Summary
HIV infection is associated with dramatic alterations in the enteric bacteria but little is known about other components of the microbiome. In this issue of Cell Host and Microbe two studies from the Virgin laboratory (2016) reveal an under-appreciated role of the enteric virome in HIV-associated gastroenteritis and disease pathogenesis.
The profound influence of the enteric microbiome, the collection of bacteria, viruses, archaea and fungi that inhabit the gut, on varied aspects of our health is becoming increasingly clear. Understanding the influence of the enteric microbiome on the health of individuals infected with Human Immunodeficiency Virus (HIV) has become a very active area of research, in part because HIV-infected individuals have higher rates of many diseases linked to alterations in the gut microbiome of non HIV-infected subjects, including dyslipidemia, cardiovascular disease, and chronic inflammation. Translocation of bacteria and their products such as LPS from the gut, driven by massive depletion of CD4+ T cells in the gut associated lymphoid tissue (GALT), is also thought to be a major driver of immune activation and HIV disease progression (Brenchley et al., 2006). Although dramatic differences in the enteric bacterial microbiome that occur in HIV-infected humans have been reported now by several research groups, little to no attention has been given to other important components of the microbiome, including viruses, fungi and other microbial eukaryotes. Furthermore, studies of the enteric microbiome of HIV-infected individuals have been dominated by studies conducted in the United States or Europe, with relatively little attention given to the regions most impacted by HIV, including Sub-Saharan Africa. In this issue of Cell Host and Microbe, two papers from the Virgin lab further expand our knowledge of differences in the enteric microbiome that occur with lentiviral infection, including both bacterial and viral components. First, Monaco et al. demonstrate that alterations in the enteric virome and bacteria microbiome are associated with progressive immunodeficiency in an understudied population of HIV-infected subjects living in Sub-Saharan Africa (Uganda). Second, Handley et al. expand upon the results of a previous study also conducted in the Virgin lab (Handley et al., 2012) that identified a link between SIV infection, the enteric virome, and disease, by evaluation of longitudinally collected samples in macaques who were or were not protected from SIV infection with successful vaccination. These papers together reveal a potentially under-appreciated role of the enteric virome in HIV-associated gastroenteritis and disease pathogenesis, and critical information on bacterial microbiome relationships with HIV-infection in a Sub-Saharan African population.
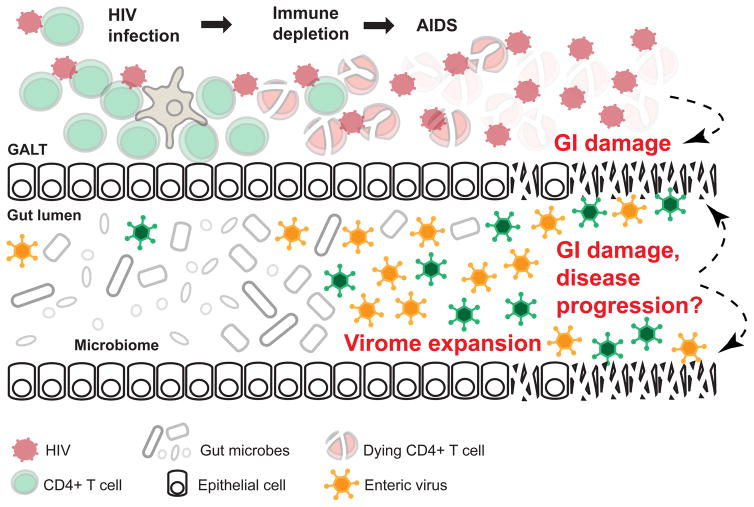
The enteric virome is usually dominated by bacteriophage, with viruses that target eukaryotic cells typically a minor component. Eukaryotic host cell targeting viruses, however, have the potential to be particularly important for HIV/SIV pathogenesis because of their involvement in gastroenteritis and colitis. Viral populations have been far less often evaluated in studies exploring associations between the enteric microbiome and disease compared to bacteria. A previous cross-sectional study conducted in macaques in the Virgin lab had demonstrated that SIV infection of macaques was associated with the expansion of eukaryotic cells targeting enteric viruses, and that adenoviruses co-localized with damaged gut epithelial cells in necropsied intestinal tissue, indicating a potential role for promoting gastroenteritis and bacterial translocation (Figure 1) (Handley et al., 2012). Monaco et al. now study the enteric virome of HIV-infected humans, and consistent with SIV-infected macaques, a significant expansion of the virome, including adenovirus, was observed.
