. RNA-seq was performed using ScriptSeq™ v2 RNA-Seq Library Preparation Kit (Epicentre). WGBS was performed as previously described (Lister et al., 2009) with minor modifications using 500 ng of genomic DNA per library. Libraries were sequenced on either NextSeq500 in single-end 75 cycles run or HiSeq2500 in paired-end 100 cycles runs. All experiments were performed in duplicates.
Computational Analysis
Sequencing raw reads were trimmed for low quality (q score < 30) and adapters, mapped to human genome (hg19) using Bowtie2 (ChIP-seq), Kallisto (RNA-seq) and BBMap (WGBS). Duplicates were merged. Gene expression values were calculated as transcripts per million (TPM). Alternative pairwise differential expression analysis of RNA-seq was performed with RankProd R package. ChIP-seq peaks were called using MACS v1.4 using the --nomodel mode. Methylation was quantified using BS-Seeker2; DMRs were identified using DSS and UMRs and LMRs using MethylSeekR.
Publically available data used in this study
DHS data for human fetal brain (18 weeks gestation) and fetal spinal cord (15 week gestation) were obtained from the Roadmap Epigenome Project, accession number GSM595913 and GSM878661. Folate-associated CpGs coordinates were obtained from Joubert et al., 2016. SNPs for all available traits were downloaded from the NHGRI-EGI GWAS Catalog (https://www.ebi.ac.uk/gwas/). RNA-seq, ChIP-seq and WGBS raw data for NPCs was obtained from Xie et al., 2013; NE and ERG from Ziller et al., 2015.
STATISTICAL ANALYSIS
Statistical significance of differences in the distribution of expression values for active versus poised or bivalent genes as well as for promoters with hyper-versus hypo-DMRs were assessed using the T-test. Statistical significance of differences between expression of NNGs for broad H3K4me1 or H3K27ac domains and all genes expressed was performed using ANOVA. Significance of the overlap of folate-associated CpGs at broad enhancers was determined using a Fisher’s test. Statistically significant overlap of trait-associated SNPs with NR enhancers was assessed using a hypergeometric test with Bonferroni correction.
Supplementary Material
Highlights.
Neural rosettes have transcriptional and epigenomic landscapes distinct from 2D NSCs
Low methylated regions in NRs predict enhancers active later in neurodevelopment
NR regulatory maps provide insights into mechanisms of neural tube defects
Acknowledgments
We thank Marjo Hakkarainen, Päivi Junni and Bogata Fezazi for the technical assistance with hESC culture. This study was supported by the National Institutes of Health (NIH), Biocenter Finland, Academy of Finland, and the Washington Life Sciences Discovery Fund.
Footnotes
ACCESSION NUMBERS
The SRA accession number for the data generated on neural rosettes is SRP094721.
All data generated on NRs as well as data for 2D NSCs used in this study can be visualized in a UCSC genome browser session from here: http://depts.washington.edu/hawklab/ucsc_sessions/public/Neural_Rosette/site/nr_page.html
Supplemental Information includes Supplemental Experimental Procedures, six figures and seven tables.
