Key Points
Full oncogenic activity of MLL-Af4 is facilitated by lymphoid commitment and is compromised in the myeloid cell context.
The CHD domain of Af4 is sufficient to confer the linkage between lymphoid lineage and leukemic transformation.
Abstract
Chromosome rearrangements involving the mixed-lineage leukemia gene (MLL) create MLL-fusion proteins, which could drive both acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML). The lineage decision of MLL-fusion leukemia is influenced by the fusion partner and microenvironment. To investigate the interplay of fusion proteins and microenvironment in lineage choice, we transplanted human hematopoietic stem and progenitor cells (HSPCs) expressing MLL-AF9 or MLL-Af4 into immunodeficient NSGS mice, which strongly promote myeloid development. Cells expressing MLL-AF9 efficiently developed AML in NSGS mice. In contrast, MLL-Af4 cells, which were fully oncogenic under lymphoid conditions present in NSG mice, displayed compromised transformation capacity in a myeloid microenvironment. MLL-Af4 activated a self-renewal program in a lineage-dependent manner, showing the leukemogenic activity of MLL-Af4 was interlinked with lymphoid lineage commitment. The C-terminal homology domain (CHD) of Af4 was sufficient to confer this linkage. Although the MLL-CHD fusion protein failed to immortalize HSPCs in myeloid conditions in vitro, it could successfully induce ALL in NSG mice. Our data suggest that defective self-renewal ability and leukemogenesis of MLL-Af4 myeloid cells could contribute to the strong B-cell ALL association of MLL-AF4 leukemia observed in the clinic.
Introduction
Acute leukemia can be characterized as acute lymphoblastic leukemia (ALL) or acute myeloid leukemia (AML) depending on the lineage markers and morphology. Chromosome rearrangements involving 11q23, fusing the N terminus of the mixed-lineage leukemia gene (MLL) to various fusion partners, can result in both AML and ALL with intermediate to poor prognosis.1,2 MLL-fusion leukemia cells of different lineages exhibit distinct properties which guide therapy.3,4 Lineage plasticity has been reported for MLL-fusion patients.4,5 Understanding how MLL-fusion leukemia makes lineage decisions could improve disease treatment. Lineage association of MLL-fusion leukemia is influenced by the fusion partner. Although some types of MLL fusions can present as both AML and ALL, MLL-AF4, the most frequent MLL fusion, is almost exclusively associated with B-cell ALL (B-ALL).1 Recently, we have established a faithful model for MLL-AF4 pro–B-ALL by expressing the MLL–murine Af4 chimeric fusion in human CD34+ hematopoietic stem and progenitor cells (HSPCs).6 The cells expressing MLL-Af4 exhibited strong predilection for lymphoid lineage and a resistance to myeloid redirection. They retained the capacity to initiate B-ALL in immunodeficient nonobese diabetic/severe combined immunodeficiency/γ (NSG) mice even after being cultured in myeloid-promoting conditions for weeks, in striking contrast to CD34+ cells expressing the MLL-AF9 fusion protein, which could only give rise to AML after such conditioning.6 We previously reported that microenvironmental cues from recipient mice can also direct the lineage decision of MLL-fusion leukemia.7 To further understand the interplay between oncogene and microenvironment in lineage choice of MLL-fusion leukemia, we investigated the possibility of fully reprogramming the MLL-Af4 cells into AML in the myeloid-biasing mouse strain (NSG mice expressing human myeloid cytokines interleukin-3, granulocyte-macrophage colony-stimulating factor, and stem cell factor [NSGS]), which has been shown to enhance AML development.8
Study design
Human CD34+ HSPCs were transduced and transplanted into conditioned NSGS mice after myeloid culture for 2 to 6 weeks or without culturing. Secondary transplantation was performed to determine the malignant nature of the disease (Figure 1A).6,9
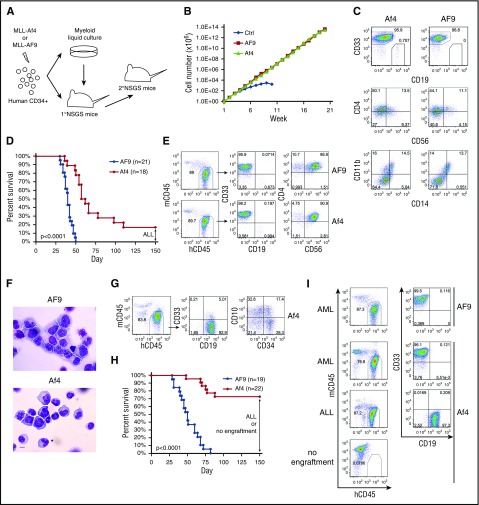
Figure 1.
