Abstract
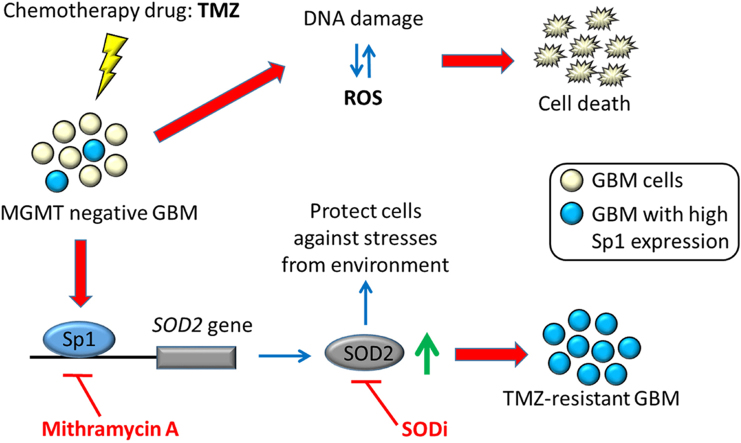
Acquisition of temozolomide (TMZ) resistance is a major factor leading to the failure of glioblastoma (GBM) treatment. The exact mechanism by which GBM evades TMZ toxicity is not always related to the expression of the DNA repair enzyme O6-methylguanine-DNA methyltransferase (MGMT), and so remains unclear. In this study, TMZ-resistant variants derived from MGMT-negative GBM clinical samples and cell lines were studied, revealing there to be increased specificity protein 1 (Sp1) expression associated with reduced reactive oxygen species (ROS) accumulation following TMZ treatment. Analysis of gene expression databases along with cell studies identified the ROS scavenger superoxide dismutase 2 (SOD2) as being disease-related. SOD2 expression was also increased, and it was found to be co-expressed with Sp1 in TMZ-resistant cells. Investigation of the SOD2 promoter revealed Sp1 as a critical transcriptional activator that enhances SOD2 gene expression. Co-treatment with an Sp1 inhibitor restored the inhibitory effects of TMZ, and decreased SOD2 levels in TMZ-resistant cells. This treatment strategy restored susceptibility to TMZ in xenograft animals, leading to prolonged survival in an orthotopic model. Thus, our results suggest that Sp1 modulates ROS scavengers as a novel mechanism to increase cancer malignancy and resistance to chemotherapy. Inhibition of this pathway may represent a potential therapeutic target for restoring treatment susceptibility in GBM.
Keywords: Specificity protein 1, Superoxide dismutase 2, Reactive oxygen species, Temozolomide, O6-methylguanine-DNA methyltransferase
Graphical abstract
Highlights
-
•
Sp1 levels increase in TMZ resistance of MGMT-deficient cells.
-
•
Sp1 upregulation enables GBM cells to escape TMZ-mediated toxicity.
-
•
Sp1 is a critical transcriptional activator to enhance SOD2 gene expression.
-
•
Sp1-SOD2 pathway raises GBM malignancy and chemotherapy resistance.
-
•
Inhibition of Sp1-SOD2 pathway restores treatment susceptibility in GBM.
1. Introduction
Glioblastoma (GBM) usually has a poor prognosis, and is never considered curable even following treatment with the most widely used first-line chemotherapeutic agent temozolomide (TMZ). In one phase III clinical trial, approximately 90% of the patients suffered from disease recurrence within two years of treatment, regardless of the initial response [1]. TMZ is known to cause lethal DNA damage (O6-methylation-mediated DNA base mismatching) and subsequent reactive oxygen species (ROS) production [2]. Resistance to TMZ treatment is well known to occur because of the presence of the DNA repair protein O6-methylguanine-DNA methyltransferase (MGMT) [3]. It is important to note that regardless of MGMT status, the acquisition of TMZ resistance inevitably occurs, suggesting that non-MGMT resistance mechanisms are operative in GBM. Compared with innate resistance, the development of acquired resistance is considered more complex, with multiple factors being involved, such as a mitochondrial adaptive response to TMZ genotoxic stress [4], and the stress-activated kinase p38, against TMZ-induced cell death [5]. It is therefore of interest to elucidate how cellular stress responses serve to protect GBMs from the effects of TMZ.