AUTHOR CONTRIBUTIONS
C.V. and R.D.H conceived the study, designed the experiments and computational analysis and wrote the manuscript. C.V performed the experiments and analyzed data. C.A. contributed to data analysis and writing of the manuscript. S.B. and R.L. contributed to DNA methylation data analysis. N.D.J. contributed to data analysis. R.J.L. provided assistance with cell culture. All authors read, commented on, and edited the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abe H, Okazawa M, Nakanishi S. The Etv1/Er81 transcription factor orchestrates activity-dependent gene regulation in the terminal maturation program of cerebellar granule cells. Proc Natl Acad Sci U S A. 2011;108:12497–12502. doi: 10.1073/pnas.1109940108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abranches E, Silva M, Pradier L, Schulz H, Hummel O, Henrique D, Bekman E. Neural Differentiation of Embryonic Stem Cells In Vitro: A Road Map to Neurogenesis in the Embryo. PLoS ONE. 2009;4:e6286. doi: 10.1371/journal.pone.0006286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold SJ, Huang G-J, Cheung AFP, Era T, Nishikawa S-I, Bikoff EK, Molnár Z, Robertson EJ, Groszer M. The T-box transcription factor Eomes/Tbr2 regulates neurogenesis in the cortical subventricular zone. Genes Dev. 2008;22:2479–2484. doi: 10.1101/gad.475408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlasi Y, Noori R, Gaspar C, Franken P, Sacchetti A, Rafati H, Mahmoudi T, Decraene C, Calin GA, Merrill BJ, Fodde R. Wnt signaling regulates the lineage differentiation potential of mouse embryonic stem cells through Tcf3 down-regulation. PLoS Genet. 2013;9:e1003424–e1003424. doi: 10.1371/journal.pgen.1003424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk M, Desai SY, Heyman HC, Colmenares C. Mice lacking the ski proto-oncogene have defects in neurulation, craniofacial patterning, and skeletal muscle development. Genes Dev. 1997;11:2029–2039. doi: 10.1101/gad.11.16.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyles AL, Billups AV, Deak KL, Siegel DG, Mehltretter L, Slifer SH, Bassuk AG, Kessler JA, Reed MC, Nijhout HF, et al. Neural tube defects and folate pathway genes: family-based association tests of gene-gene and gene-environment interactions. Environ Health Perspect. 2006;114:1547–1552. doi: 10.1289/ehp.9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger L, Gaidatzis D, Schuebeler D, Stadler MB. Identification of active regulatory regions from DNA methylation data. Nucleic Acids Res. 2013;41:e155. doi: 10.1093/nar/gkt599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho G, Lim Y, Cho I-T, Simonet JC, Golden JA. Arx together with FoxA2, regulates Shh floor plate expression. Dev Biol. 2014;393:137–148. doi: 10.1016/j.ydbio.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti L, Cattaneo E. Neural stem cell systems: physiological players or in vitro entities? Nat Rev Neurosci. 2010;11:176–187. doi: 10.1038/nrn2761. [DOI] [PubMed] [Google Scholar]
- Conti L, Pollard SM, Gorba T, Reitano E, Toselli M, Biella G, Sun Y, Sanzone S, Ying Q-L, Cattaneo E, Smith A. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 2005;3:e283. doi: 10.1371/journal.pbio.0030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copp AJ, Greene NDE. Neural tube defects-disorders of neurulation and related embryonic processes. WIREs Dev Biol. 2012;2:213–227. doi: 10.1002/wdev.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradin O, Scacheri PC. Enhancer variants: evaluating functions in common disease. Genome Medicine. 2014;6:85. doi: 10.1186/s13073-014-0085-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proceedings of the National Academy of Sciences. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deglincerti A, Etoc F, Ozair MZ, Brivanlou AH. Essays on Developmental Biology Part A. Elsevier Inc; 2016. Chapter Six: Self-Organization of Spatial Patterning in Human Embryonic Stem Cells; pp. 99–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan APA, Basson MA. The neuroanatomy of autism - a developmental perspective. J Anat. 2016 doi: 10.1111/joa.12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkabetz Y, Panagiotakos G, Shamy Al G, Socci ND, Tabar V, Studer L. Human ES cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev. 2008;22:152–165. doi: 10.1101/gad.1616208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmailpour T, Huang T. TBX3 Promotes Human Embryonic Stem Cell Proliferation and Neuroepithelial Differentiation in a Differentiation Stage-dependent Manner. Stem Cells. 