Transformation capacity of MLL-Af4 is compromised in the myeloid microenvironment. (A) Schematic of experiment. (B) Growth curve of human HSPCs expressing MLL-AF9/MLL-Af4 in myeloid culture. One representative experiment of 3 is shown. (C) Flow cytometry analysis of cell surface marker of week 5 myeloid cultures. (D) Survival curve of primary NSGS mice received MLL-AF9/MLL-Af4 cells. The BM of the remaining living Af4 mice was examined at day 150 and showed ALL. Five independent experiments were included. (E) Cell surface maker analysis by flow cytometry of BM from MLL-AF9/MLL-Af4 primary NSGS mice developing myeloid disease. (F) Wright-Giemsa–stained BM cytospins of primary mice. The images were obtained using a Motic BA310 microscope with ×40 objective. Scale bar, 10 µm. (G) Flow cytometry analysis of BM from MLL-Af4 primary NSGS mice developing lymphoid disease. (H) Survival curve of secondary NSGS mice receiving primary myeloid disease. The BM of the remaining living Af4 mice was examined at day 150 and showed ALL or no human engraftment. Five independent experiments were included. (I) Representative flow cytometry analysis of BM from secondary recipients. P values were calculated using the log-rank test (see also supplemental Table 1). hCD45, human CD45; mCD45, murine CD45.
Results and discussion
MLL-Af4 cells maintained long-term growth under myeloid culture condition, showing similar proliferation rate and immunophenotype as MLL-AF9 cells (Figure 1B-C). However, CD19+CD33− lymphoid cells were consistently observed only in MLL-Af4 but not MLL-AF9 cultures (Figure 1C).6 When transplanted into NSGS mice, MLL-AF9 cells initiated myeloid disease with 100% penetrance (Figure 1D). The engrafted cells were CD33+CD19−, with aberrant expression of CD56 and CD4 (Figure 1E), as reported for MLL-fusion AML patients.10 The myeloid blasts were confirmed in the bone marrow (BM) (Figure 1F). The majority (15 of 18) of NSGS mice receiving MLL-Af4 cells developed rapid disease, though with a slightly longer latency compared with MLL-AF9 (Figure 1D). The disease also showed AML features, with the immunophenotype of CD33+CD19−CD56+CD4+ and myeloid blast morphology (Figure 1E-F). Interestingly, 3 MLL-Af4 mice remained alive at 150 days posttransplant (Figure 1D). BM examination showed an expansion of CD19+CD33− lymphoid cells variably expressing CD34 and CD10 (Figure 1G), representing an early stage of pro–B-ALL.6 Accordingly, these ALL cells displayed upregulation of lymphoid-signature genes and downregulation of myeloid-signature genes identified in MLL-fusion leukemia patients when compared with MLL-Af4 myeloid cells (supplemental Figure 1, available on the Blood Web site).11 Thus, MLL-Af4 cells can induce B-ALL even in a BM environment favoring myeloid lineage development, emphasizing their lymphoid bias.