Specificity protein 1 (Sp1) is a nuclear transcription factor that is ubiquitously expressed in mammalian cells, and regulates multiple genes and cellular functions. It has been demonstrated previously that Sp1 is upregulated in most cancer cell types, such as breast, gastric, cervical, and GBM [6], [7], [8]. We have previously shown that Sp1 is a stress-sensitive transcription factor that increases the expression of genes that can protect against stress-induced cellular damage [9], [10]. Chemotherapy is commonly performed after surgery in order to target cancer cells, but whether TMZ-induced environmental stress affects Sp1 levels and its functions, remains unclear.
Strict regulation of ROS levels is essential for cells to maintain viability and to avoid oxidative damage from stress overload [11]. Multiple antioxidant enzymes including superoxide dismutases (SODs), catalase, and glutathione peroxidases (GPxs) are important, and have individual roles in the steps required to convert superoxide into water and oxygen [12]. Under certain circumstances, tumors have been shown to upregulate antioxidant enzyme expression to promote survival and resistance to certain anticancer agents [12]. For example, SOD2-overexpressing human ovarian and prostate cancer cells are prone to survive radiation toxicity, demonstrating increased resistance to the radiation treatment compared with control cells [13], [14]. Notably, TMZ is known to induce ROS production in glioma cells, thus leading to the activation of cell death signaling pathways [2]. The question remains, however, whether antioxidant enzymes promote the development of TMZ resistance in GBM, and especially in MGMT-deficient cells.
In order to determine the major factors involved in acquired TMZ drug resistance, we hypothesized that exposure of GBM to TMZ would activate the antioxidant defensive system in malignant cell populations that are prone to survive TMZ-induced cytotoxicity. By using MGMT-negative cell lines, we identified Sp1 as a key factor. Sp1 expression was induced in TMZ-resistant cells. Further exploration of this process identified SOD2 as a downstream and critical target of Sp1 action. Inhibition of Sp1 restored the TMZ effect in both TMZ-resistant cells and xenografts.
2. Materials and methods
2.1. Cells culture
Three patient-derived GBM lines, P#3, P#5, and P#11, were obtained according to the Taipei Medical University IRB protocol (201006011). These cell lines as well as human U87MG (ATCC) and A172 (ATCC) GBM cells, human MCF7 breast cancer cells (ATCC), and human Hone-1 nasopharyngeal carcinoma cells [15] were cultured in DMEM medium (Invitrogen) containing 10% FBS, 100 μg/mL streptomycin sulfate, and 100 U/mL penicillin-G sodium at 37 °C and 5% CO2. TMZ-resistant cells were maintained in the same culture medium containing 50 μM or 100 μM TMZ as indicated.
2.2. MTT assay
Cells were plated onto 24-well culture plates at an initial density of 1 × 105 cells/well. After one day of incubation, cells were treated with different doses of drugs as indicated for various time intervals. Subsequently, fresh medium containing 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reagent (final concentration of 0.5 mg/mL, Sigma Aldrich) was added to each well, and cells were then incubated for 1 h at 37 °C. Finally, the MTT medium was removed and the resultant formazan crystals were dissolved in 200 μL of DMSO. The absorbance readings of the DMSO extracts were measured at 570 nm by using an iMark Microplate Absorbance Reader (Bio-Rad).
2.3. Databases
To investigate the transcriptional levels of Sp1, SODs, catalase, and GPxs, human brain tumor gene expression profiles and the associated clinical pathological parameters were obtained from publicly available cancer microarray databases ONCOMINE (www.oncomine.org) [16]. In ONCOMINE, gene expression data from similar studies using the same methodology were used to compare gene expression for a tumor type with its normal counterpart. All data were log2-transformed and median-centered per array, and the standard deviation was normalized to one per array data [16]. We considered a gene to be overexpressed in a brain tumor when its mean expression level in the tumor samples was higher than the mean expression value in the normal tissue counterpart (Threshold: p-value < 0.05, fold change > 1.5).
2.4. Western blotting analysis
Protein samples were separated by SDS-PAGE, and transferred onto a PVDF membrane (Bio-Rad Laboratories). Subsequently, the PVDF membrane was blocked for 1 h by using 5% nonfat milk in TBST buffer and incubated with primary antibodies, including anti-Sp1 (1:1000, Millipore), anti-SOD2 (1:1000, GeneTex), anti-MGMT (1:1000, Santa Cruz Biotechnology), anti-phospho-Rad50 (Ser635) (1:1000, Cell Signaling), and anti-tubulin (1:3000, Proteintech) antibodies, for 2 h. After incubation, the membranes were washed with TBST buffer and then incubated with the appropriate secondary antibodies (1:3000, Santa Cruz Biotechnology) for 1 h. Finally, the membranes were washed again and the peroxidase was detected using chemiluminescence with Amersham Hyperfilm ECL (GE Healthcare).
