Abstract
Smoking is the leading cause of death in the United States. It exerts its effects by increasing susceptibility to a variety of complex disorders among those who smoke, and if pregnant, to their unborn children. In prior efforts to understand the epigenetic mechanisms through which this increased vulnerability is conveyed, a number of investigators have conducted genome wide methylation analyses. Unfortunately, secondary to methodological limitations, these studies were unable to examine methylation in gene regions with significant amounts of genetic variation. Using genome wide genetic and epigenetic data from the Framingham Heart Study, we re-examined the relationship of smoking status to genome wide methylation status. When only methylation status is considered, smoking was significantly associated with differential methylation in 310 genes that map to a variety of biological process and cellular differentiation pathways. However, when SNP effects on the magnitude of smoking associated methylation changes are also considered, cis and trans-interaction effects were noted at a total of 266 and 4353 genes with no marked enrichment for any biological pathways. Furthermore, the SNP variation participating in the significant interaction effects is enriched for loci previously associated with complex medical illnesses. The enlarged scope of the methylome shown to be affected by smoking may better explicate the mediational pathways linking smoking with a myriad of smoking related complex syndromes. Additionally, these results strongly suggest that combined epigenetic and genetic data analyses may be critical for a more complete understanding of the relationship between environmental variables, such as smoking, and pathophysiological outcomes.
Keywords: smoking, DNA methylation, genetic variation, meQTL, AHRR, schizophrenia, attention deficit hyperactivity disorder, panic disorder
INTRODUCTION
Smoking is the leading preventable cause of mortality in the United States and most of the industrialized world.(Centers for Disease Control and Prevention 2008) Smoking does not exert its effects on mortality directly. Instead, smoking increases the likelihood that other “more traditional” causes of death, such as coronary heart disease (CHD), chronic obstructive pulmonary disease or stroke, will develop. Due to steady advances in genomics, and the unwavering efforts of a large number of investigators, the major pathophysiological mechanisms of each of these more traditional causes of death are fairly well established, allowing mechanistic connections between smoking and cause of death to be developed. With respect to cancer, the major mechanism through which smoking exerts its effects, benzo[a] pyrene mediated mutagenesis, on disease pathogenesis is well established.(Pfeifer 2016) However, for other complex disorders, the degree of our understanding through which smoking exerts its effects varies.
To better understand how smoking exerts its effects on each of these outcomes, a number of investigators have published genome wide analyses of the relationship of cigarette consumption to DNA methylation in peripheral white blood cells (WBC).(Joubert and others 2012; Monick and others 2012; Shenker and others 2012; Zeilinger and others 2013) These studies have advanced our understanding of disease pathogenesis. For example, for stroke, the prominent changes in the methylation signatures of clotting factors associated with smoking coupled with prior observations that smoking increases the activity of clotting factors in the serum provides a very plausible rationale to explain the two-fold increase risk for stroke conveyed by smoking.(Breitling and others 2012; Dogan and others 2014; Wannamethee and others 2005) Yet for other disorders, such as ectopic pregnancy (Bouyer and others 2003) and a host of psychiatric disorders (Aubin and others 2012; Bouyer and others 2003; Isensee and others 2003; Kendler and others 2015; Moylan and others 2013), these genome wide studies have been less illuminating with the result that the connection between smoking and increased risk for many other disorder is not well understood.
There is reason to believe that part of this failure of methylation approaches to explain the effects of smoking on disease risk may be secondary to the presence of confounding interaction effects involving genetic variation with methylation. An initial indication of the existence of such effects was our finding that the degree of smoking induced differential methylation at the most highly ranked methylation probes is strongly influenced by ethnicity, which in this instance is presumably ethnic specific genetic variation.(Dogan and others 2015; Smith and others 2014) In a series of earlier publications that examined the effect of smoking on DNA methylation at Monoamine Oxidase A (MAOA), for example, we found that genotype at the well-characterized promoter associated variable nucleotide repeat (VNTR) had a strong effect on DNA methylation across both of the promoter associated CpG islands, resulting in smoking contextual genetic effects on methylation (SNP*Smoke).(Philibert and others 2010; Philibert and others 2008) Subsequently, several genome wide methylation analyses that included some degree of genetic information have noted SNP*Smoke effects.(Dogan and others 2015; Tsaprouni and others 2014) However, to date, no studies have attempted to quantify the magnitude of those effects of smoking at the genome wide level.
The lack of attempts to conduct SNP*Smoke analyses to better explicate the impact of smoking on genome wide methylation results from two primary obstacles. First, in contrast to straightforward genome wide methylation analyses that can be conducted on a desktop computer, analyses of genetically contextual methylation effects typically require high performance computing environments that may be less readily available. Second, and perhaps more importantly, these analyses require larger datasets with both genome wide genetic and epigenetic data for each subject, along with reliable data on smoking. Fortunately, over the past several years both of these obstacles have become less daunting.
In this communication, we report an integrated examination of the effects of smoking on DNA methylation in the presence and absence of genetic context using the resources of the largest publically available cohort of subjects with both genome wide genetic and epigenetic data, the Framingham Heart Study.(Dawber and others 1963)
METHODS
Framingham Heart Study
The data used in this study is derived from participants in the Framingham Heart Study (FHS).(Dawber and others 1963) FHS is a population-based longitudinal study aimed at understanding the risks of cardiovascular disease (CVD) and consists of several cohorts including the Original Cohort, Offspring Cohort, Omni Cohort, Third Generation Cohort, New Offspring Spouse Cohort and Second Generation Omni Cohort.(Mahmood and others 2014) Specifically, the Offspring Cohort, initiated in 1971, consisting of the offspring of the Original Cohort and their spouses was used in this study. This cohort consists of 2,483 males and 2,641 females (total of 5,124). Data were obtained via dbGaP (https://dbgap.ncbi.nlm.nih.gov). The University of Iowa Institutional Review Board approved the specific analyses described in this communication.
Genome wide DNA Methylation
Of the 5,124 individuals in the Offspring Cohort, only 2,567 individuals (duplicates removed) with DNA methylation data were considered. These individuals were included in the DNA methylation study because they attended the Framingham Offspring 8th exam, provided consent for genetic research, had a buffy coat (WBC) sample, and had sufficient DNA quantity and quality for methylation profiling. Exam 8 took place between 2005 and 2008. Genomic DNA extracted from the buffy coat was bisulfite converted and genome wide DNA methylation was profiled using the Illumina HumanMethylation450 BeadChip (San Diego, CA) at either the University of Minnesota or Johns Hopkins University. The intensity data (IDAT) files of the samples alongside their slide and array information were used to perform the DASEN normalization using the MethyLumi, WateRmelon and IlluminaHumanMethylation450k.db R packages. (Pidsley and others 2013) The DASEN normalization performs probe filtering, background correction and adjustment for probe types. Samples were removed if they contained >1% of CpG sites with a detection p-value >0.05. CpG sites were removed if they had a bead count of <3 and/or >1% of samples had a detection p-value >0.05. After DASEN normalization, there were 2,560 samples and 484,241 sites remaining (484,125 CpG sites). CpG sites were grouped by chromosome. Methylation beta values were converted to M values using the beta2m R function in the Lumi package and subsequently converted to z-scores using an R script. (Du and others 2010) Because a growing number of large scale studies have shown that the effects of smoking on DNA methylation are robust to the effects of cellular heterogeneity(Bauer and others 2016; Dogan and others 2014; Joubert and others 2016) and current mechanism for correction are computationally prohibitive for the current analyses, we have followed the recommendations of Bauer and colleagues(Bauer and others 2016) and not corrected for cellular composition.
