Abstract
Triclosan (TCS) is widely used as broad-spectrum antibacterial agent. However, it may threaten the health of human offspring if the mother is exposed to TCS during pregnancy. The present study aimed to identify potential mechanisms behind the toxic effect of TCS on the offspring of Caenorhabditis elegans (C. elegans), using this nematode as a suitable animal model. The results of the current study demonstrated that the locomotory behavior and reproductive capacity of C. elegans offspring was severely affected by prenatal exposure to different concentrations of TCS. A high-throughput gene microarray was performed to investigate molecular alterations in C. elegans offspring following TCS exposure during pregnancy. Microarray results indicated that 113 genes were differentially expressed following TCS treatment compared with the control group. Gene ontology analysis demonstrated that these dysregulated genes were primarily associated with neuron development, muscular strength and reproduction. Pathway analysis results demonstrated that differentially expressed genes participated in several signaling pathways, including arginine, proline, and purine metabolism, progesterone-mediated oocyte maturation and neuroactive ligand-receptor interaction. Finally, 7 TCS toxicity-associated genes were confirmed by reverse transcription-quantitative polymerase chain reaction. The present study indicates that TCS exposure during pregnancy may disturb the locomotory behavior and reproductive capacity of C. elegans offspring, primarily through 7 TCS toxicity-associated genes, which merits further study from an environmental health perspective.
Keywords: triclosan, locomotory behavior, reproduction, Caenorhabditis elegans, gene microarray
Introduction
Triclosan (TCS), a man-made spectrum antimicrobial agent, has been widely used in various types of personal care and industrial products, including hand soap and shampoo, due to its antibacterial properties and low acute toxicity (1). Until the 1990s, TCS was considered to be safe. However, TCS has since been associated with various pathologies and disorders, including obesity, thyroid dysfunction and breast cell hyper-proliferation (2–5). Humans are exposed to TCS via direct contact with household products, as well as via exposure to water, soil and other organisms that contain TCS, including fish (6,7). Previous epidemiology studies have demonstrated that TCS exists in human milk (8,9), blood plasma (10,11) and urine (12,13). The potential toxic effects of TCS may threaten the health of offspring if the mother is exposed during pregnancy. Paul et al (14) identified that perinatal maternal TCS exposure resulted in maternal and early neonatal hypothyroxinemic rats. A previous study also demonstrated that TCS exposure reduced thyroxine levels in pregnant and lactating rat dams, and in directly exposed offspring (15). However, the underlying mechanisms responsible for the observed disorders or pathologies are not well elucidated.
Caenorhabditis elegans (C. elegans) is a well-established animal model for the investigation of the molecular basis of fundamental biological processes (16). The worms are easily handled and sensitive to environmental stimuli, and highly similar to mammals with regard to pharmacological mechanisms, with 60–80% matching human genetic homologs having been identified (17). C. elegans has been previously used to investigate the molecular mechanisms underlying the initiation and development of various diseases (18–20). Therefore, C. elegans is an excellent candidate for investigating the mechanisms involved in toxicity following prenatal TCS exposure.
The present study exposed adult wild-type C. elegans N2 to TCS, collected the C. elegans filial 1 (F1) generation and performed high-throughput gene microarray analysis to determine the gene expression levels in the offspring in order to analyze the effect of TCS on the systematic gene expression and investigate the potential mechanisms underlying the toxicity of prenatal TCS exposure. The present study may help to improve an understanding of the toxic effects of TCS and provide evidence against the use of TCS in daily life.
Materials and methods
Chemicals and reagents
TCS was purchased from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany) and dissolved in dimethyl sulfoxide (DMSO) at a concentration of 50 mg/ml and stored at 20°C.
Culture conditions
C. elegans wild-type N2 was provided by the Caenorhabditis Genetics Center of Southeast University (Nanjing, China), C. elegans were cultured in standard nematode growth medium (NGM) (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) with 2.0×108 living Escherichia coli OP50 bacteria (American Type Culture Collection, Manassas, VA, USA) at 20°C (21).
C. elegans activity assay
C. elegans were seeded in 24-well plates and treated with different doses of TCS (0, 100, 200, 300, 400 and 500 µmol/l); subsequently, an equal volume of DMSO (dissolved in M9 minimal medium, Sigma-Aldrich; Merck KGaA; cat. no. M6030) was added to the culture medium and set as the solvent control. For each group, 10 young adult C. elegans were seeded into the well and exposed to TCS for 24 h at 20°C. The treated C. elegans were observed and recorded by stereo microscope (SZ61-SET; Olympus Corporation, Tokyo, Japan) at ×50 magnification. The death ratio was scored by failure to move after being prodded with a platinum wire (n=10/group).