2.5. Immunohistochemistry (IHC)
All immunohistochemical staining was performed on 10-μm-thick sections of paraformaldehyde-fixed, paraffin-embedded xenograft tumor tissues as well as on a human Glioma Tissue Microarray (US Biomax). The IHC method has been described previously [8]. The slides were incubated with a primary antibody at a dilution of 1:200 after antigen retrieval, and then developed using the EnVision+ staining kit (DAKO). The antibodies used were Sp1 (Millipore) and SOD2 (Cell Signaling).
2.6. Soft agar colony formation assay
Cells (2 × 104) were suspended in 2 mL of a 37 °C, 0.4% SeaKem LE Agarose (Lonza) solution in DMEM containing 10% FBS and the same culture medium supplements as described above. This suspension formed the upper layer, which was poured onto a lower layer formed of 0.8% agarose in a composition otherwise identical to the upper layer, in one 6-cm dish. Subsequently, the cells were incubated at 37 °C in a 5% CO2 atmosphere. A few drops of fresh medium were added twice a week. After fourteen days, the cells were treated with different doses of drugs as indicated for three days. The colonies were then stained with 0.05% crystal violet and photographed. All cultures were performed in duplicate.
2.7. Measurement of cellular ROS
Intracellular ROS levels were measured using dihydroethidium (DHE) [17]. Cells were collected and incubated with 10 µM DHE at 37 °C for 30 min. After incubation, cells were washed and DHE fluorescence was detected by flow cytometry (BD FACSCalibur), and analyzed using BD CellQuest software.
2.8. Real-time PCR
Complementary DNA was converted from total mRNA by reverse-transcription PCR [18]. The level of SOD2 cDNA were determined using 2 × SYBR real time master mix (Applied Biosystems) and primers specific for SOD2 (F: 5′- GGCCTACGTGAACAACCTGAA; R: 5′- CTGTAACATCTCCCTTGGCCA), or the control gene GAPDH (F: 5′- GAAGGTGAAGGTCGGAGTC; R: 5′- GAAGATGGTGATGGGATTC). SYBR green fluorescence was then monitored using an ABI 7000 Sequence Detection System (Applied Biosystems).
2.9. DNA affinity precipitation assay (DAPA)
The DAPA method has been described previously [9], [19]. A 5′-biotin end-labeled double-stranded probe, derived from the human SOD2 promoter (sequence -103 to -75: 5′- CCGCGGGGGGGGGGGGCGGGGCGGCGGTGC containing two Sp1 binding elements), was synthesized by Mission Biotech, Inc.
2.10. Experimental animals
Male NOD-SCID mice (5–6 weeks old, BioLASCO, Taiwan) were maintained at the animal facility of the National Health Research Institutes (Taiwan). For subcutaneous inoculation, 2 × 106 TMZ-resistant cells were injected into the right flank. Tumor volume was measured twice a week based on the following formula provided by the National Cancer Institute: tumor volume = length × width2 × 3.14/6. When the volume reached 200 mm3, animals were randomly assigned at the start of treatment. For the orthotropic model, a skull burr hole was first created in the right frontal brain area. An ultra-fine needle was then inserted to a depth of 3 mm using sterostatic guiding and 5 × 105 TMZ-resistant cells were injected. Treatment was initiated 10 days later. The scheduled treatment was interrupted when body weight loss became greater than 10% and was reinitiated after weight recovery. The endpoint was death or profound tumor impact leading to sacrifice. With regard to the treatment, mithromycin A (MA) or vehicle (DMSO) was dosed intraperitoneally, and TMZ was dosed by oral gavage. In the combined treatment group, TMZ was administered 3–4 h after MA injection. The survival curve was plotted using SPSS (IBM).
2.11. Statistics
Similar results were obtained from more than three independent experiments, and are expressed as the mean ± standard deviation. Comparisons among multiple groups were performed using a one-way ANOVA with appropriate post hoc tests, whereas comparisons between two groups were performed using unpaired, two-tailed Student's t-test with a significance level set at p < 0.05.