Genome wide Genotype
Of the 2,560 remaining individuals after DNA methylation quality control, 2,406 (1,100 males and 1,306 females) had genome wide genotype data from the Affymetrix GeneChip HumanMapping 500K Array Set (Santa Clara, CA). This array is capable of profiling 500,568 SNPs in the genome. Quality control was performed at both the sample and SNP probe levels in PLINK. (Purcell and others 2007)The initial quality control step involved identifying individuals with discordant sex information. None were identified. Next, individuals with a heterozygosity rate of greater or smaller than the mean ± 2SD and with a proportion of missing SNPs >0.03 were excluded. Population stratification was performed and no individuals were removed. Related individuals were also excluded if the identity by descent value was >0.185 (approximately halfway between second and third degree relatives). After performing these sample level quality control steps, 1,599 individuals remained (722 males and 877 females). On the probe level, SNPs with a minor allele frequency >1%, Hardy-Weinberg equilibrium p-value >10−5 and SNP missing rate of <5% were retained. A total of 403,192 SNPs remained after these quality control steps. Using the recode option in PLINK, genotypes were coded as 0, 1 or 2.
Phenotypes
In the analyses, phenotypes that were considered include age, gender, batch, and smoke exposure. The age used was the age of an individual at exam 8. Batch represented the laboratory batch and smoke exposure was the self-reported current smoking status of an individual at exam 8. Smoking status was coded as a binary variable. The demographics of the 1599 individuals included in this study are summarized in Table I.
Table I.
Clinical and Demographic Characteristics of the Subjects
| Smoker | Non-smoker | |
|---|---|---|
| Gender | ||
| Male | 52 | 668 |
| Female | 69 | 808 |
| Age | ||
| Male | 62.0±6.5 | 67.7±8.6 |
| Female | 63.0±7.9 | 67.3±8.7 |
Smoking status not available for 2 individuals
Linear Regression of Genome wide DNA Methylation
Our first analysis was to identify smoking associated DNA methylation (CpG) sites. To achieve that, a linear model was fitted in R:
| (1) |
For every CpG site, the methylation was regressed against smoking status (smoke), controlling for age, gender and batch. Correction for multiple comparisons was performed using Bonferroni correction.(Fleiss 1981) A genome wide significance level of 0.05 was used. A total of 484,125 independent tests were conducted. Therefore, only CpG sites with a nominal p-value of 1e-07 or less (0.05/484125) were considered to be significantly associated with smoking status.
SNPxSmoke (GxE) Interaction
To understand the effect of SNP on methylation given smoking status, the following model was interrogated:
| (2) |
Specifically, the interaction term (SNPj*Smoke) was of interest because it allows the understanding of the interplay between genotype and smoke exposure (environment) on DNA methylation. A cis distance of 1Mb was used to distinguish cis-interaction effects (SNP in SNPxSmoke interaction term cis of methylation site) from trans-interaction effects (SNP in SNPxSmoke interaction term trans of methylation site). In the MatrixeQTL package, this was achieved using the modelLINEAR_CROSS model type. (Shabalin 2012) Correction for multiple comparisons at a 0.05 significance level was performed using the Bonferroni method. (Fleiss 1981) A total of 126,369,511 cis and 195,068,554,297 trans independent interaction tests were performed, which implies that cis-interactions with a nominal p-value of 3.96e-10 or less (0.05/126369511) and trans-interactions with a nominal p-value of 2.60e–13 or less (0.05/195068554297) are significant. Genomic inflation factors for each chromosome for cis-interactions and Y chromosome for trans-interactions were calculated using the GenABEL R package.
Protein-protein Interaction Networks
To better visualize and compare the connectivity and gene ontology (GO) enrichment between significant genes in the regression and interaction analyses, we generated protein-protein interaction (PPI) networks. To generate these networks, STRING Version 10 was used.(Szklarczyk and others 2010) This database contains information of known and predicted direct and indirect PPIs. A total of three networks were produced. The first network was for the methylation regression analysis, where the input genes were those that had at least one significant CpG site with respect to smoking status after correction for multiple comparisons. The second (cis-interaction) network was generated using genes of methylation probes with at least one significant SNP*Smoke interaction effect where the SNP and methylation probe are in cis of each other. Finally, the third (trans-interaction) network was generated using genes of methylation probes with at least one significant SNPxSmoke interaction effect where the SNP and methylation probe are in trans of each other.
Functional SNP Mapping
To complement the PPI analysis and obtain a holistic understanding of smoking contextual effects on disease outcomes, we performed an analysis to determine and map functional SNPs. Specifically, the significant cis- and trans-interaction SNPs were used in this analysis. The freely available, user-friendly SNPnexus functional annotation web server was used.(Dayem Ullah and others 2012)
RESULTS
Epigenetic and genetic data from 1599 subjects were included in this study (Table I). All subjects were of northern European ancestry and tended to be in their early to mid-sixties. The rate of self-reported smoking was 7.6%, which is slightly lower than the national rate of smoking for subjects > 65 yrs of age, and markedly less than the national rate of 21% for those between 45 and 64 years of age. (Centers for Disease Control and Prevention 2011) For both men and women, self-reported smokers were significantly younger than self-reported non-smokers (p<0.0001 for each). Consequently, all analyses were controlled for age, as well as batch and gender.
Linear Regression of Genome wide DNA Methylation
A total of 484,125 methylation probes survived data cleaning and the implementation of quality control measures. The first set of analyses regressed each of these probe values against self-reported smoking status, controlling for age, gender and batch. A total of 525 methylation sites remained significant after correction for genome wide comparisons at an α of 0.05. The top 30 of these CpG sites are shown in Table II while the complete list of 525 sites is given in Supplemental Table I. Consistent with prior studies, cg05575921 was the top ranked probe with a p-value <0.001 (7.65 × 10−155) with 8 of the top 30 probes localizing to AHRR. Other notable consistencies include the third ranked probe cg03636183 (F2RL3) with a p value <0.001 (3.5 × 10−60) and the 13th ranked probe cg19859270 (GPR15) with a p-value <0.001 (2.9 × 10−39). Interestingly, there was a relationship between slope of the regression (i.e. demethylation or hypermethylation) in response to smoking and probe rank. Overall, 386 of the 525 probes (74%) manifested demethylation in response to smoking. All of the top 30 sites shown in Table II manifested demethylation in response to smoking. However, as illustrated in Supplementary Table I, as probe rank decreased, the likelihood of having a positive slope (i.e. hypermethylation) in response to smoking increased (Wilcoxon rank sum p<0.0001).