Detection of toxicity of TCS on the behavioral characteristics of C. elegans F1 generation
In order to detect the potential toxicity of exposure to TCS during pregnancy, the present study observed the locomotory behavior, brood number, generation time and fat accumulation of the TCS-treated C. elegans F1 generation. Generally, it is recommended to detect the toxicity of chemicals by exposure to doses that are more than one-tenth of the IC50 dose (i.e., the half-maximal inhibitory concentration). Following exposure to different doses of TCS (100 nmol/l, and 1, 10 and 20 µmol/l) for 24 h at 20°C, 10 worms were selected randomly and transferred to normal NGM plates. The locomotory behavior, brood number, body length/width and generation time of the C. elegans F1 generation were calculated when the F1 generation synchronously grew to the larval stage 4 and were subjected to RNA detection subsequently (22).
Extraction of total RNA of C. elegans
Total RNA was extracted from 1,000 F1 generation of the TCS-treated (20 µmol/l) C. elegans (24 h at 20°C) and the solvent control group by using a total RNA kit (cat. no. R6688-00, Omega Bio-Tek, Inc., Norcross, GA, USA), according to the manufacturer's protocol. The purity and concentration of total RNA were determined by measuring the absorbance at 260 and 280 nm (260/280), and the integrity was checked using a NanoDrop 2000 (Thermo Fisher Scientific, Inc., Waltham, MA, USA) (23).
Gene microarray analysis
Based on the activity assay of TCS-treated C. elegans (F1 generation were calculated when the F1 generation synchronously grew to the larval stage 4), the median lethal concentration (LC50) was calculated and 1/10th the concentration of LC50 was selected as the experimental exposure concentration group for gene microarray analysis, and 1,000 F1 generation of TCS-treated (20 µmol/l) C. elegans (24 h at 20°C) and the solvent control group were selected for gene microarray analysis. Global gene expression was detected using an Affymetrix GeneChip™ C. elegans Gene 1.0 ST Array (Affymetrix, Inc., Santa Clara, CA, USA). Total RNA (~500 ng) was employed in each experiment. Briefly, total RNA from each sample was amplified using the Ambion WT Expression kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol (24). Subsequently, the generated cDNA (~5.5 mg) was separated into fragments and labeled using the GeneChip™ WT Terminal Labeling and Controls kit (Affymetrix, Inc., cat. no. G2519F), according to the manufacturer's protocol. Labeled cDNA target was hybridized to the Affymetrix GeneChip™ C. elegans Gene 1.0 ST Array using the GeneChip™ Hybridization, Wash and Stain kit (Affymetrix, Inc.) at 45°C for 16 h; subsequently, hybridized arrays were washed and stained on the Affymetrix GeneChip™ Command Console (Affymetrix, Inc.), according to the manufacturer's protocol, and scanned by a GeneChip™ Scanner 3000 7G (Affymetrix, Inc.). The acquired array images were analyzed by Affymetrix GeneChip™ Operating software (version GCOS1.4; Affymetrix, Inc.). Affymetrix Expression Console software (Affymetrix, Inc.) was used to perform quantile normalization and subsequent data processing. Data transformation was applied to set all negative raw values at 1.0, followed by quantile normalization. A filter on low gene expression was used to keep only the probes expressed in at least one sample. Differentially regulated genes were identified via fold change filtering.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated and reverse-transcribed into cDNA at 42°C by using the PrimeScript RT Master Mix (Perfect Real Time; Takara Bio, Inc., Otsu, Japan), and qPCR was performed on an ABI 7900HT Fast Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.) using Power SYBR Green PCR Master Mix (2X; Applied Biosystems; Thermo Fisher Scientific, Inc.) (25). Briefly, samples were incubated at 95°C for 10 min for initial denaturation, followed by 40 cycles of amplification that were performed at 95°C for 15 sec and at 60°C for 1 min. All data were calculated using standard relative quantification 2−ΔΔCq methods (26) and experiments were performed in triplicate. GAPDH was used as a normalization control. Primers for amplification were listed in Table I.
Table I.
Primers for reverse transcription-quantitative polymerase chain reaction of differentially expressed genes.
| Primer (5′-3′) | ||
|---|---|---|
| Gene symbol | Forward | Reverse |
| OSTR158D11_1 | AGGCGACAACACATTTCGTC | TCCCTCAGCCACTCTTATGC |
| F14F8.14 | ATACGACGAAATTTGCGGCA | CGGTCGAATATTGTGCCCTC |
| C48B6.4 | ATCGGCTCGCAACGTATTTC | GTCACAAACACCGAACACGA |
| skr-7 | ATGTCGCTCCTGAGAATCGT | TGCTCGGATGCCTCGATAAT |
| fip-1 | TGGCTGTCTTCTGTGCTGTA | TCGACCAGCTCCAGACAATT |
| B0563.9 | GTTGATGCGACCCCAAGATC | CGTCTCCAAGTCGATCCAGA |
| ent-5 | GCAAGAGTTCCAGTGATGGC | TGCTCCAAACGAACAGCATC |
| GAPDH | CACCAGATGTTTCCGTCGTT | GGCGAGGATTCCCTTCAT |
OSTR158D11_1, an expressed sequence tag for protein T13F3.4 (www.ncbi.nlm.nih.gov/nucest/30737247; www.ncbi.nlm.nih.gov/UniGene/clust.cgi?UGID=3496801&TAXID=6239); F14F8.14, hypothetical protein; C48B6.4, hypothetical protein; skr-7, SKp1 related (ubiquitin ligase complex component); fip-1, fungus-induced protein 1; B0563.9, hypothetical protein; ent-5, equilibrative nucleoside transporter.