3. Results
3.1. TMZ resistance in MGMT-deficient GBM cells is not associated with de novo expression of MGMT
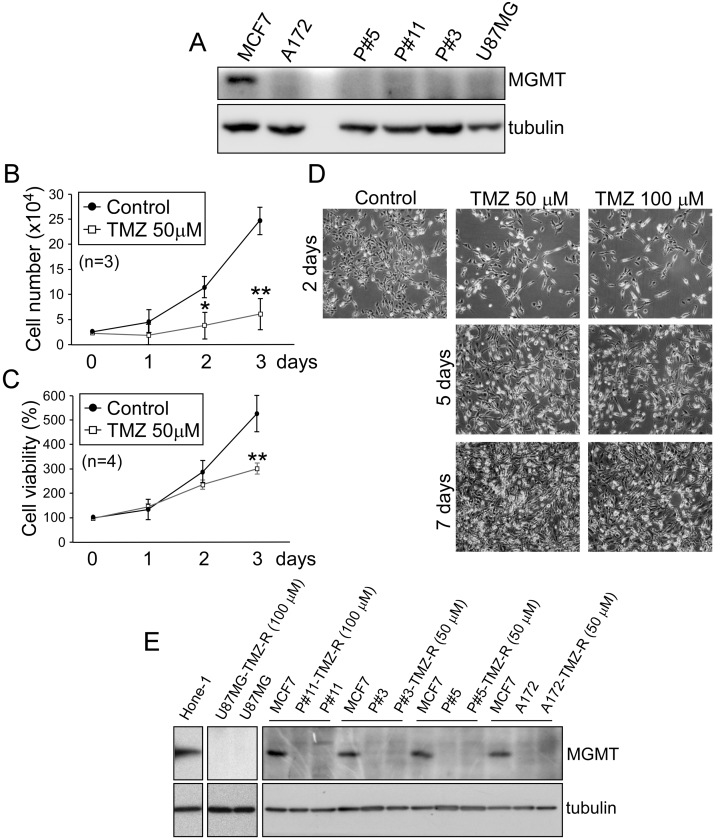
To investigate the MGMT-independent mechanisms of chemo-resistance in GBM, we treated MGMT-negative GBM cells, including U87MG, A172, P#3, P#5, and P#11 cells (Fig. 1A), with TMZ. Although the drug significantly reduced cellular proliferation and survival immediately after treatment (U87MG cells, Fig. 1B–D; other cell lines, data not shown), cell viability recovered after approximately one week (U87MG and P#11) to one month (A172, P#3 and P#5) of TMZ treatment. The resistance to TMZ was not related to MGMT expression, which remained undetectable (Fig. 1E). This finding confirmed that the survival of the several GBM cells treated with TMZ was MGMT-independent.
Fig. 1.
MGMT does not contribute to TMZ resistance in MGMT-negative GBM cells. (A) MGMT expression in MCF7 breast cancer cells, two GBM cell lines (U87MG, A172), and three patient-derived GBM cell lines (P#3, P#5, P#11). (B to D) U87MG cells were treated with either 50 µM or 100 µM TMZ for different time intervals as indicated. After treatment, (B) the number of viable cells was measured using the trypan blue dye exclusion method, (C) cell viability was assessed using the MTT colorimetric assay, and (D) representative images of cell morphology were acquired by microscopy. (E) Expression of MGMT in five GBM cell lines and TMZ-resistant cells (TMZ-R), formed after treatment with 50 µM or 100 µM TMZ for 1 month. HONE-1 cells and MCF7 cells were used as representative MGMT-positive cell lines. (t-test: *p < 0.05; **p < 0.01).