Table II.
Top 30 methylation probes associated with smoking status
| Rank | CpG | Smoke Beta | Gene | Position | Island Status | Corrected p-value |
|---|---|---|---|---|---|---|
| 1 | cg05575921 | −2.30 | AHRR | Body | N Shore | 7.7E–155 |
| 2 | cg21566642 | −1.86 | Island | 1.04E–90 | ||
| 3 | cg03636183 | −1.84 | F2RL3 | Body | N Shore | 3.51E–90 |
| 4 | cg01940273 | −1.80 | Island | 3.28E–86 | ||
| 5 | cg21161138 | −1.63 | AHRR | Body | 2.04E–69 | |
| 6 | cg05951221 | −1.58 | Island | 2.32E–61 | ||
| 7 | cg25648203 | −1.49 | AHRR | Body | 3.29E–54 | |
| 8 | cg26703534 | −1.44 | AHRR | Body | S Shelf | 1.84E–50 |
| 9 | cg06126421 | −1.31 | 4.12E–46 | |||
| 10 | cg24859433 | −1.28 | 1.58E–42 | |||
| 11 | cg09935388 | −1.33 | GFI1 | Body | Island | 4.18E–42 |
| 12 | cg19859270 | −1.30 | GPR15 | 1stExon | 2.89E–39 | |
| 13 | cg15342087 | −1.26 | 4.77E–37 | |||
| 14 | cg23079012 | −1.24 | 2.69E–35 | |||
| 15 | cg12806681 | −1.18 | AHRR | Body | N Shore | 5.73E–31 |
| 16 | cg14817490 | −1.16 | AHRR | Body | 4.58E–30 | |
| 17 | cg12876356 | −1.15 | GFI1 | Body | Island | 3.63E–29 |
| 18 | cg03991871 | −1.14 | AHRR | Body | N Shore | 4.18E–29 |
| 19 | cg05284742 | −1.11 | ITPK1 | Body | 2.32E–27 | |
| 20 | cg18316974 | −1.11 | GFI1 | Body | Island | 5.17E–27 |
| 21 | cg23576855 | −1.11 | AHRR | Body | N Shore | 8.95E–27 |
| 22 | cg03329539 | −1.10 | N Shore | 1.27E–26 | ||
| 23 | cg09662411 | −1.03 | GFI1 | Body | Island | 2.08E–22 |
| 24 | cg03450842 | −0.95 | ZMIZ1 | 5′UTR | 1.13E–21 | |
| 25 | cg07339236 | −1.02 | ATP9A | Body | 1.41E–21 | |
| 26 | cg11660018 | −0.96 | PRSS23 | TSS1500 | N Shore | 5.72E–21 |
| 27 | cg18146737 | −1.00 | GFI1 | Body | Island | 1.5E–20 |
| 28 | cg21322436 | −0.95 | CNTNAP2 | TSS1500 | N Shore | 2.11E–20 |
| 29 | cg02451831 | −0.97 | KIAA0087 | Body | 8.65E–20 | |
| 30 | cg08709672 | −0.93 | AVPR1B | 5′UTR | S Shore | 3.53E–19 |
Using the Illumina (San Diego, CA) annotation file, we attempted to map the significant probes to specific genes. A total of 412 CpG of the 525 probes mapped to 310 unique genes. Several genes had at least three or more significant probes mapping to their locus including AHRR (22 probes) and GFL1 (8 probes). The 310 unique genes were subsequently used as the input to generate the first protein-protein interaction (PPI) network.
SNPxSmoke (GxE) Interactions
Based on formula (2) listed in methods, a total of 126,369,511 independent cis- and 195,068,554,297 independent trans-interaction regression analyses were conducted. Using the p-value thresholds stated in methods, there were a total of 827 (0.00065%) and 448,342 (0.00023%) significant cis- and trans-interactions, respectively, after correction for multiple comparisons (0.05 significance level). The 827 significant cis-interactions mapped to 388 unique CpG sites (0.08%) and 785 unique SNPs (0.2%) whereas the 448,342 trans-interactions mapped to 9,566 unique CpG sites (2%) and 78,362 unique SNPs (19%). Methylation sites of significant cis-interactions and trans-interactions mapped to 266 and 4,353 genes, respectively. Almost 90% (238 of 266) of the genes in the cis group overlapped with those in trans group (Chi Square p<0.0001). In contrast, the degree of overlap between the significant cis (10 of 266 or 3.8%) and trans (83 of 4353 or 1.9%) genes with the direct effect group (n=310) only 10 and 83 genes having significant of the cis and trans genes, respectively, overlapped with the group of 310 genes with significant direct effects (Chi Square p<0.003 and p<0.04, respectively). The complete list of significant cis- and trans-interactions are provided in Supplemental Tables II and III, respectively. To determine if SNPxSmoke effects were present for any of the top 30 smoking associated DNA methylation sites shown in Table II, the presence of significant cis- and trans-interactions were examined. None were identified.
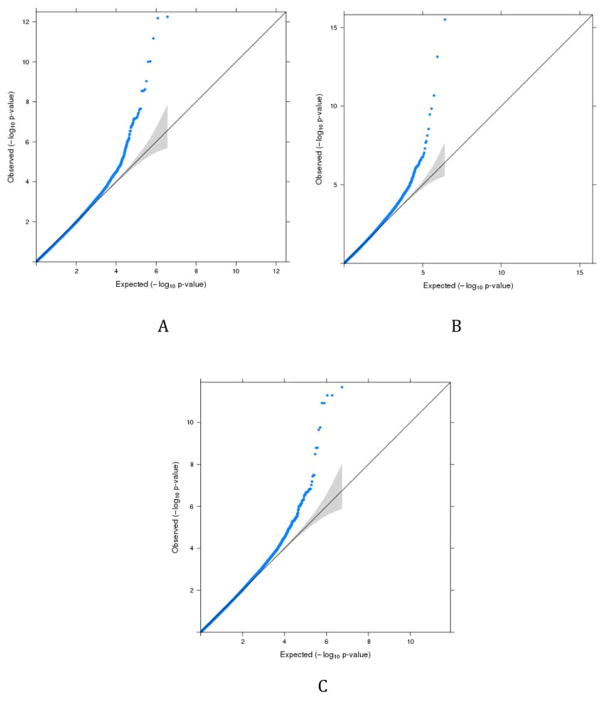
None of the calculated genomic inflation factors were greater than 1, and therefore, the p-values of interactions were not corrected. Considering the trans-interactions genomic inflation factors were not calculated for all chromosomes, it is likely that the number of significant loci may decrease. However, it should not affect the most significant loci identified through our analysis. Figure 1 shows example QQ plots generated for several chromosomes. Also, to understand if smoking status did contribute to the observed significance of the interactions, we permuted the smoking status of the 1599 individuals included in this study and re-ran the interaction analysis for one of the smaller chromosomes (chromosome 18) to ensure a reasonable runtime. With the permuted smoking status, no significant cis-interactions were observed after Bonferroni correction for multiple comparisons (vs. five significant cis-interactions after Bonferroni correction for multiple comparisons without permutation) and eight trans-interactions were observed after Bonferroni correction for multiple comparisons (vs. 2508 significant trans-interactions after Bonferroni correction for multiple comparisons without permutation). This suggests that smoking does play a role in the reported results.