Gene ontology (GO) and pathway analysis
GO function (www.geneontology.org) and Kyoto Encyclopedia of Genes and Genomes pathway analysis (www.kegg.jp) for differentially expressed genes were used to identify the significantly enriched biological terms and pathways. The database for annotation, visualization and integrated discovery (DAVID; david.abcc.ncifcrf.gov) was applied to perform GO function enrichment for differentially expressed genes. Putative genes that were upregulated >2-fold with P<0.05 following TCS exposure were considered to be differentially expressed and selected and submitted to DAVID Bioinformatics Resources 6.7 (david.abcc.ncifcrf.gov). The overall functions regulated by TCS-modulated genes were identified by functional annotation clustering and ranked by enrichment scores.
Statistical analysis
Statistical analysis was performed by SPSS 20.0 software (IBM Corp., Armonk, NY, USA). All data are presented as the mean ± standard deviation. Comparisons between groups were performed by one-way analysis of variance, followed by Tukey's post hoc test. P<0.05 was considered to indicate a statistically significant difference. Experiments were repeated at least three times independently.
Results
TCS exposure reduces the activity of C. elegans
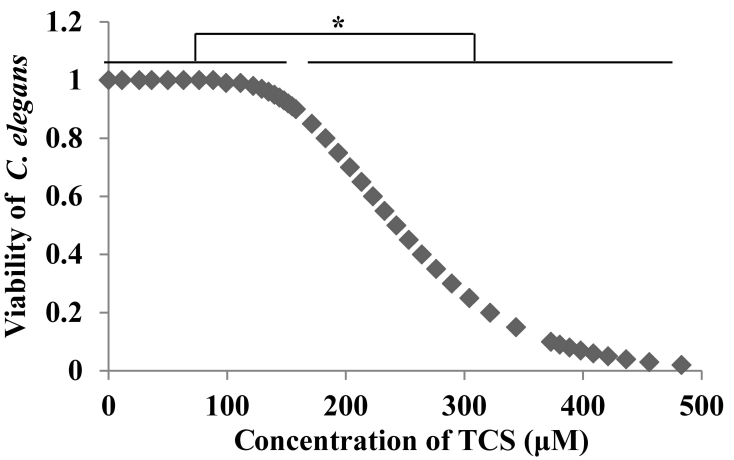
Young adult C. elegans were plated in 24-well plates and exposed to different doses of TCS (0, 100, 200, 300, 400 and 500 µmol/l) for 24 h. An equal volume of DMSO was added to the culture medium and used as the solvent control. The results demonstrated that TCS reduced the viability of C. elegans in a dose-dependent manner (Fig. 1). The LC50 (LC=242.639 µmol/l) was calculated using SPSS software and four doses of TCS were selected (100 nmol/l, and 1, 10, and 20 µmol/l), corresponding to the <1/10th LC50 for subsequent experiments.
Figure 1.
TCS exposure reduced the viability of C. elegans. C. elegans were exposed to different doses of TCS (0, 100, 200, 300, 400 and 500 µmol/l) for 24 h and the viability was calculated. The results indicated that TCS reduce the viability of C. elegans in a dose-dependent manner. The median lethal concentration (LC=242.639 µmol/l) was calculated with the SPSS software. Each experiment was performed at least three times independently. *P<0.05 vs. 0 µM TCS. TCS, triclosan; C. elegans, Caenorhabditis elegans.