2012;30:2152–2163. doi: 10.1002/stem.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flames N, Hobert O. Gene regulatory logic of dopamine neuron differentiation. Nature. 2009;458:885–889. doi: 10.1038/nature07929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- Greene NDE, Copp AJ. Development of the vertebrate central nervous system: formation of the neural tube. Prenat Diagn. 2009;29:303–311. doi: 10.1002/pd.2206. [DOI] [PubMed] [Google Scholar]
- Hagey DW, Zaouter C, Combeau G, Lendahl MA, Andersson O, Huss M, Muhr J. Distinct transcription factor complexes act on a permissive chromatin landscape to establish regionalized gene expression in CNS stem cells. Genome Research. 2016;26:908–917. doi: 10.1101/gr.203513.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, Antosiewicz-Bourget J, Ye Z, Espinoza C, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010a;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Ren B. Next-generation genomics: an integrative approach. Nat Rev Genet. 2010b;11:476–486. doi: 10.1038/nrg2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Yang C, Antosiewicz-Bourget JE, Lee LK, Ngo Q-M, Klugman S, Ching KA, Edsall LE, Ye Z, Kuan S, Yu P, et al. Dynamic chromatin states in human ES cells reveal potential regulatory sequences and genes involved in pluripotency. Cell Res. 2011;21:1393–1409. doi: 10.1038/cr.2011.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins RD, Larjo A, Tripathi SK, Wagner U, Luu Y, Lönnberg T, Raghav SK, Lee LK, Lund R, Ren B, Lähdesmäki H, Lahesmaa R. Global Chromatin State Analysis Reveals Lineage-Specific Enhancers during the Initiation of Human T helper 1 and T helper 2 Cell Polarization. Immunity. 2013;38:1271–1284. doi: 10.1016/j.immuni.2013.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon GC, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Hirata H, Tomita K, Bessho Y, Kageyama R. Hes1 and Hes3 regulate maintenance of the isthmic organizer and development of the mid/hindbrain. EMBO J. 2001;20:4454–4466. doi: 10.1093/emboj/20.16.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, Young RA. Super-Enhancers in the Control of Cell Identity and Disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg J, Clarke DL, Frisén J. Regulation of repulsion versus adhesion by different splice forms of an Eph receptor. Nature. 2000;408:203–206. doi: 10.1038/35041577. [DOI] [PubMed] [Google Scholar]
- Huang Z, Wu Q, Guryanova OA, Cheng L, Shou W, Rich JN, Bao S. Deubiquitylase HAUSP stabilizes REST and promotes maintenance of neural progenitor cells. Nat Cell Biol. 2011;13:142–152. doi: 10.1038/ncb2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imayoshi I, Sakamoto M, Yamaguchi M, Mori K, Kageyama R. Essential roles of Notch signaling in maintenance of neural stem cells in developing and adult brains. J Neurosci. 2010;30:3489–3498. doi: 10.1523/JNEUROSCI.4987-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubert BR, Dekker den HT, Felix JF, Bohlin J, Ligthart S, Beckett E, Tiemeier H, van Meurs JB, Uitterlinden AG, Hofman A, et al. Maternal plasma folate impacts differential DNA methylation in an epigenome-wide meta-analysis of newborns. Nature Communications. 2016;7:10577–10577. doi: 10.1038/ncomms10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelava I, Lancaster MA. Protocol Review. Stem Cell. 2016;18:736–748. doi: 10.1016/j.stem.2016.05.022. [DOI] [PubMed] [Google Scholar]
- Koch P, Opitz T, Steinbeck JA, Ladewig J, Brüstle O. A rosette-type, self-renewing human ES cell-derived neural stem cell with potential for in vitro instruction and synaptic integration. Proceedings of the National Academy of Sciences. 