Figure 1.
Protein-protein Interaction (PPI) Networks
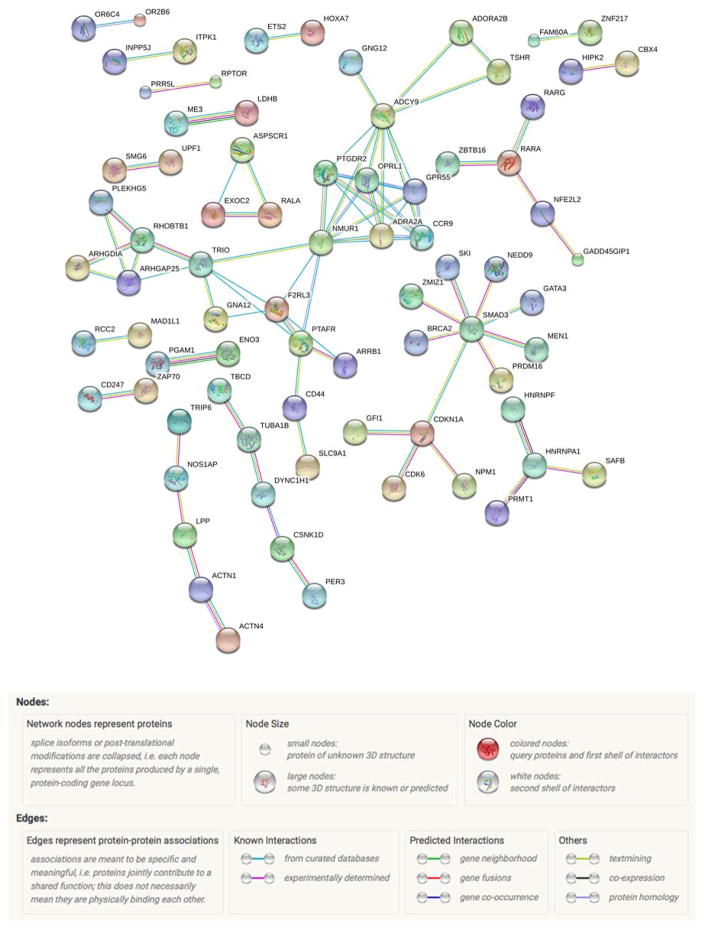
To identify coherent networks of genes influenced directly by smoking, we examined the potential interconnections of the 310 genes identified in the genome wide methylation analysis using PPI information in the STRING database. 302 of the 310 genes matched with a protein in the database. This network is shown in Figure 2. In this figure, only nodes with edges are shown. Also, only edges with the highest confidence interaction score of 0.9 or larger were retained to focus on patterns of known significance. The colors of the edges represent interaction types and are elaborated in the figure legend. This network consisted of 302 nodes, 84 edges (61 expected by chance, PPI enrichment p-value=0.00355) with an average node degree of 0.556. This suggests that smoking is producing changes relevant to protein expression that may have coordinated functions due to their potential to interact with each other physically. Using GO miner analyses, the network portrayed in Figure 1 mapped to 141 biological process GO pathways at an FDR of 5%. The top 10 pathways are listed are listed in Table III. Of those 141 GO pathways, the largest number of proteins, 124 (41%), was observed in the regulation of metabolic process pathway (GO:0019222) with an FDR p-value of 0.000393.
Figure 2.
Table III.
Top Ten Gene Ontology Biological Process Pathways from Main Effect Analyses
| GO Pathway ID | Pathway Description | Observed Gene Count | False Discovery Rate p-value |
|---|---|---|---|
| GO.0009893 | positive regulation of metabolic process | 83 | 0.000223 |
| GO.0010604 | positive regulation of macromolecule metabolic process | 68 | 0.000223 |
| GO.0048518 | positive regulation of biological process | 108 | 0.000223 |
| GO.1903707 | negative regulation of hematopoiesis | 13 | 0.000223 |
| GO.0019222 | regulation of metabolic process | 124 | 0.000393 |
| GO.0045596 | negative regulation of cell differentiation | 28 | 0.000393 |
| GO.0048522 | positive regulation of cellular process | 94 | 0.00054 |
| GO.0040007 | growth | 21 | 0.000644 |
| GO.0045638 | negative regulation of myeloid cell differentiation | 10 | 0.000953 |
| GO.0000122 | negative regulation of transcription from RNA polymerase II promoter | 29 | 0.00104 |
Cis- and Trans-Interaction PPI Networks
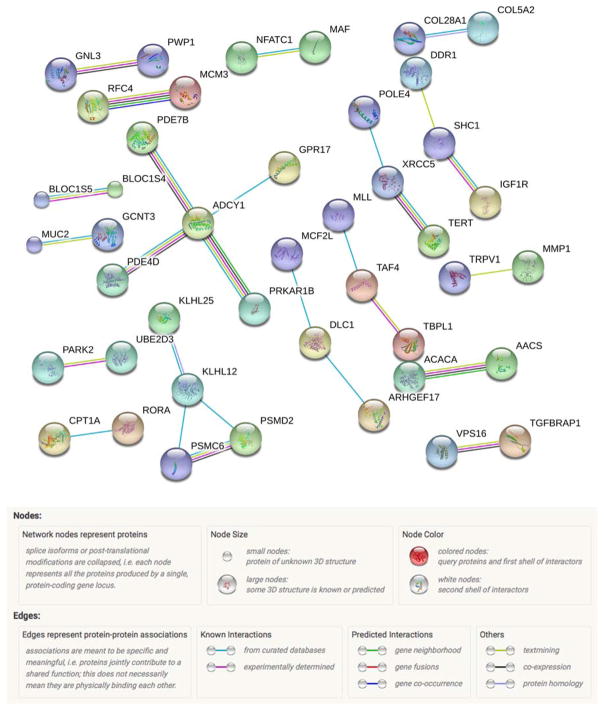
To obtain a comprehensive understanding on the interplay between genotype, environment and DNA methylation, cis- and trans-interaction networks were generated. Significant cis-interactions were associated with 266 unique genes and mapped to 249 proteins. This network consisted of 249 nodes, 27 edges (20 expected, PPI enrichment p-value=0.0664) with an average node degree of 0.217. Of these genes, 10 (4%) were also identified in the methylation regression analysis. There were two biological process GO pathways at an FDR of 5%, and are shown in Table IV. This network is shown in Figure 3. In this figure, only nodes with edges are shown. Also, only edges with the highest confidence interaction score of 0.9 or larger were retained to focus on patterns of known significance. The colors of the edges represent interaction types and are elaborated in the figure legend.