Figure 1. Expansion of the enteric virome contributes to HIV/SIV disease progression.
HIV infection leads to massive depletion of CD4+ T cells in the gut-associated lymphoid tissue (GALT), and is associated with gastrointestinal (GI) damage and disease. The current studies show that severe immunodeficiency caused by HIV/SIV infection is linked with expansion of the viral component of the gut microbiome. Enteric viruses may contribute to further GI damage and HIV/SIV disease progression by direct infection of cells along the GI tract.
Together these studies shed light on the role of HIV disease progression/severity on the expansion of the enteric virome. The longitudinal studies conducted by Handley et al. allowed for the assessment of microbiome change over the course of disease progression, and also in animals that succumbed to AIDS. In Monaco et al., the HIV-infected population included individuals over a broad range of disease severity. These studies of human and macaques share the observation that virome expansion, particularly of those viruses that also associated with GI disease, was greatly pronounced with AIDS compared to with chronic (clinically latent) lentiviral infection. Taken together, these results suggest a role for the virome in progression to AIDS, but not necessarily in increased inflammation and gut permeability that occurs in chronic untreated infection or in populations on successful ART.
Monaco et al. are also use next generation sequencing to report the adult HIV-associated microbiome in Sub-Saharan Africa rather than in the United States or Europe as done in previous studies. Performing HIV/microbiome research in Sub-Saharan Africa is crucial because of a high disease burden in those regions, and because strong baseline gut microbiome differences between regions, particularly in agrarian populations compared to those with Western diets, may lead to different responses to HIV (Yatsunenko et al., 2012). Also, the demographics of individuals infected with HIV differs substantially across populations, with transmission among men who have sex with men (MSM) and IV-drug users being common in most populations in the US and Europe, and heterosexual transmission being dominant in Sub-Saharan Africa.
Similar to their results for the virome, Monoco et al. found the most dramatic HIV-associated bacterial microbiome differences in subjects with CD4+ T numbers < 200 cells/ul. These individuals had significantly decreased bacterial richness compared to subjects with CD4+ T cell numbers > 200 cells/ul. Most prior studies of the HIV-associated gut microbiome had included few individuals with AIDS, but studies conducted in Europe that have included individuals with CD4+ T cell counts <200 had a consistent result of reduced alpha diversity (Marc Noguera-Julian a et al., 2016; Nowak et al., 2015).
Perhaps the most surprising findings of this work, however, is the lack of a pronounced difference in enteric bacteria composition in HIV-infected subjects with CD4+ T cell numbers > 200 cells/ul in individuals in Uganda. This finding is in stark contrast to studies of the enteric bacterial microbiome in the US and Europe which have reported substantial differences in the bacterial microbiome in HIV-infected subjects with CD4+ T cell numbers >200 and in those on successful ART. A likely cause of this difference is revealed by a recent study that found that sexual behavior (men who have sex with men; MSM) and not HIV status, explained the strong differences in bacterial composition that had been previously associated with HIV status by several research groups. Most notably an increase in the bacterial genus Prevotella and decrease in Bacteroides in HIV-infected individuals was associated with MSM populations independently of HIV status (Marc Noguera-Julian a et al., 2016). The Ugandan cohort studied Monoco et al. was greater than 60% female indicating that transmission in this cohort was dominated by heterosexual activity and accordingly no changes in Prevotella and Bacteroides were found. Furthermore, even an MSM-associated microbiome would likely be less pronounced in a Ugandan population because studies of HIV-negative individuals from Sub-Saharan Africa have shown high frequency of an MSM-resembling Prevotella-rich and Bacteroides-poor microbiome type independent of sexual behavior or HIV status. These findings highlight the importance of performing studies in diverse populations of HIV-infected subjects and in including proper control populations for sexual behavior.