To confirm the malignant nature of the myeloid disease, we performed secondary transplantation. As expected, all MLL-AF9 secondary recipients developed AML (Figure 1H-I). In contrast, only some of the MLL-Af4 myeloid disease was transplantable, with only 6 of 22 mice showing signs of AML after a relatively short latency. Strikingly, no human cells were detected in the majority of MLL-Af4 secondary recipients at 150 days posttransplant. Intriguingly, a myeloid-lymphoid lineage switch was observed in 4 MLL-Af4 secondary recipients (Figure 1H-I; supplemental Table 1). These data suggest that MLL-Af4 myeloid cells showed compromised long-term self-renewal in vivo, in stark contrast to MLL-Af4–induced ALL, which was reproducibly transplantable.6
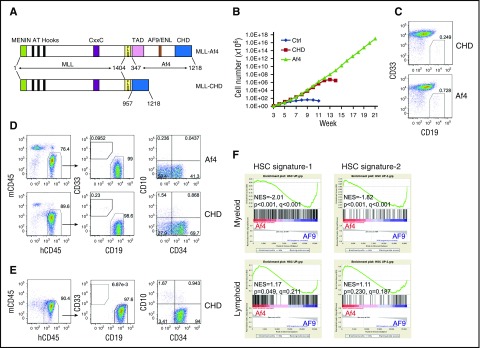
AF5q31 (AFF4) is a rare fusion partner of MLL.12 It belongs to the same family as AF4 and shares extensive sequence similarity. The C-terminal homology domain (CHD) of AF5q31 is critical for the function of MLL-AF5q31, and MLL fused with the CHD of AF5q31 is sufficient to immortalize murine HSPCs.13 Whether the CHD of the Af4 protein plays a similar role in MLL-Af4–mediated transformation of human HSPCs has not been successfully explored. Therefore, we fused MLL with the Af4 CHD (MLL-CHD) (Figure 2A) and investigated its transforming capacity. Human CD34+ cells expressing MLL-CHD failed to establish long-term myeloid cultures, although a transient proliferative advantage was evident (Figure 2B), indicating that MLL-CHD has reduced self-renewal ability in myeloid conditions compared with MLL-Af4. However, we found that MLL-CHD cultures contained a small percentage of CD19+CD33− cells, similar to MLL-Af4 cultures (Figure 2C), suggesting that these cells retain the same lymphoid lineage preference. To test their potential for lymphoid transformation, we transplanted MLL-CHD cells into NSG mice. Indeed, the injected MLL-CHD cells initiated CD19+CD34+CD10− pro–B-ALL, behaving the same as MLL-Af4 cells (Figure 2D). Importantly, the MLL-CHD–induced pro–B-ALL could be reproducibly transplanted into secondary recipients and maintained the same immunophenotype (Figure 2E; supplemental Table 2). These results demonstrate that the CHD is a significant functional unit for MLL-Af4–mediated leukemogenesis and also confers lymphoid lineage preference.
Figure 2.
The full activation of the self-renewal program by MLL-Af4 depends on lymphoid lineage. (A) Structure of MLL-Af4 and MLL-CHD constructs. (B) Growth curve of human HSPCs expressing MLL-Af4/MLL-CHD in myeloid culture. One representative experiment of 3 is shown. (C) Flow cytometry analysis of CD33/CD19 expression of MLL-Af4/MLL-CHD cultures at week 5. (D) Cell surface maker analysis by flow cytometry of BM from primary NSG mice receiving MLL-Af4/MLL-CHD myeloid cells. (E) Representative flow cytometry analysis of secondary MLL-CHD ALL (see also supplemental Table 2). (F) Gene set enrichment analysis to assess the activation of HSC signature comparing MLL-Af4 cells with MLL-AF9 cells in myeloid or lymphoid lineage. NES, normalized enrichment score; TAD, transactivation domain.