Table IV.
Gene Ontology Biological Process Pathways from Cis-Interaction Analysis
| GO Pathway ID | Pathway Description | Observed Gene Count | False Discovery Rate p value |
|---|---|---|---|
| GO.0009891 | positive regulation of biosynthetic process | 41 | 0.0455 |
| GO.0031328 | positive regulation of cellular biosynthetic process | 40 | 0.0455 |
Figure 3.
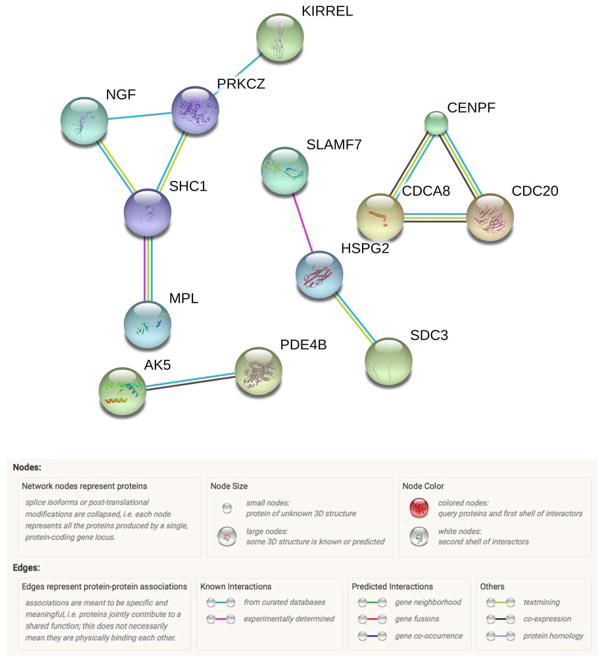
Due to a large number of trans-interactions, only the genes of the top 1,000 significant interactions were considered. These interactions were associated with 158 unique genes and 148 proteins. Of those, 9 (6%) were also identified in the methylation analysis. The network had 148 nodes, 11 edges (8 expected, PPI enrichment p-value=0.189) with an average node degree of 0.149. At an FDR of 5%, this network did not map to any biological process GO pathways. This network is shown in Figure 4. In this figure, only nodes with edges are shown. Also, only edges with the highest confidence interaction score of 0.9 or larger were retained to focus on patterns of known significance. The colors of the edges represent interaction types and are elaborated in the figure legend.
Figure 4.
Functional SNP Mapping
All significant cis- and trans-interactions mapped to 786 and 78,362 SNPs, respectively. Of those 687 cis-SNPs and 60,504 trans-SNPs had confirmed associations to complex diseases and disorders as curated in the Genetic Association Database (GAD).(Becker and others 2004) These SNPs map to a wide range of disease classes including cardiovascular, metabolic, immune, psychiatry, neurological, cancer, developmental, aging, and pharmacogenomics.
To determine if the number of observed associations is due to chance, a goodness of fit chi-square test was performed for the observed count of disease associated SNPs in cis-interactions (687 of 786) compared to the expected count based on proportion of all the SNPs in the Affymetrix chip that survived data cleaning that were previously associated with disease in the GAD archive (312,736 of 403,192). The same was performed for the trans-interaction functional SNPs (60,504 of 78,362). For the cis-interactive SNPs, the chi-squared value was 43.4 with a p-value<0.0001 whereas for the trans-interactive SNPs, the chi-squared value was 7.1 with a p-value of 0.0078.
DISCUSSION
In summary, we report that when cis and trans effects of genetic variation are considered, smoking has a significant effect on DNA methylation signature of peripheral blood DNA at 4379 genes and that the SNP variation participating in significant interactions preferentially mapping to loci previously associated with complex illness. Still, before considering the current findings it is important to note that FHS cohort is exclusively White and late mid-life. Therefore, further examination of the current findings in subjects of other ethnicities and age groups will be necessary before generalizability of the findings can be accepted. In addition, it should be noted that although both the epigenetic and genetic platforms used herein are considered “genome wide”, it is generally appreciated that large portions of both the epigenome and genome are not interrogated by these measures.
The current findings will be particularly useful for those seeking a better understanding of the relationship between smoking and illnesses not classically associated with inflammation. The prominence of inflammatory related genes reported in prior analyses of the effects of smoking on DNA methylation from infants and adults has clearly outlined a plausible molecular explanation for the relationship of smoking to the increased rates of asthma and respiratory tract infections observed in infants and adults.(Dogan and others 2014; Gunawardhana and others 2014; Zhang and others 2014) However, these more limited prior findings do not readily explain a connection between smoking and syndromes such as mild intellectual delay or attention deficit hyperactivity disorder in the offspring of mothers who smoke (DiFranza and others 2004; Langley and others 2005; Mick and others 2002; Milberger and others 1996) or the increased risk for later onset neuropsychiatric disorders, namely, panic disorder and schizophrenia in adults who smoke. (Gurillo and others 2015; Kendler and others 2015) The current findings showing a non-preferential mapping “hidden” genetic contextual effects in a variety of non-immune system genes allows the generation of other potential mechanisms to connect smoking to these syndromes. In essence, using strict Bonferroni correction methods, over 20% of all genes show evidence of abnormal methylomic regulation. Should we have used less stringent correction methodologies (e.g. FDR), the number of genes would have been substantially larger. In any case, since the vast majority of pathways in the Gene Ontology database contain at least 5 genes and the effects of smoking on DNA methylation between at least some tissues are correlated,(Novakovic and others 2013; Teschendorff and others 2015) it is likely that cellular pathways in many if not all tissues contain at least one gene potentially affected by smoking. Since the list of smoking associated illnesses is large and encompasses most, if not all, organ systems,(Office of The Surgeon General 2014) this is not unexpected.
The marked enrichment of those SNPs with significant cis or trans interactions with those SNPs already associated disease in the GAD archive suggests the potential for the creation of greater insight into the biology of complex smoking associated illnesses. Unfortunately, exactly reconciling these methylation findings to prior genetic findings with respect to smoking associated illnesses will be difficult for a number of reasons. For example, with respect to a syndrome of critical interest to behavioral geneticists, using genome wide SNP data from over 36,989 cases and 113,075 controls, the Psychiatric Genetics consortium has published a list of 108 unique loci, representing 349 individual genes, that contain genetic vulnerability to schizophrenia.(Ripke and others 2014) Unfortunately, because the area of linkage disequilibrium surrounding many of these SNPs may cover several genes and our network analyses are methylation probe centric, a direct comparison of the genes implicated in our network analyses and those of the Psychiatric Genetics consortium using traditionally associated approaches is not possible. Still, given the wealth of epidemiological data that implicates smoking in the etiology of schizophrenia,(de Leon and Diaz 2005; Dickerson and others 2013; Kendler and others 2015) these data suggest that genetically informed epigenetic analyses of DNA from cohorts informative for schizophrenia may be useful in generating new hypotheses and refining existing hypotheses with respect to the biological underpinnings of psychosis.