TCS exposure affects the behavior characteristics of C. elegans F1 generation
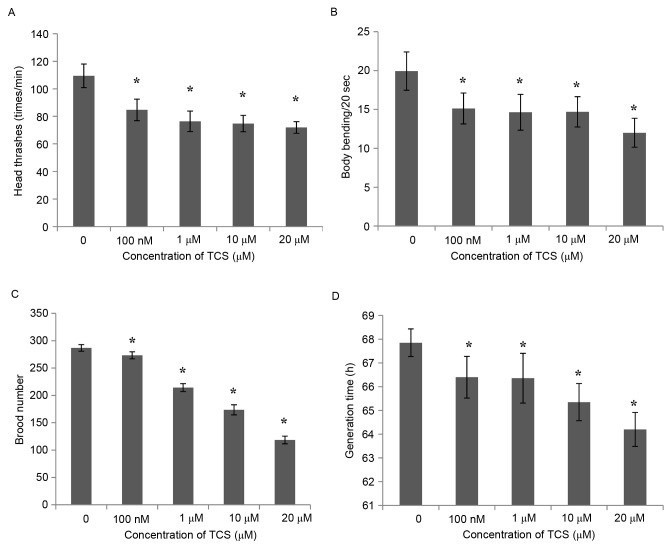
In order to investigate the genetic effect of TCS exposure on C. elegans, the present study evaluated several behavioral characteristics of the C. elegans F1 generation, including locomotory ability, reproduction, growth time, body length and width. The results demonstrated that, compared with the control group, the number of head thrashes of the F1 generation was inhibited significantly following TCS treatment (Fig. 2A), and the same was observed for body bending every 20 sec (Fig. 2B). These results indicate that TCS exposure may reduce the exercise capacity of the F1 generation of C. elegans. Subsequently, the brood number of the F1 generation was calculated, and the results demonstrated that the reproductive ability of the F1 generation was reduced in a dose-dependent manner (by 4.71, 25.60, 39.45 and 58.67% for the 100 nmol/l, and 1, 10, and 20 µmol/l treatments, respectively) compared with the control group (Fig. 2C). In addition to locomotory and reproductive ability, the growth time, body length and width of the F1 generation were also detected. The results indicated that the generation time was shortened by 2.14–5.38% following TCS exposure, and appeared to be regulated in a dose-dependent manner (Fig. 2D), while the body length and width were not affected by TCS exposure (data not shown).
Figure 2.
TCS exposure affected the behavioral characteristics of the C. elegans F1 generation. (A) Number of head thrashes per min, (B) body bending per 20 sec, (C) brood number and (D) generation time of the F1 C. elegans generation were measured. Each experiment was performed at least three times independently. *P<0.05 vs. 0 µM TCS. TCS, triclosan; C. elegans, Caenorhabditis elegans; F1, filial 1.
Microarray analysis following TCS treatment
To investigate the potential mechanisms of the effect of TCS on the behavioral characteristics of C. elegans F1 generation, gene microarray analysis was performed to analyze the systematic gene expression pattern of the C. elegans F1 generation following TCS exposure (data not shown). The results demonstrated that, compared with the control group, 113 genes were dysregulated following TCS treatment, including 25 that were upregulated and 88 that were downregulated (the fold change threshold was 2.0; P<0.05; Fig. 3).
Figure 3.
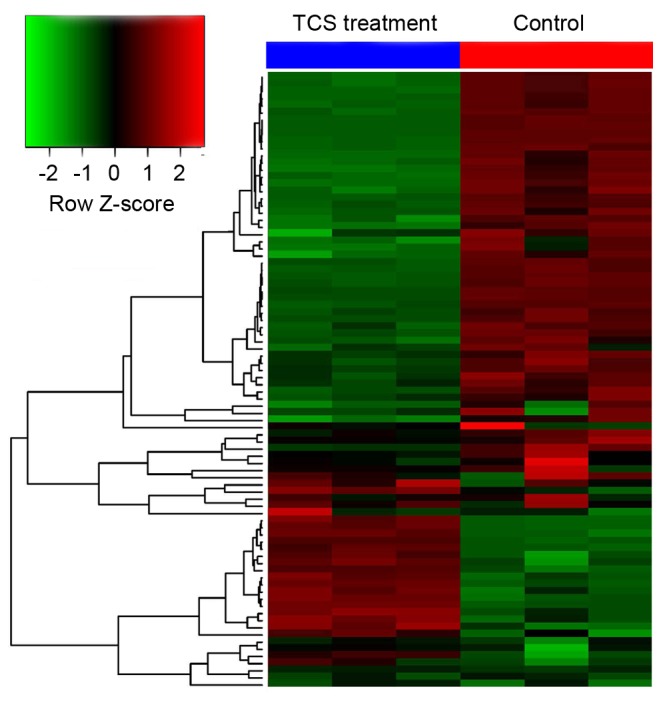
Gene microarray analysis of the filial 1 C. elegans generation following TCS exposure. Compared with the control group, 113 genes were dysregulated following TCS exposure, including 25 upregulated and 88 downregulated genes. The fold change threshold was 2.0 and P<0.05. Each experiment was performed at least three times independently. C. elegans, Caenorhabditis elegans; TCS, triclosan.