Before the emergence of these more recent papers, it appeared that SIV infection of macaques was driving distinctive and less pronounced microbiome changes than HIV infection of humans, which raised the question of how well macaques model the human population. The observation of little to no bacterial microbiome differences in the absence of severe disease in SIV-infected macaques reported here by Handley et al. was consistent with these previous studies of macaques (although it may be important to note that more pronounced bacterial microbiome changes have also been observed in macaques in the acute phase of infection (Glavan et al., 2015), which was not evaluated by Handley et al.). Although the results of human and macaque studies may be more consistent than had been previously thought in terms of a lack of a very pronounced phenotype, it is also important to note that all of these studies have used feces to evaluate microbiome composition. Human studies that have evaluated microbiome changes with HIV using both feces and mucosal biopsies from the same individuals have shown more pronounced bacterial microbiome phenotypes in the biopsies (Dillon et al., 2014; Mutlu et al., 2014) and particularly in the terminal Ileum (Mutlu et al., 2014), which presumably would be less likely affected by MSM behavior. Furthermore, mucosal biopsies allow for the examination of microbes that are most closely associated with the immune system. Unfortunately both of the aforementioned studies suffered the limitation of not controlling for sexual behavior. Evaluating bacterial microbiome and virome using biopsies in human studies that are controlled for MSM sexual behavior will be an important future direction. Also, the DNA-based shotgun metagenomic sequencing performed in these studies does not evaluate RNA viruses, and detailed analysis of enteric fungi (the mycobiome) and other microbial eukaryotes using next generation sequencing have not yet been done. Evaluation of the complete microbiome in properly controlled cohorts will help to further our understanding of the degree to which the enteric microbiome is altered with HIV. This knowledge coupled with detailed mechanistic follow-up studies will reveal the degree to which the enteric microbiome may play a direct role in HIV pathogenesis and co-morbidity.
References
- Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, Kazzaz Z, Bornstein E, Lambotte O, Altmann D, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12:1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- Dillon SM, Lee EJ, Kotter CV, Austin GL, Dong Z, Hecht DK, Gianella S, Siewe B, Smith DM, Landay AL, et al. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol. 2014;7:983–994. doi: 10.1038/mi.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glavan TW, Gaulke CA, Santos Rocha C, Sankaran-Walters S, Hirao LA, Raffatellu M, Jiang G, Baumler AJ, Goulart LR, Dandekar S. Gut immune dysfunction through impaired innate pattern recognition receptor expression and gut microbiota dysbiosis in chronic SIV infection. Mucosal Immunol. 2015 doi: 10.1038/mi.2015.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handley SA, Thackray LB, Zhao G, Presti R, Miller AD, Droit L, Abbink P, Maxfield LF, Kambal A, Duan E, et al. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell. 2012;151:253–266. doi: 10.1016/j.cell.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguera-Julian M, et al. Gut Microbiota Linked to Sexual Preference and HIV Infection. EBioMedicine. 2016 doi: 10.1016/j.ebiom.2016.01.032. http://dx.doi.org/10.1016/j.ebiom.2016.01.032. [DOI] [PMC free article] [PubMed]
- Mutlu EA, Keshavarzian A, Losurdo J, Swanson G, Siewe B, Forsyth C, French A, Demarais P, Sun Y, Koenig L, et al. A compositional look at the human gastrointestinal microbiome and immune activation parameters in HIV infected subjects. PLoS Pathog. 2014;10:e1003829. doi: 10.1371/journal.ppat.1003829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak P, Troseid M, Avershina E, Barqasho B, Neogi U, Holm K, Hov JR, Noyan K, Vesterbacka J, Svard J, et al. Gut microbiota diversity predicts immune status in HIV-1 infection. AIDS. 2015;29:2409–2418. doi: 10.1097/QAD.0000000000000869. [DOI] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]