Even so, interpretation of the strength of individual interactions should be done with caution for several reasons. First, what matters most for the cell is the amount of protein translation. Unfortunately, the relationship between DNA methylation at a given CpG site with both local and regional gene transcription, and more distally, protein translation, is not well constrained. Second, the number of significant comparisons is dependent on the manner in which the analyses are conducted. Since long range interactions are known to occur,(Smallwood and Ren 2013) we used a 500,000 bp window on either side of the probe in question in our cis interaction analyses. But the ideal window for these analyses is not known. In prior work, the distance between the CpG site (or probe) and the SNP participating in the interactions has been reported to vary from an average of 15 kb to 1 kb, a discrepancy that is likely secondary to differences in methodology.(Shoemaker and others 2010; Smith and others 2014) This uncertainty in the distance between the two partners in the interaction is made even more difficult by the observation that with respect to gene expression, over 20% of all eQTLs exert their effects on non-adjacent gene regions.(Li and others 2016) As a result of this lack of knowledge of the best window for comparison, the significance of our cis comparisons may be over corrected or under corrected. Third, the SNP that was used in the analyses may not be the polymorphism that is driving the strength of the interaction term. The 403,192 SNPs used in our analyses are merely the tag SNP(s) for a given region that may be in full or partial equilibrium with another polymorphism that is driving SNP*Smoke effects. Should it be necessary to identify the actual variation driving an interaction term, the region will need to be sequenced and carefully controlled experiments similar to those used to identify eQTLs (Albert and Kruglyak 2015) will need to be conducted.
The current study utilized data from mature adults. However, it is very possible that many of the effects of smoking are also developmentally contextual. With respect to the latter, in a follow up to their 2012 work, Joubert and colleagues recently conducted a meta-analysis of genome wide DNA methylation cord blood from 6685 newborns with respect to maternal smoking. (Joubert and others 2016) Although the ranking of the top probes was very similar to that found in adults, in their listing of 6073 FDR probes, they did not find a significant differential methylation at F2RL3 or GPR15, which are loci repeatedly implicated in most all studies of adult samples.(Andersen and others 2015; Gao and others 2015) Also, they found that over half (52%) of all the 6073 FDR significant probes were hypermethylated in response to maternal smoking which is in direct contrast to the current and prior findings that the vast majority of probes are de-methylated in response to smoking.(Andersen and others 2015) Although there are a number of possible explanations for the latter discrepancy, it may well be as the number of probes deemed “significantly” affected by smoking increases, the percentage of probes manifesting hypermethylation increases. Our analyses of our lower ranked non-significant probes listed in Supplementary Table I supports this hypothesis. Still the differences at key loci such as F2RL3 and GPR15 suggest a differing biological response of in utero fetuses as compared to adolescents and adults to smoking.
The next few steps in the fuller understanding of these findings will be challenging. First, the findings need to be replicated and extended in several ethnically informative populations with good substance use characterization. Although the FHS is one of premier clinical resources in the world, it is not well characterized for substance use and biochemical validation of smoking status was not performed. Unreliable self-report is a challenge to these analyses of the biology of smoking, particularly in the high-risk populations in which the effects are most evident.(Caraballo and others 2004; Caraballo and others 2001; Shipton and others 2009) Second, the relationship of individual program methylation to gene transcription needs to be better understood. Unfortunately, the methylation (Illumina 450K) and gene expression platforms used in the FHS (GeneChip Human Exon 1.0 ST Array) study are already no longer state-of-the-art. In that regard, it probably makes more sense to approach the problem using more up to date methods such whole bisulfitome and RNAseq data that better capture the total methylome and transcriptome, respectively. Third, it is important to note that almost all studies conducted to date, including the current study, use DNA prepared from peripheral white blood cells or saliva. However, for understanding the effect on smoking on vulnerability to non-hematologic disorders, it will be necessary to study biomaterial from the affected tissues, such as the brain, in both humans and model organisms. In that regard, we note the accumulation of high quality biomaterials and data being conducted by the GTEx Consortium, which is conducting integrated genetic, epigenetic and gene expression studies on fresh post mortem sample from a variety of tissues.(Carithers and others 2015) Given recent studies showing the strong role of complement activation in the pathogenesis of schizophrenia (Nsaiba and others 2015; Sekar and others 2016) and the strong effects of smoking on the methylation of complement cascade genes (Dogan and others 2014), it is quite possible that studies of these or similar resources could lead to interventions that could prevent or mitigate the severity of several types of smoking related mental illness.
In summary, we show that integrated genetic and epigenetic analyses demonstrate the profound effects of smoking on methylation status throughout the genome and an enrichment of SNPs with significant smoking contextual methylation effects with those known to be associated with disease. We suggest further integrated analyses could lead to new insights for better prevention and treatment of a variety of smoking related illnesses.
Supplementary Material
Acknowledgments
This work was supported by NIH Grant R01DA037648 (Philibert Multiple PI).
Footnotes
CONFLICTS OF INTEREST
The use of DNA methylation to assess smoking status is covered by US patent 8,637,652, 9,273,358 and other pending claims. Dr. Philibert is a potential royalty recipient and the Chief Executive Officer of Behavioral Diagnostics (www.bdmethylation.com).