GO and pathway analysis of differentially expressed genes
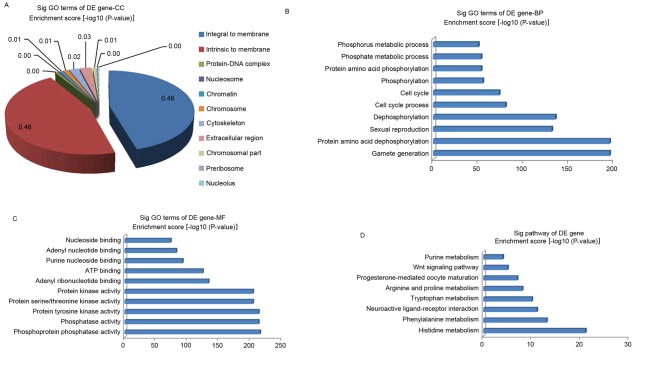
GO analysis was performed in the present study to clarify the functions of the dysregulated genes. The GO project (http://www.geneontology.org) primarily covers three areas (encompassing biological processes, molecular function, and the cellular component), and the GO-analyzed results indicated that these gene products are primarily associated with the cellular component terms ‘integral to membrane’, ‘intrinsic to membrane’, ‘protein-DNA complex’, ‘nucleosome’ and ‘chromatin’ (Fig. 4A). In addition, the genes were enriched in the biological processes of ‘phosphorus metabolic process’, ‘phosphate metabolic process’, ‘protein amino acid dephosphorylation’, ‘protein amino acid phosphorylation’, and others not presented in Fig. 4B. The molecular functions of these genes included phosphoprotein phosphatase activity, protein tyrosine kinase activity and protein kinase activity (Fig. 4C). At the same time, the pathway analysis indicated that these gene products participated in several signaling pathways, including arginine and proline metabolism (cel00330), purine metabolism (cel00230), progesterone-mediated oocyte maturation (cel04914), neuroactive ligand-receptor interaction (cel04080), the Wnt signaling pathway (cel04310), phenylalanine metabolism (cel00360), histidine metabolism (cel00340) and tryptophan metabolism (cel00380; Fig. 4D). The P-value denotes the significance of the GO term enrichment and the pathway associated with the conditions: The lower the P-value, the more significant the GO term and pathway.
Figure 4.
GO and pathway analysis of DE genes (Database for Annotation, Visualization and Integrated Discovery). GO analysis indicated the (A) CC, (B) BP and (C) MF terms that the DE genes were enriched in. (D) Pathways that DE genes were enriched in, as determined by pathway analysis. GO, gene ontology; DE, differentially expressed; CC, cellular component; BP, biological process; MF, molecular function; Sig, significant.
Discovery of TCS toxicity-associated genes
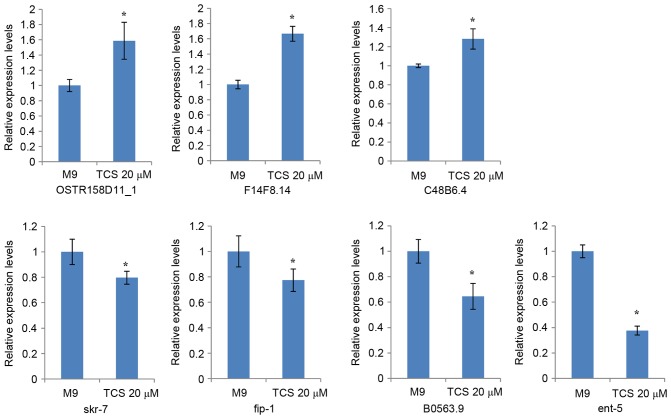
The present study validated the expression levels of seven dysregulated genes, which were associated with motor nerves and selected according to the following criteria: Fold-change filtering, absolute fold-change >2.0; standard Student t-test, P<0.05; multiple hypotheses testing, false discovery rate <0.05; and at least 1 out of 2 groups have flags in Present or Marginal. The RT-qPCR results demonstrated that 7 genes (OSTR158D11_1, F14F8.14, C48B6.4, skr-7, fip-1, B0563.9, and ent-5) had differential expression levels in the TCS-treated F1 generation compared with the controls (Fig. 5).
Figure 5.
Discovery of TCS toxicity-associated genes. Results of reverse transcription-quantitative polymerase chain reaction demonstrated that 7 genes had differential expression in the TCS-treated filial 1 generation compared with the solvent control (dimethyl sulfoxide dissolved in M9 minimal medium). Each experiment was performed at least three times independently. *P<0.05 vs. solvent control. TCS, triclosan; OSTR158D11_1, an expressed sequence tag for protein T13F3.4; F14F8.14, hypothetical protein; C48B6.4, hypothetical protein; skr-7, SKp1 related (ubiquitin ligase complex component); fip-1, fungus-induced protein 1; B0563.9, hypothetical protein; ent-5, equilibrative nucleoside transporter; M9, solvent control.