3.2. Upregulation of Sp1 enables GBM cells to escape TMZ-mediated toxicity
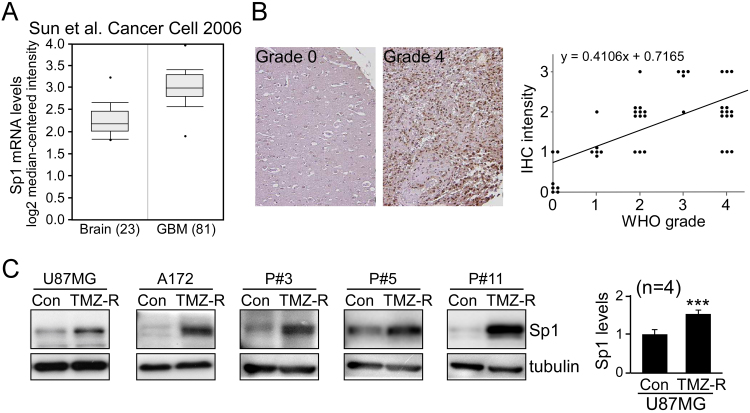
TMZ is also known to induce ROS generation, leading to cell death [2], [20]. Our previous studies reported that Sp1 in brain acts as a pleiotropic oxidative stress response protein [9], [10], providing protection to cells from peroxide damage. Using the ONCOMINE database (Fig. 2A and Table 1) and immunostaining of a tissue array (Fig. 2B), we found that Sp1 expression was increased in GBM compared with normal controls. Subsequent analysis of Sp1 expression in TMZ resistance revealed that Sp1 protein levels in TMZ-resistant cells were significantly higher than their parental tumor lines, including two established GBM lines, and three patient-derived GBM lines (Fig. 2C).
Fig. 2.
Sp1 is upregulated in GBM and enriched in TMZ-resistant cells. (A) A database study from Sun et al. in ONCOMINE (www.oncomine.org) showed significantly increased Sp1 mRNA expression in GBM [40]. (B) Sp1 protein expression in samples from brain tumor tissue array blocks was analyzed by IHC. The correlation between Sp1 level and tumor grade is shown in the right panel. (C) The expression of Sp1 in five TMZ-resistant GBM lines (TMZ-R) and their parental (Con) cells was measured by Western blotting. Quantitation of Sp1 levels in control and TMZ-resistant U87MG cells is shown in the right panel. (t-test: ***p < 0.001).
Table 1.
Studies in the ONCOMINE database showing an up-regulated expression of Sp1 in GBMs as compared with normal brain tissues.
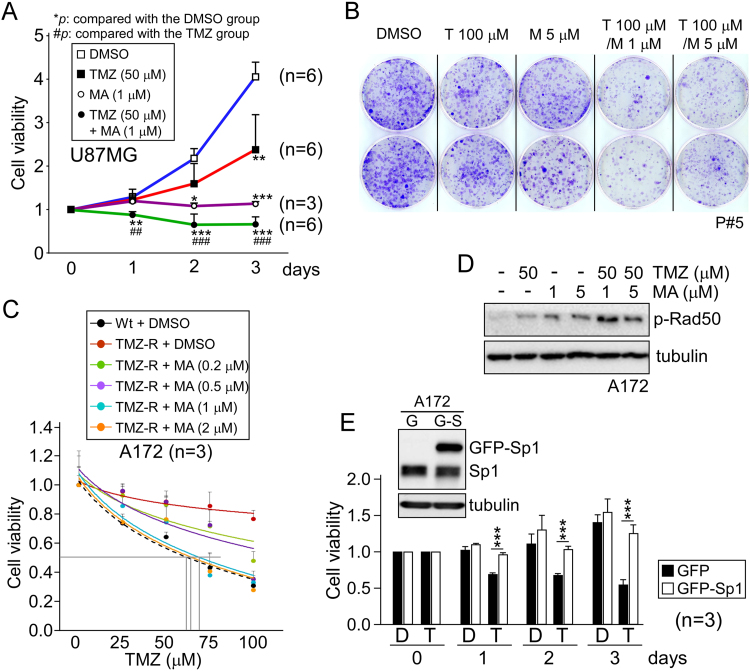
We next hypothesized that reducing Sp1 functionality may sensitize these cells to the anti-neoplastic agent. The Sp1 inhibitor mithromycin A (MA), was thus used in combination with TMZ in U87MG and P#5 GBM cells, which resulted in a significant inhibition of cell viability (Fig. 3A) and proliferation (Fig. 3B) compared to either drug alone. Additionally, we examined the viability of parental and TMZ-resistant A172 cells following TMZ exposure, and showed that the IC50 value for TMZ-inhibited growth in resistant cells (approximately 380 μM) was higher than in wild-type cells (approximately 60 μM, Fig. 3C). However, co-treatment with MA strongly sensitized resistant cells to TMZ (Fig. 3C) and increased TMZ-induced genotoxic stress, as shown by increased radiation-sensitive mutant 50 (Rad50) phosphorylation in response to DNA damage (Fig. 3D). Furthermore, we enhanced Sp1 expression by transient transfection, and showed that Sp1 overexpression reduced the inhibitory effect of TMZ on the growth of A172 cells (Fig. 3E). These findings collectively suggest that Sp1 protects GBM cells against TMZ-mediated cytotoxicity.