References
- Albert FW, Kruglyak L. The role of regulatory variation in complex traits and disease. Nat Rev Genet. 2015;16(4):197–212. doi: 10.1038/nrg3891. [DOI] [PubMed] [Google Scholar]
- Andersen AM, Dogan MV, Beach SR, Philibert RA. Current and Future Prospects for Epigenetic Biomarkers of Substance Use Disorders. Genes. 2015;6(4):991–1022. doi: 10.3390/genes6040991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubin H-J, Rollema H, Svensson TH, Winterer G. Smoking, quitting, and psychiatric disease: A review. Neuroscience & Biobehavioral Reviews. 2012;36(1):271–284. doi: 10.1016/j.neubiorev.2011.06.007. [DOI] [PubMed] [Google Scholar]
- Bauer M, Fink B, Thürmann L, Eszlinger M, Herberth G, Lehmann I. Tobacco smoking differently influences cell types of the innate and adaptive immune system—indications from CpG site methylation. Clinical Epigenetics. 2016;8(1):83. doi: 10.1186/s13148-016-0249-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker KG, Barnes KC, Bright TJ, Wang SA. The genetic association database. Nature genetics. 2004;36(5):431–432. doi: 10.1038/ng0504-431. [DOI] [PubMed] [Google Scholar]
- Bouyer J, Coste J, Shojaei T, Pouly J-L, Fernandez H, Gerbaud L, Job-Spira N. Risk Factors for Ectopic Pregnancy: A Comprehensive Analysis Based on a Large Case-Control, Population-based Study in France. American Journal of Epidemiology. 2003;157(3):185–194. doi: 10.1093/aje/kwf190. [DOI] [PubMed] [Google Scholar]
- Breitling LP, Salzmann K, Rothenbacher D, Burwinkel B, Brenner H. Smoking, F2RL3 methylation, and prognosis in stable coronary heart disease. European Heart Journal. 2012 doi: 10.1093/eurheartj/ehs091. [DOI] [PubMed] [Google Scholar]
- Caraballo RS, Giovino GA, Pechacek TF. Self-reported cigarette smoking vs. serum cotinine among U.S. adolescents. Nicotine & Tobacco Research. 2004;6(1):19–25. doi: 10.1080/14622200310001656821. [DOI] [PubMed] [Google Scholar]
- Caraballo RS, Giovino GA, Pechacek TF, Mowery PD. Factors associated with discrepancies between self-reports on cigarette smoking and measured serum cotinine levels among persons aged 17 years or older: Third National Health and Nutrition Examination Survey, 1988–1994. Am J Epidemiol. 2001;153(8):807–14. doi: 10.1093/aje/153.8.807. [DOI] [PubMed] [Google Scholar]
- Carithers LJ, Ardlie K, Barcus M, Branton PA, Britton A, Buia SA, Compton CC, DeLuca DS, Peter-Demchok J, Gelfand ET, et al. A Novel Approach to High-Quality Postmortem Tissue Procurement: The GTEx Project. Biopreservation and Biobanking. 2015;13(5):311–319. doi: 10.1089/bio.2015.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Smoking-attributable mortality, years of potential life lost, and productivity losses--United States, 2000–2004. MMWR Morb Mortal Wkly Rep. 2008;57(45):1226–8. [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Vital signs: current cigarette smoking among adults aged >/=18 years--United States, 2005–2010. MMWR. 2011;60:1207–12. [PubMed] [Google Scholar]
- Dawber TR, Kannel WB, Lyell LP. An approach to longitudinal studies in a community: the Framingham Study. Annals of the New York Academy of Sciences. 1963;107(2):539–556. doi: 10.1111/j.1749-6632.1963.tb13299.x. [DOI] [PubMed] [Google Scholar]
- Dayem Ullah AZ, Lemoine NR, Chelala C. SNPnexus: a web server for functional annotation of novel and publicly known genetic variants (2012 update) Nucleic Acids Research. 2012;40(Web Server issue):W65–W70. doi: 10.1093/nar/gks364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leon J, Diaz FJ. A meta-analysis of worldwide studies demonstrates an association between schizophrenia and tobacco smoking behaviors. Schizophrenia research. 2005;76(2):135–157. doi: 10.1016/j.schres.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Dickerson F, Stallings CR, Origoni AE, Vaughan C, Khushalani S, Schroeder J, Yolken RH. Cigarette Smoking Among Persons With Schizophrenia or Bipolar Disorder in Routine Clinical Settings, 1999–2011. Psychiatric Services. 2013;64(1):44–50. doi: 10.1176/appi.ps.201200143. [DOI] [PubMed] [Google Scholar]
- DiFranza JR, Aligne CA, Weitzman M. Prenatal and postnatal environmental tobacco smoke exposure and children’s health. Pediatrics. 2004;113(Supplement 3):1007–1015. [PubMed] [Google Scholar]
- Dogan MV, Shields B, Cutrona C, Gao L, Gibbons FX, Simons R, Monick M, Brody G, Tan K, Philibert R. The effect of smoking on DNA methylation of peripheral blood mononuclear cells from African American women. BMC Genomics. 2014;15:151. doi: 10.1186/1471-2164-15-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogan MV, Xiang J, Beach SR, Cutrona C, Gibbons FX, Simons RL, Brody GH, Stapleton JT, Philibert RA. Ethnicity and Smoking-Associated DNA Methylation Changes at HIV Co-Receptor GPR15. Frontiers in Psychiatry. 2015:6. doi: 10.3389/fpsyt.2015.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P, Zhang X, Huang C-C, Jafari N, Kibbe W, Hou L, Lin S. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11(1):587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleiss JL. Statistical Methods for Rates and Proportions. New York, NY: John Wiley & Sons Inc; 1981. [Google Scholar]
- Gao X, Jia M, Zhang Y, Breitling LP, Brenner H. DNA methylation changes of whole blood cells in response to active smoking exposure in adults: a systematic review of DNA methylation studies. Clinical Epigenetics. 2015;7(1):113. doi: 10.1186/s13148-015-0148-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs JR, van der Brug MP, Hernandez DG, Traynor BJ, Nalls MA, Lai S-L, Arepalli S, Dillman A, Rafferty IP, Troncoso J, et al. Abundant Quantitative Trait Loci Exist for DNA Methylation and Gene Expression in Human Brain. PLoS Genet. 2010;6(5):e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardhana LP, Gibson PG, Simpson JL, Benton MC, Lea RA, Baines KJ. Characteristic DNA methylation profiles in peripheral blood monocytes are associated with inflammatory phenotypes of asthma. Epigenetics. 2014;9(9):1302–1316. doi: 10.4161/epi.33066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurillo P, Jauhar S, Murray RM, MacCabe JH. Does tobacco use cause psychosis? Systematic review and meta-analysis. The Lancet Psychiatry. 2015;2(8):718–725. doi: 10.1016/S2215-0366(15)00152-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isensee B, Wittchen HU, Stein MB, Hofler M, Lieb R. Smoking increases the risk of panic: findings from a prospective community study. Arch Gen Psychiatry. 2003;60(7):692–700. doi: 10.1001/archpsyc.60.7.692. [DOI] [PubMed] [Google Scholar]
- Joubert Bonnie R, Felix Janine F, Yousefi P, Bakulski Kelly M, Just Allan C, Breton C, Reese SE, Markunas Christina A, Richmond Rebecca C, Xu C-J, et al. DNA Methylation in Newborns and Maternal Smoking in Pregnancy: Genome-wide Consortium Meta-analysis. The American Journal of Human Genetics. 2016;98(4):680–696. doi: 10.1016/j.ajhg.2016.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubert BR, Håberg SE, Nilsen RM, Wang X, Vollset SE, Murphy SK, Huang Z, Hoyo C, Midttun Ø, Cupul-Uicab LA, et al. 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environmental Health Perspectives. 2012 doi: 10.1289/ehp.1205412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Lönn SL, Sundquist J, Sundquist K. Smoking and Schizophrenia in Population Cohorts of Swedish Women and Men: A Prospective Co-Relative Control Study. American Journal of Psychiatry. 2015;172(11):1092–1100. doi: 10.1176/appi.ajp.2015.15010126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley K, Rice F, Van den Bree M, Thapar A. Maternal smoking during pregnancy as an environmental risk factor for attention deficit hyperactivity disorder behaviour. A review. Minerva pediatrica. 2005;57(6):359–371. [PubMed] [Google Scholar]