Discussion
TCS has been widely used in the environment around human beings and has been identified to be associated with various types of diseases, including thyroid dysfunction and obesity (3,4). The mechanism that is responsible for TCS-induced disorders remains unclear, and TCS exposure may threaten the health of offspring if exposed during pregnancy. The current study aimed to investigate the potential mechanisms underlying the effect of TCS on offspring by using C. elegans.
In order to exclude the effect of TCS exposure on C. elegans viability, the toxicity of TCS on C. elegans was investigated. The results demonstrated that TCS reduced the viability of C. elegans at doses >100 µmol/l in a dose-dependent manner (Fig. 1) and the LC50 was calculated to be 242.639 µmol/l. Subsequently, four doses of TCS were selected for further investigation.
In addition, the toxic effect of TCS on offspring (F1 generation) during pregnancy exposure was investigated by comparing the behavioral characteristics of the TCS-treated F1 generation with those of the control group. The results demonstrated that, compared with the control group, the head thrashes, body bending, brood number and generation time of the F1 generation were inhibited significantly by treatment with TCS. These results indicated that TCS clearly induced genetic toxicity in the F1 generation. Previously, Jung et al (27) demonstrated that TCS was able to decrease total serum triiodothyronine and thyroxine in pregnant rats, and decrease sex ratio and pup body weights, and delayed vaginal opening in offspring, which also demonstrated the toxicity of TCS exposure on the next generation.
To investigate the potential mechanisms of the effect of TCS on the behavioral characteristics of the C. elegans F1 generation, gene microarray analysis was performed to analyze the systematic gene expression pattern of the C. elegans F1 generation following TCS exposure. Compared with the control group, 113 genes were dysregulated after TCS treatment, including 25 that were up-regulated, and 88 that were down-regulated (the fold change threshold was 2.0; P<0.05). In order to elucidate the basic function of these dysregulated genes, GO analysis was performed. The GO project (http://www.geneontology.org) primarily covers three areas (encompassing biological processes, molecular function, and the cellular component), and the GO-analyzed results indicated that these gene products are primarily associated with the cellular component terms ‘integral to membrane’, ‘intrinsic to membrane’, ‘protein-DNA complex’, ‘nucleosome’ and ‘chromatin’. In addition, the genes were primarily enriched in the biological processes of ‘phosphorus metabolic process’, ‘phosphate metabolic process’, ‘protein amino acid dephosphorylation’ and others not mentioned in Fig. 4B. The molecular functions of these genes included phosphoprotein phosphatase activity, protein tyrosine kinase activity, protein kinase activity, pattern binding, and polysaccharide binding and chitin binding. At the same time, the pathway analysis indicated that these gene products participated in several signaling pathways, including arginine and proline metabolism (cel00330), purine metabolism (cel00230), progesterone-mediated oocyte maturation (cel04914), neuroactive ligand-receptor interaction (cel04080), the Wnt signaling pathway (cel04310), phenylalanine metabolism (cel00360), histidine metabolism (cel00340) and tryptophan metabolism (cel00380).
A previous report demonstrated that neuroactive ligand-receptors are associated with neuron development (28). In the present study, it was observed that TCS decreased the ability of head thrashes and body bending in C. elegans, meaning that TCS may possibly affect neuronal development in C. elegans through regulation of neuroactive ligand-receptor interaction. In an additional study, Tribulo et al (29) demonstrated that the Wnt signaling pathway was involved in the regulation of embryonic development, through which TCS may decrease the brood number of C. elegans. The bioinformatic analysis results from the present study demonstrated that TCS was able to regulate the behaviors of C. elegans through the dysregulation of certain genes. In order to investigate the underlying mechanism of the toxic effect of TCS, motor nerve and reproduction-associated genes were selected for examination. The results of the present study demonstrated that 7 genes (OSTR158D11_1, F14F8.14, C48B6.4, skr-7, fip-1, B0563.9, and ent-5) were differentially expressed in the TCS-treated F1 generation compared with the controls. These 7 genes may be important in explaining the effect of TCS exposure during pregnancy. Coincidentally, Nayak et al (30) demonstrated that the SKp1-related gene family in C. elegans may have critical roles in regulating cell proliferation, meiosis and morphogenesis. This finding may explain the toxicity of TCS exposure observed on brood number and generation time of C. elegans progeny in the present study.
In conclusion, the present study demonstrated that locomotory behavior and reproductive capacity of C. elegans offspring was severely affected by prenatal exposure to different concentrations of TCS, and 7 genes were confirmed to be associated with toxicity induced by TCS exposure, which merits further investigation from an environmental health perspective.
Acknowledgements
The present study was financially supported by the National Key Basic Research Program of China (grant no. 2013CB530604), the Key Project of the National Natural Science Foundation of China (grant no. 81330067), the Key Project of Medical Science and Technology Development Foundation, Nanjing Department of Health (grant no. JQX13012) and the Program for Innovative Research Teams of Jiangsu Province (grant no. LJ201108).