Fig. 3.
Sp1 inhibition enhances the TMZ anti-tumor effect. (A) U87MG cells were treated with TMZ (50 µM) alone, with the Sp1 inhibitor MA (1 µM) alone, or a combination of TMZ + MA and cell viability was assessed using the MTT assay. (B) P#5 GBM cells were grown in soft agar for two weeks, followed by treatment with TMZ (T) and/or MA (M) at the indicated concentrations for three days. Colonies were then fixed and stained with crystal violet. (C) Wild-type (Wt, dotted line) and resistant (TMZ-R, solid lines) A172 cells were treated with TMZ and/or MA at the indicated concentrations for two days, and the number of viable cells was measured using the trypan blue dye exclusion method. (D) One day after treatment with TMZ and/or MA, the activation of Rad50 in A172 cells was measured by Western blotting. (E) A172 cells were transfected with pEGFP or pEGFP-Sp1. One day after protein expression (upper panel), cells were treated with DMSO (D) or 100 µM TMZ (T) for the different time intervals as indicated, and cell viability was analyzed using the MTT assay. (*p < 0.05; **,##p < 0.01; ***,###p < 0.001).
3.3. TMZ-resistant cells exhibit low levels of intracellular ROS, but Sp1 inhibition abolishes this
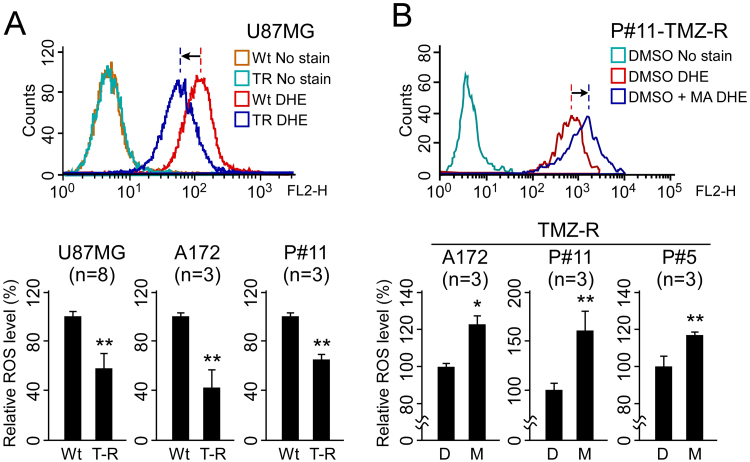
Since Sp1 is an oxidative stress-inducible, anti-death transcription factor [21], upregulated Sp1 may regulate the antioxidant system altering the production and/or removal of ROS in cells. Therefore, we examined intracellular ROS levels in TMZ-resistant cells using the superoxide indicator DHE. As shown in Fig. 4A background intracellular ROS levels in TMZ-resistant cells were significantly lower compared with parental cells. To further investigate whether Sp1 plays a role in the reduction of ROS levels, we treated TMZ-resistant cells with the Sp1 inhibitor MA and found that Sp1 inhibition induced a significant accumulation of intracellular ROS levels in resistant cells (Fig. 4B). These data suggest that increased Sp1 activity confers low levels of intracellular ROS, and that the antioxidant defensive system may be driven by Sp1.
Fig. 4.
Inhibition of Sp1 by MA increases intracellular ROS levels in resistant cells. (A) The intracellular ROS levels of wild-type (Wt) and TMZ-resistant (T-R) U87MG, A172, and P#11 cells were detected by labeling cells with DHE followed by flow cytometry. (B) TMZ-resistant cells (A172, P#11, P#5) were treated with DMSO (D) or 1 µM MA (M) for one day, and the intracellular ROS levels were then measured using DHE. (*p < 0.05; **p < 0.01).