2009;106:3225–3230. doi: 10.1073/pnas.0808387106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwahara A, Sakai H, Xu Y, Itoh Y, Hirabayashi Y, Gotoh Y. Tcf3 Represses Wnt-beta-Catenin Signaling and Maintains Neural Stem Cell Population during Neocortical Development. PLoS ONE. 2014;9:e94408. doi: 10.1371/journal.pone.0094408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KE, Seo J, Shin J, Ji EH, Roh J, Kim JY, Sun W, Muhr J, Lee S, Kim J. Positive feedback loop between Sox2 and Sox6 inhibits neuronal differentiation in the developing central nervous system. Proc Natl Acad Sci U S A. 2014;111:2794–2799. doi: 10.1073/pnas.1308758111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon GC, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- Mansour AA, Nissim-Eliraz E, Zisman S, Golan-Lev T, Schatz O, Klar A, Ben-Arie N. Foxa2 regulates the expression of Nato3 in the floor plate by a novel evolutionarily conserved promoter. Mol Cell Neurosci. 2011;46:187–199. doi: 10.1016/j.mcn.2010.09.002. [DOI] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhardt A, Eberle D, Tazaki A, Ranga A, Niesche M, Wilsch-Bräuninger M, Stec A, Schackert G, Lutolf M, Tanaka EM. 3D reconstitution of the patterned neural tube from embryonic stem cells. Stem Cell Reports. 2014;3:987–999. doi: 10.1016/j.stemcr.2014.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill BJ, Pasolli HA, Polak L, Rendl M, Garcia-Garcia MJ, Anderson KV, Fuchs E. Tcf3: a transcriptional regulator of axis induction in the early embryo. Development. 2004;131:263–274. doi: 10.1242/dev.00935. [DOI] [PubMed] [Google Scholar]
- Morin I, Platt R, Weisberg I, Sabbaghian N, Wu Q, Garrow TA, Rozen R. Common variant in betaine-homocysteine methyltransferase (BHMT) and risk for spina bifida. Am J Med Genet. 2003;119A:172–176. doi: 10.1002/ajmg.a.20115. [DOI] [PubMed] [Google Scholar]
- Murdoch JN, Copp AJ. The relationship between sonic Hedgehog signaling, cilia, and neural tube defects. Birth Defects Research Part A: Clinical and Molecular Teratology. 2010;88:633–652. doi: 10.1002/bdra.20686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazki FH, Sameer AS, Ganaie BA. Folate: Metabolism, genes, polymorphisms and the associated diseases. Gene. 2014;533:11–20. doi: 10.1016/j.gene.2013.09.063. [DOI] [PubMed] [Google Scholar]
- Niwa H, Ogawa K, Shimosato D, Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature. 2009;460:118–122. doi: 10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- Oksenberg N, Ahituv N. The role of AUTS2 in neurodevelopment and human evolution. Trends in Genetics. 2013;29:600–608. doi: 10.1016/j.tig.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankratz MT, Li XJ, LaVaute TM, Lyons EA, Chen X, Zhang SC. Directed Neural Differentiation of Human Embryonic Stem Cells via an Obligated Primitive Anterior Stage. Stem Cells. 2007;25:1511–1520. doi: 10.1634/stemcells.2006-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker SCJ, Stitzel ML, Taylor DL, Orozco JM, Erdos MR, Akiyama JA, van Bueren KL, Chines PS, Narisu N, Black BL, et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc Natl Acad Sci U S A. 2013;110:17921–17926. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson M, Stamataki D, Welscher te P, Andersson E, Böse J, Rüther U, Ericson J, Briscoe J. Dorsal-ventral patterning of the spinal cord requires Gli3 transcriptional repressor activity. Genes Dev. 2002;16:2865–2878. doi: 10.1101/gad.243402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanoski CE, Glass CK, Stunnenberg HG, Almouzni LWG. Epigenomics: Roadmap for regulation. Nature. 2015;518:314–316. doi: 10.1038/518314a. [DOI] [PubMed] [Google Scholar]
- Sansom SN, Griffiths DS, Faedo A, Kleinjan DJ, Ruan Y, Smith J, van Heyningen V, Rubenstein JL, Livesey FJ. The Level of the Transcription Factor Pax6 Is Essential for Controlling the Balance between Neural Stem Cell Self-Renewal and Neurogenesis. PLoS Genet. 2009;5:e1000511. doi: 10.1371/journal.pgen.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A, Hochedlinger K. The Sox Family of Transcription Factors: Versatile Regulators of Stemand Progenitor Cell Fate. Stem Cell. 2013;12:15–30. doi: 10.1016/j.stem.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa A, Mao CA, Hadjantonakis AK, Klein WH, Broccoli V. Tbr2 directs conversion of radial glia into basal precursors and guides neuronal amplification by indirect neurogenesis in the developing neocortex. Neuron. 2008;60:56–69. doi: 10.1016/j.neuron.2008.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug J, Schuller WP, Kappen C, Salbaum JM, Bucan M, Stoeckert CJ. Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biology. 