References
- 1.Dhillon GS, Kaur S, Pulicharla R, Brar SK, Cledón M, Verma M, Surampalli RY. Triclosan: Current status, occurrence, environmental risks and bioaccumulation potential. Int J Environ Res Public Health. 2015;12:5657–5684. doi: 10.3390/ijerph120505657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jung EM, An BS, Choi KC, Jeung EB. Potential estrogenic activity of triclosan in the uterus of immature rats and rat pituitary GH3 cells. Toxicol Lett. 2012;208:142–148. doi: 10.1016/j.toxlet.2011.10.017. [DOI] [PubMed] [Google Scholar]
- 3.Geens T, Dirtu AC, Dirinck E, Malarvannan G, van Gaal L, Jorens PG, Covaci A. Daily intake of bisphenol A and triclosan and their association with anthropometric data, thyroid hormones and weight loss in overweight and obese individuals. Environ Int. 2015;76:98–105. doi: 10.1016/j.envint.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Xue J, Wu Q, Sakthivel S, Pavithran PV, Vasukutty JR, Kannan K. Urinary levels of endocrine-disrupting chemicals, including bisphenols, bisphenol A diglycidyl ethers, benzophenones, parabens, and triclosan in obese and non-obese Indian children. Environ Res. 2015;137:120–128. doi: 10.1016/j.envres.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Lee HR, Hwang KA, Nam KH, Kim HC, Choi KC. Progression of breast cancer cells was enhanced by endocrine-disrupting chemicals, triclosan and octylphenol, via an estrogen receptor-dependent signaling pathway in cellular and mouse xenograft models. Chem Res Toxicol. 2014;27:834–842. doi: 10.1021/tx5000156. [DOI] [PubMed] [Google Scholar]
- 6.Symsaris EC, Fotidis IA, Stasinakis AS, Angelidaki I. Effects of triclosan, diclofenac, and nonylphenol on mesophilic and thermophilic methanogenic activity and on the methanogenic communities. J Hazard Mater. 2015;291:45–51. doi: 10.1016/j.jhazmat.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 7.Yang YS, Kwon JT, Shim I, Kim HM, Kim P, Kim JC, Lee K. Evaluation of toxicity to triclosan in rats following 28 days of exposure to aerosol inhalation. Regul Toxicol Pharmacol. 2015;71:259–268. doi: 10.1016/j.yrtph.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 8.Adolfsson-Erici M, Pettersson M, Parkkonen J, Sturve J. Triclosan, a commonly used bactericide found in human milk and in the aquatic environment in Sweden. Chemosphere. 2002;46:1485–1489. doi: 10.1016/S0045-6535(01)00255-7. [DOI] [PubMed] [Google Scholar]
- 9.Dayan AD. Risk assessment of triclosan [Irgasan] in human breast milk. Food Chem Toxicol. 2007;45:125–129. doi: 10.1016/j.fct.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 10.Hovander L, Malmberg T, Athanasiadou M, Athanassiadis I, Rahm S, Bergman A, Wehler EK. Identification of hydroxylated PCB metabolites and other phenolic halogenated pollutants in human blood plasma. Arch Environ Contam Toxicol. 2002;42:105–117. doi: 10.1007/s002440010298. [DOI] [PubMed] [Google Scholar]
- 11.Allmyr M, McLachlan MS, Sandborgh-Englund G, Adolfsson-Erici M. Determination of triclosan as its pentafluorobenzoyl ester in human plasma and milk using electron capture negative ionization mass spectrometry. Anal Chem. 2006;78:6542–6546. doi: 10.1021/ac060666x. [DOI] [PubMed] [Google Scholar]
- 12.Calafat AM, Ye X, Wong LY, Reidy JA, Needham LL. Urinary concentrations of triclosan in the U.S. population: 2003–2004. Environ Health Perspect. 2008;116:303–307. doi: 10.1289/ehp.10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X, Ying GG, Zhao JL, Chen ZF, Lai HJ, Su HC. 4-Nonylphenol, bisphenol-A and triclosan levels in human urine of children and students in China, and the effects of drinking these bottled materials on the levels. Environ Int. 2013;52:81–86. doi: 10.1016/j.envint.2011.03.026. [DOI] [PubMed] [Google Scholar]
- 14.Paul KB, Hedge JM, Devito MJ, Crofton KM. Developmental triclosan exposure decreases maternal and neonatal thyroxine in rats. Environ Toxicol Chem. 2010;29:2840–2844. doi: 10.1002/etc.339. [DOI] [PubMed] [Google Scholar]
- 15.Axelstad M, Boberg J, Vinggaard AM, Christiansen S, Hass U. Triclosan exposure reduces thyroxine levels in pregnant and lactating rat dams and in directly exposed offspring. Food Chem Toxicol. 2013;59:534–540. doi: 10.1016/j.fct.2013.06.050. [DOI] [PubMed] [Google Scholar]