3.4. SOD2 is highly expressed in TMZ-resistant GBM
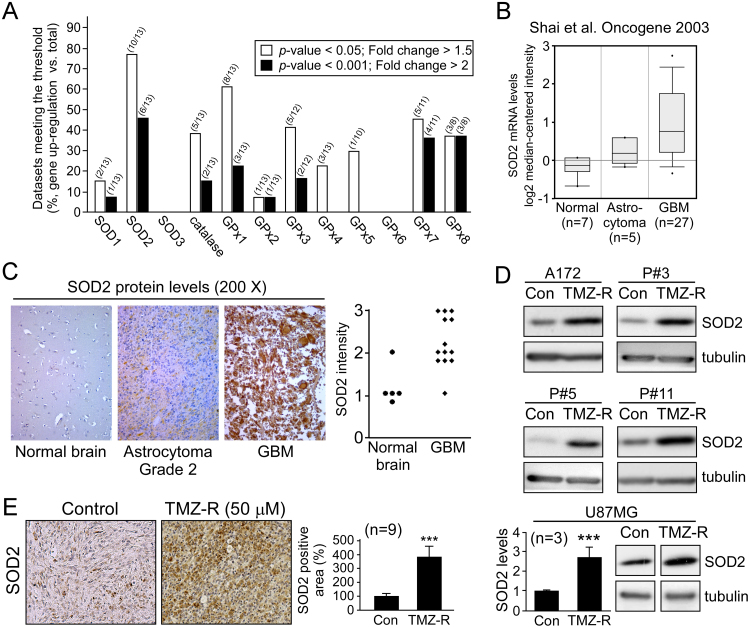
Investigation of the expression of individual ROS scavenging factors in GBM was first conducted by examining the ONCOMINE [16] database of clinical samples, in which the gene expression of the major cellular antioxidants including SOD 1–3, catalase, and GPx 1–8 was analyzed. In thirteen microarray datasets, SOD2 was increased to the greatest extent (Fig. 5A), with 10 of the 13 datasets showing a more than 1.5-fold difference (threshold: p-value < 0.05, fold change > 1.5) and 6 of the 13 showing a more than 2-fold difference (threshold: p-value < 0.001, fold change > 2; Table 2). Subsequent analysis of SOD2 gene expression revealed that the increased expression was associated with higher tumor grade (Fig. 5B). Increased SOD2 expression in GBM was confirmed by immunostaining of brain tissue arrays from GBM patients (Fig. 5C). Next, to determine the role of SOD2 in resistant GBM, the aforementioned TMZ-resistant cells (Fig. 5D), as well as recurrent tumors from GBM xenografts following long-term treatment with TMZ (Fig. 5E), were analyzed for SOD2 expression revealing that SOD2 levels were significantly overexpressed in these TMZ resistance models.
Fig. 5.
SOD2 is associated with acquired resistance to TMZ. (A) Gene expression profiles of SODs, catalase, and GPxs in brain tumors were analyzed using the ONCOMINE database. Fractions in parentheses indicate the number of upregulated (numerator) and total (denominator) datasets. (B) The correlation between SOD2 mRNA levels and tumor grade, as shown by the ONCOMINE database [44]. SOD2 significantly increased in GBM (p = 3.14E-6; fold change = 2.132). (C) SOD2 protein levels in different grades of brain tumor from tissue array blocks were analyzed using IHC. Dot plots represent the staining intensity of the individual samples (0–3+, t-test: p < 0.05). (D) SOD2 protein levels in five TMZ-resistant GBM lines (TMZ-R) and their parental (Con) cells were assessed by Western blotting. (E) SOD2 expression levels in subcutaneous U87MG (control and resistance) xenografts were analyzed by IHC. The resistant xenografts are recurrences of GBM xenografts following long-term treatment with TMZ (5 mg/kg, 5 days/week). (***p < 0.001).
Table 2.
Studies in the ONCOMINE database showing a significantly increased expression of SOD2 in GBMs as compared with normal brain tissues.