- Li YI, van de Geijn B, Raj A, Knowles DA, Petti AA, Golan D, Gilad Y, Pritchard JK. RNA splicing is a primary link between genetic variation and disease. Science. 2016;352(6285):600–604. doi: 10.1126/science.aad9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmood SS, Levy D, Vasan RS, Wang TJ. The Framingham Heart Study and the epidemiology of cardiovascular disease: a historical perspective. The Lancet. 2014;383(9921):999–1008. doi: 10.1016/S0140-6736(13)61752-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mick E, Biederman J, Faraone SV, Sayer J, Kleinman S. Case-Control Study of Attention-Deficit Hyperactivity Disorder and Maternal Smoking, Alcohol Use, and Drug Use During Pregnancy. Journal of the American Academy of Child & Adolescent Psychiatry. 2002;41(4):378–385. doi: 10.1097/00004583-200204000-00009. [DOI] [PubMed] [Google Scholar]
- Milberger S, Biederman J, Faraone SV, Chen L, Jones J. Is maternal smoking during pregnancy a risk factor for attention deficit hyperactivity disorder in children? The American Journal of Psychiatry. 1996;153(9):1138. doi: 10.1176/ajp.153.9.1138. [DOI] [PubMed] [Google Scholar]
- Monick MM, Beach SR, Plume J, Sears R, Gerrard M, Brody GH, Philibert RA. Coordinated changes in AHRR methylation in lymphoblasts and pulmonary macrophages from smokers. Am J Med Genet. 2012;159B(2):141–51. doi: 10.1002/ajmg.b.32021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moylan S, Jacka FN, Pasco JA, Berk M. How cigarette smoking may increase the risk of anxiety symptoms and anxiety disorders: a critical review of biological pathways. Brain and Behavior. 2013;3(3):302–326. doi: 10.1002/brb3.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novakovic B, Ryan J, Pereira N, Boughton B, Craig JM, Saffery R. Postnatal stability and tissue-and time-specific effects of AHRR methylation change in response to maternal smoking in pregnancy. Epigenetics. 2013;9(3):0–1. doi: 10.4161/epi.27248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nsaiba MJ, Lapointe M, Mabrouk H, Douki W, Gaha L, Pérusse L, Bouchard C, Jrad BBH, Cianflone K. C3 polymorphism influences circulating levels of C3, ASP and lipids in schizophrenic patients. Neurochemical research. 2015;40(5):906–914. doi: 10.1007/s11064-015-1543-z. [DOI] [PubMed] [Google Scholar]
- Office of The Surgeon General; Services USDoHaH, editor. The Health Consequences of Smoking—50 Years of Progress. Rockville, MD: 2014. [Google Scholar]
- Pfeifer GP. How tobacco smoke changes the (epi)genome. Science. 2016;354(6312):549–550. doi: 10.1126/science.aal2114. [DOI] [PubMed] [Google Scholar]
- Philibert RA, Beach SR, Gunter TD, Brody GH, Madan A, Gerrard M. The effect of smoking on MAOA promoter methylation in DNA prepared from lymphoblasts and whole blood. American Journal of Medical Genetics. 2010;153B(2):619–28. doi: 10.1002/ajmg.b.31031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philibert RA, Gunter TD, Beach SR, Brody GH, Madan A. MAOA methylation is associated with nicotine and alcohol dependence in women. American Journal of Medical Genetics. 2008;147B(5):565–70. doi: 10.1002/ajmg.b.30778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidsley R, Wong YCC, Volta M, Lunnon K, Mill J, Schalkwyk LC. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics. 2013;14(1):1–10. doi: 10.1186/1471-2164-14-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, De Bakker PI, Daly MJ. PLINK: a tool set for whole-genome association and population-based linkage analyses. The American Journal of Human Genetics. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, Neale BM, Corvin A, Walters JT, Farh K-H, Holmans PA, Lee P, Bulik-Sullivan B, Collier DA, Huang H. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekar A, Bialas AR, de Rivera H, Davis A, Hammond TR, Kamitaki N, Tooley K, Presumey J, Baum M, Van Doren V, et al. Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530(7589):177–183. doi: 10.1038/nature16549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28(10):1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenker NS, Polidoro S, van Veldhoven K, Sacerdote C, Ricceri F, Birrell MA, Belvisi MG, Brown R, Vineis P, Flanagan JM. Epigenome-wide association study in the European Prospective Investigation into Cancer and Nutrition (EPIC-Turin) identifies novel genetic loci associated with smoking. Human Molecular Genetics. 2012:dds488. doi: 10.1093/hmg/dds488. [DOI] [PubMed] [Google Scholar]
- Shipton D, Tappin DM, Vadiveloo T, Crossley JA, Aitken DA, Chalmers J. Reliability of self reported smoking status by pregnant women for estimating smoking prevalence: a retrospective, cross sectional study. BMJ. 2009;339:B4347. doi: 10.1136/bmj.b4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker R, Deng J, Wang W, Zhang K. Allele-specific methylation is prevalent and is contributed by CpG-SNPs in the human genome. Genome Research. 2010;20(7):883–889. doi: 10.1101/gr.104695.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood A, Ren B. Genome organization and long-range regulation of gene expression by enhancers. Current Opinion in Cell Biology. 2013;25(3):387–394. doi: 10.1016/j.ceb.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AK, Kilaru V, Kocak M, Almli LM, Mercer KB, Ressler KJ, Tylavsky FA, Conneely KN. Methylation quantitative trait loci (meQTLs) are consistently detected across ancestry, developmental stage, and tissue type. BMC Genomics. 2014;15(1):1–11. doi: 10.1186/1471-2164-15-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic acids research. 2010;39(suppl 1):561–568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff AE, Yang Z, Wong A, et al. Correlation of smoking-associated dna methylation changes in buccal cells with dna methylation changes in epithelial cancer. JAMA Oncology. 2015;1(4):476–485. doi: 10.1001/jamaoncol.2015.1053. [DOI] [PubMed] [Google Scholar]
- Tsaprouni LG, Yang T-P, Bell J, Dick KJ, Kanoni S, Nisbet J, Viñuela A, Grundberg E, Nelson CP, Meduri E, et al. Cigarette smoking reduces DNA methylation levels at multiple genomic loci but the effect is partially reversible upon cessation. Epigenetics. 2014;9(10):1382–1396. doi: 10.4161/15592294.2014.969637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wannamethee SG, Lowe GDO, Shaper AG, Rumley A, Lennon L, Whincup PH. Associations between cigarette smoking, pipe/cigar smoking, and smoking cessation, and haemostatic and inflammatory markers for cardiovascular disease. European Heart Journal. 2005;26(17):1765–1773. doi: 10.1093/eurheartj/ehi183. [DOI] [PubMed] [Google Scholar]
- Zeilinger S, Kühnel B, Klopp N, Baurecht H, Kleinschmidt A, Gieger C, Weidinger S, Lattka E, Adamski J, Peters A, et al. Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS ONE. 2013;8(5):e63812. doi: 10.1371/journal.pone.0063812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yang R, Burwinkel B, Breitling LP, Brenner H. F2RL3 methylation as a biomarker of current and lifetime smoking exposures. Environmental Health Perspectives (Online) 2014;122(2):131. doi: 10.1289/ehp.1306937. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.