- 16.Chalasani SH, Chronis N, Tsunozaki M, Gray JM, Ramot D, Goodman MB, Bargmann CI. Corrigendum: Dissecting a circuit for olfactory behaviour in Caenorhabditis elegans. Nature. 2016;533:130. doi: 10.1038/nature16515. [DOI] [PubMed] [Google Scholar]
- 17.Chatterjee N, Yang JS, Park K, Oh SM, Park J, Choi J. Screening of toxic potential of graphene family nanomaterials using in vitro and alternative in vivo toxicity testing systems. Environ Health Toxicol. 2015;30:e2015007. doi: 10.5620/eht.e2015007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tedesco P, Visone M, Parrilli E, Tutino ML, Perrin E, Maida I, Fani R, Ballestriero F, Santos R, Pinilla C, et al. Investigating the role of the host multidrug resistance associated protein transporter family in burkholderia cepacia complex pathogenicity using a Caenorhabditis elegans infection model. PLoS One. 2015;10:e0142883. doi: 10.1371/journal.pone.0142883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao Y, Yu X, Jia R, Yang R, Rui Q, Wang D. Lactic acid bacteria protects Caenorhabditis elegans from toxicity of graphene oxide by maintaining normal intestinal permeability under different genetic backgrounds. Sci Rep. 2015;5:17233. doi: 10.1038/srep17233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Srivastava S, Pant A, Trivedi S, Pandey R. Curcumin and β-caryophellene attenuate cadmium quantum dots induced oxidative stress and lethality in Caenorhabditis elegans model system. Environ Toxicol Pharmacol. 2016;42:55–62. doi: 10.1016/j.etap.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 21.Warnhoff K, Kornfeld K. New links between protein N-terminal acetylation, dauer diapause and the insulin/IGF-1 signaling pathway in Caenorhabditis elegans. Worm. 2015;4:e1023498. doi: 10.1080/21624054.2015.1023498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roussel N, Sprenger J, Tappan SJ, Glaser JR. Robust tracking and quantification of C. elegans body shape and locomotion through coiling, entanglement, and omega bends. Worm. 2015;3:e982437. doi: 10.4161/21624054.2014.982437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lv J, Xia K, Xu P, Sun E, Ma J, Gao S, Zhou Q, Zhang M, Wang F, Chen F, et al. miRNA expression patterns in chemoresistant breast cancer tissues. Biomed Pharmacother. 2014;68:935–942. doi: 10.1016/j.biopha.2014.09.011. [DOI] [PubMed] [Google Scholar]
- 24.Lv J, Fu Z, Shi M, Xia K, Ji C, Xu P, Lv M, Pan B, Dai L, Xie H. Systematic analysis of gene expression pattern in has-miR-760 overexpressed resistance of the MCF-7 human breast cancer cell to doxorubicin. Biomed Pharmacother. 2015;69:162–169. doi: 10.1016/j.biopha.2014.11.028. [DOI] [PubMed] [Google Scholar]
- 25.Zhang W, Wang F, Xu P, Miao C, Zeng X, Cui X, Lu C, Xie H, Yin H, Chen F, et al. Perfluorooctanoic acid stimulates breast cancer cells invasion and up-regulates matrix metalloproteinase-2/−9 expression mediated by activating NF-κB. Toxicol Lett. 2014;229:118–125. doi: 10.1016/j.toxlet.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Jung EM, An BS, Choi KC, Jeung EB. Potential estrogenic activity of triclosan in the uterus of immature rats and rat pituitary GH3 cells. Toxicol Lett. 2011;208:142–148. doi: 10.1016/j.toxlet.2011.10.017. [DOI] [PubMed] [Google Scholar]
- 28.Xiao J, Vemula SR, Xue Y, Khan MM, Kuruvilla KP, Marquez-Lona EM, Cobb MR, LeDoux MS. Motor phenotypes and molecular networks associated with germline deficiency of Ciz1. Exp Neurol. 2016;283:110–120. doi: 10.1016/j.expneurol.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tribulo P, Moss JI, Ozawa M, Jiang Z, Tian XC, Hansen PJ. WNT regulation of embryonic development likely involves pathways independent of nuclear CTNNB1. Reproduction. 2017;153:405–419. doi: 10.1530/REP-16-0610. [DOI] [PubMed] [Google Scholar]
- 30.Nayak S, Santiago FE, Jin H, Lin D, Schedl T, Kipreos ET. The Caenorhabditis elegans Skp1-related gene family: Diverse functions in cell proliferation, morphogenesis, and meiosis. Curr Biol. 2002;12:277–287. doi: 10.1016/S0960-9822(02)00682-6. [DOI] [PubMed] [Google Scholar]