These observations suggest that the lymphoid cellular context is necessary for MLL-Af4 to elicit full oncogenic activity. It has been shown that the MLL-fusion protein promotes leukemia development by activating or maintaining a hematopoietic stem cell (HSC) molecular network.14 We extracted the human HSC gene signature from 2 independent data sets (supplemental Figure 2A-B). Gene set enrichment analysis15 showed that the HSC gene signatures derived by us and others were underrepresented in MLL-Af4 myeloid cells compared with MLL-AF9 myeloid cells, potentially accounting for the diminished AML transformation of MLL-Af4 cells. In contrast, this incomplete activation of the HSC program by MLL-Af4 was not seen in ALL cells (Figure 2F; supplemental Figure 2C). Similar results were observed using pathway enrichment analysis by LRpath (supplemental Figure 2D).16
Taken together, our study reveals that the full transforming capacity of the MLL-Af4 fusion protein is interlinked with lymphoid lineage identity and the CHD portion of the protein is sufficient to provide this linkage. Although MLL-Af4 cells may be reprogrammed into AML by microenvironmental pressure, these cells appear unstable and vulnerable, with most cells either losing long-term self-renewal ability or undergoing lineage switch back to ALL. The relatively defective self-renewal ability and leukemogenesis of MLL-Af4 myeloid cells may explain the selective advantage for lymphoid commitment, contributing to the strong B-ALL association of MLL-AF4 leukemia. It has been reported that partial myelomonocytic differentiation is required for MLL-AF9–induced AML,17 likely due to myeloid transcription factor contribution to the oncogenic program.18,19 Thus, there is the possibility that an essential cooperation exists between lymphoid-specific factors and MLL-Af4, which provides the lymphoid preference of MLL-Af4 cells. It was reported that 2 t(4;11) ALL patients displayed a myeloid shift after CD19-directed therapy.4,20 These 2 patients then received myeloid-directed therapy followed by allogeneic HSC transplantation. One infant patient, whose myeloid lineage transition was triggered by blinatumomab, achieved complete remission and remained alive at the time of publication.4 In contrast, the adult patient who showed a myeloid shift after CD19 chimeric antigen receptor T therapy, although achieving complete remission, died of AML relapse.20 The ALL-to-AML switch can also be induced by chemotherapy and is usually associated with poor outcome.5 Acquisition of additional genetic events enhancing the self-renewal program may give rise to the occasional MLL-Af4 AML in our model and t(4;11) AML in patients. Future investigation of our MLL-Af4 AML may help to identify critical cooperating factors driving the myeloid transition and potential therapeutic targets for this poor-outcome disease (or for preventing escape from CD19-directed therapy).
Supplementary Material
The online version of this article contains a data supplement.
Acknowledgments
The authors thank members of the Mulloy and Thirman laboratories for experimental support.
This work was supported by an Institutional Clinical and Translational Science Award, National Institutes of Health, National Institute of General Medical Sciences grant number 1UL1RR026314-01, a Translational Trials Development and Support Laboratory Award (US Public Health Service grant number MO1 RR 08084), a National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases Center of Excellence in Molecular Hematology P30 award (DK090971), a CancerFree KIDS Research Award (S.L.), an American Society of Hematology Bridge grant (M.J.T.), National Institutes of Health, National Cancer Institute grant CA215504 (J.C.M. and M.J.T.), and a Leukemia & Lymphoma Society Translational Research grant (M.J.T.). J.C.M. is a Leukemia & Lymphoma Society Scholar.
Footnotes
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: S.L. and J.C.M. conceived and designed the study, performed experiments, analyzed data, and wrote the manuscript; M.S. performed experiments; and R.T.L. and M.J.T. conceived the study and built the constructs.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: James C. Mulloy, Cancer and Blood Disease Institute, Cincinnati Children’s Hospital Medical Center, 3333 Burnet Ave, ML 7013, Cincinnati, OH 45229; e-mail: james.mulloy@cchmc.org; and Shan Lin, Cancer and Blood Disease Institute, Cincinnati Children’s Hospital Medical Center, 3333 Burnet Ave, ML 7013, Cincinnati, OH 45229; e-mail: shan.lin@cchmc.org.