2005;6:R33–R33. doi: 10.1186/gb-2005-6-4-r33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield NC, Thurman RE, Song L, Safi A, Stamatoyannopoulos JA, Lenhard B, Crawford GE, Furey TS. Patterns of regulatory activity across diverse human cell types predict tissue identity, transcription factor binding, and long-range interactions. Genome Research. 2013;23:777–788. doi: 10.1101/gr.152140.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: fromproperties to genome-wide predictions. Nat Rev Genet. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- Sie KKY, Li J, Ly A, Sohn KJ, Croxford R, Kim YI. Effect of maternal and postweaning folic acid supplementation on global and gene-specific DNA methylation in the liver of the rat offspring. Mol Nutr Food Res. 2013;57:677–685. doi: 10.1002/mnfr.201200186. [DOI] [PubMed] [Google Scholar]
- Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- Stachowiak MK, Kucinski A, Curl R, Syposs C, Yang Y, Narla S, Terranova C, Prokop D, Klejbor I, Bencherif M, et al. Schizophrenia: a neurodevelopmental disorder--integrative genomic hypothesis and therapeutic implications from a transgenic mouse model. Schizophr Res. 2013;143:367–376. doi: 10.1016/j.schres.2012.11.004. [DOI] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Schöler A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, Tiwari VK, Schübeler D. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- Stamataki D, Ulloa F, Tsoni SV, Mynett A, Briscoe J. A gradient of Gli activity mediates graded Sonic Hedgehog signaling in the neural tube. Genes Dev. 2005;19:626–641. doi: 10.1101/gad.325905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiss VA, Casaccia P. Cell-Context Specific Role of the E2F/Rb Pathway in Development and Disease. Glia. 2010;58:377–390. doi: 10.1002/glia.20933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt P, Tee W-W, Reinberg D. A double take on bivalent promoters. Genes Dev. 2013;27:1318–1338. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallingford JB, Niswander LA, Shaw GM, Finnell RH. The continuing challenge of understanding, preventing, and treating neural tube defects. Science. 2013;339:1222002–1222002. doi: 10.1126/science.1222002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Kam RKT, Shi W, Xia Y, Chen X, Cao Y, Sun J, Du Y, Lu G, Chen Z, Chan WY, Chan SO, Deng Y, Zhao H. The Proto-oncogene Transcription Factor Ets1 Regulates Neural Crest Development through Histone Deacetylase 1 to Mediate Output of Bone Morphogenetic Protein Signaling. J Biol Chem. 2015;290:21925–21938. doi: 10.1074/jbc.M115.644864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master Transcription Factors and Mediator Establish Super-Enhancers at Key Cell Identity Genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Schultz MD, Lister R, Hou Z, Rajagopal N, Ray P, Whitaker JW, Tian S, Hawkins RD, et al. Epigenomic Analysis of Multilineage Differentiation of Human Embryonic Stem Cells. Cell. 2013;153:1134–1148. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon K, Gaiano N. Notch signaling in the mammalian central nervous system: insights from mouse mutants. Nat Neurosci. 2005;8:709–715. doi: 10.1038/nn1475. [DOI] [PubMed] [Google Scholar]
- Zaganjor I, Sekkarie A, Tsang BL, Williams J, Razzaghi H, Mulinare J, Sniezek JE, Cannon MJ, Rosenthal J. Describing the Prevalence of Neural Tube Defects Worldwide: A Systematic Literature Review. PLoS ONE. 2015;11:e0151586–e0151586. doi: 10.1371/journal.pone.0151586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SC, Wernig M, Ducan ID, Brüstle O, Thomson JA. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat Biotechnol. 2001;19:1129. doi: 10.1038/nbt1201-1129. [DOI] [PubMed] [Google Scholar]
- Zhu X, Li B, Ai Z, Xiang Z, Zhang K, Qiu X, Chen Y, Li Y, Rizak JD, Niu Y, Hu X, Sun YE, Ji W, Li T. A Robust Single Primate Neuroepithelial Cell Clonal Expansion System for Neural Tube Development and Disease Studies. Stem Cell Reports. 2016;6:228–242. doi: 10.1016/j.stemcr.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller MJ, Edri R, Yaffe Y, Donaghey J, Pop R, Mallard W, Issner R, Gifford CA, Goren A, Xing J, et al. Dissecting neural differentiation regulatory networks through epigenetic footprinting. Nature. 2015;258:355–359. doi: 10.1038/nature13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.