References
- 1.Meyer C, Hofmann J, Burmeister T, et al. The MLL recombinome of acute leukemias in 2013. Leukemia. 2013;27(11):2165-2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7(11):823-833. [DOI] [PubMed] [Google Scholar]
- 3.Liu H, Westergard TD, Cashen A, et al. Proteasome inhibitors evoke latent tumor suppression programs in pro-B MLL leukemias through MLL-AF4. Cancer Cell. 2014;25(4):530-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rayes A, McMasters RL, O’Brien MM. Lineage switch in MLL-rearranged infant leukemia following CD19-directed therapy. Pediatr Blood Cancer. 2016;63(6):1113-1115. [DOI] [PubMed] [Google Scholar]
- 5.Rossi JG, Bernasconi AR, Alonso CN, et al. Lineage switch in childhood acute leukemia: an unusual event with poor outcome. Am J Hematol. 2012;87(9):890-897. [DOI] [PubMed] [Google Scholar]
- 6.Lin S, Luo RT, Ptasinska A, et al. Instructive role of MLL-fusion proteins revealed by a model of t(4;11) pro-B acute lymphoblastic leukemia. Cancer Cell. 2016;30(5):737-749. [DOI] [PubMed] [Google Scholar]
- 7.Wei J, Wunderlich M, Fox C, et al. Microenvironment determines lineage fate in a human model of MLL-AF9 leukemia. Cancer Cell. 2008;13(6):483-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wunderlich M, Chou FS, Link KA, et al. AML xenograft efficiency is significantly improved in NOD/SCID-IL2RG mice constitutively expressing human SCF, GM-CSF and IL-3. Leukemia. 2010;24(10):1785-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Link KA, Lin S, Shrestha M, et al. Supraphysiologic levels of the AML1-ETO isoform AE9a are essential for transformation. Proc Natl Acad Sci USA. 2016;113(32):9075-9080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baer MR, Stewart CC, Lawrence D, et al. Acute myeloid leukemia with 11q23 translocations: myelomonocytic immunophenotype by multiparameter flow cytometry. Leukemia. 1998;12(3):317-325. [DOI] [PubMed] [Google Scholar]
- 11.Kohlmann A, Schoch C, Dugas M, et al. New insights into MLL gene rearranged acute leukemias using gene expression profiling: shared pathways, lineage commitment, and partner genes. Leukemia. 2005;19(6):953-964. [DOI] [PubMed] [Google Scholar]
- 12.Taki T, Kano H, Taniwaki M, Sako M, Yanagisawa M, Hayashi Y. AF5q31, a newly identified AF4-related gene, is fused to MLL in infant acute lymphoblastic leukemia with ins(5;11)(q31;q13q23). Proc Natl Acad Sci USA. 1999;96(25):14535-14540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yokoyama A, Lin M, Naresh A, Kitabayashi I, Cleary ML. A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell. 2010;17(2):198-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krivtsov AV, Twomey D, Feng Z, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442(7104):818-822. [DOI] [PubMed] [Google Scholar]
- 15.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545-15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sartor MA, Leikauf GD, Medvedovic M. LRpath: a logistic regression approach for identifying enriched biological groups in gene expression data. Bioinformatics. 2009;25(2):211-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye M, Zhang H, Yang H, et al. Hematopoietic differentiation is required for initiation of acute myeloid leukemia. Cell Stem Cell. 2015;17(5):611-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohlsson E, Hasemann MS, Willer A, et al. Initiation of MLL-rearranged AML is dependent on C/EBPα. J Exp Med. 2014;211(1):5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou J, Wu J, Li B, et al. PU.1 is essential for MLL leukemia partially via crosstalk with the MEIS/HOX pathway. Leukemia. 2014;28(7):1436-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gardner R, Wu D, Cherian S, et al. Acquisition of a CD19-negative myeloid phenotype allows immune escape of MLL-rearranged B-ALL from CD19 CAR-T-cell therapy. Blood. 2016;127(20):2406-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.