| Data bank or Author | Normal brain | GBM | Fold | t-test | P-value |
|---|---|---|---|---|---|
| The Cancer Genome Atlas (TCGA)/NCI/NIH | 10 | 542 | 2.356 | 13.77 | 1.95E−18 |
| Liang et al. [43] | 2 | 29 | 3.24 | 6.404 | 4.6E−7 |
| Sun, et al. [40] | 23 | 81 | 4.73 | 11.903 | 2.73E−21 |
| Bredel et al. [41] | 4 | 27 | 3.119 | 7.936 | 5.08E−9 |
| Shai et al. [44] | 7 | 27 | 2.132 | 5.407 | 3.14E−6 |
| Lee et al. [45] | 3 | 22 | 4.638 | 6.76 | 3.4E−7 |
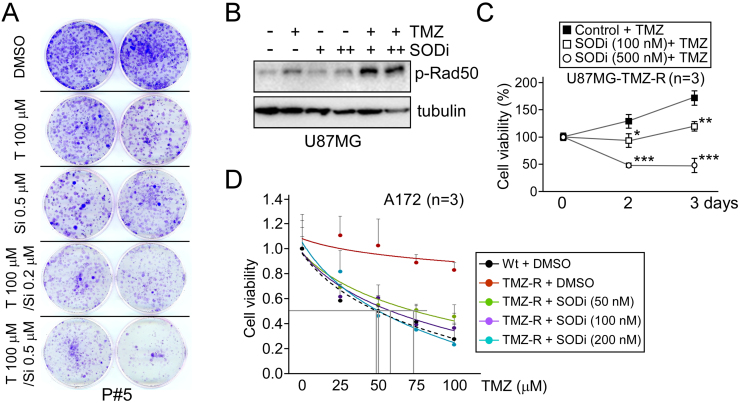
An SOD inhibitor (SODi), diethyldithiocarbamate, in combination with TMZ was subsequently applied to both GBM cells and TMZ-resistant GBM cells to investigate the effect of antioxidants on chemotherapy resistance. The data, shown in Fig. 6A, revealed a significant reduction in colony formation in GBM P#5 cells after treatment with the TMZ/SODi combination compared with either drug alone. Furthermore, we verified that SODi co-treatment strongly increased TMZ-induced DNA damage in U87MG cells (Fig. 6B), suppressed cell viability (Fig. 6C), and restored TMZ-induced cytotoxicity (Fig. 6D) in resistant cells as well. These findings suggest that increased SOD2 expression is an essential event in the process by which GBM cells become resistant to the anti-neoplastic agent TMZ.
Fig. 6.
SOD2 inhibition by diethyldithiocarbamate significantly reduces the TMZ resistance of GBM cells. (A) A colony formation assay was performed on P#5 GBM cells treated with TMZ (T) and/or SODi (Si) at the indicated concentrations. (B) One day after treatment with TMZ and/or SODi, the activation of Rad50 in U87MG cells was measured by Western blotting. (C) U87MG TMZ-resistant cells were treated with 50 µM TMZ alone or co-treated with SODi at different doses as indicated for two and three days. Cell proliferation was then assessed using a colorimetric MTT assay. (D) The wild-type (Wt, dotted line) and resistant (TMZ-R, solid lines) A172 cells were exposed to various concentrations of TMZ and/or SODi as indicated for two days, and cell viability was measured by the trypan blue exclusion method. (*p < 0.05; **p < 0.01; ***p < 0.001).
3.5. Sp1 activates SOD2 transcription in TMZ-resistant cells
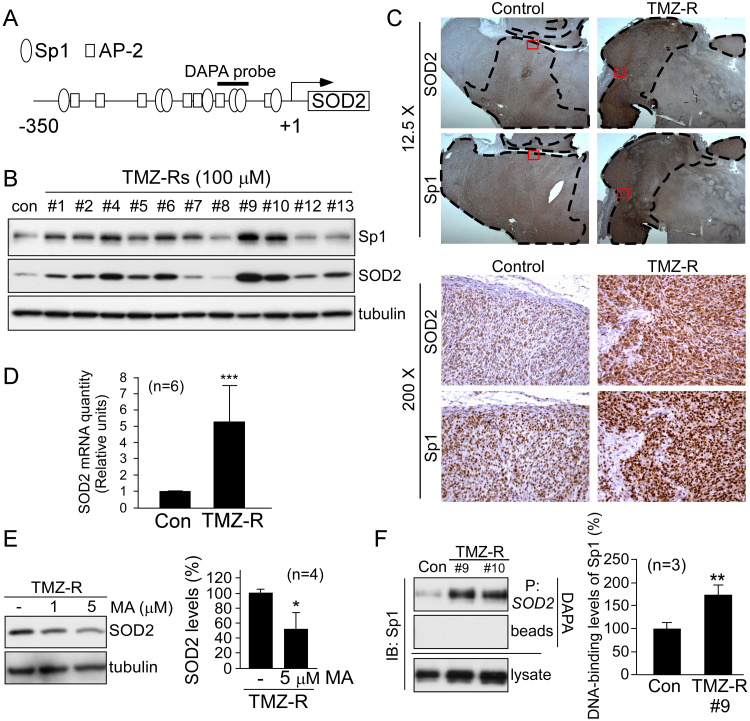
As shown in Fig. 7A, the human SOD2 promoter contains several Sp1 binding sites [22], indicating that Sp1 likely regulates SOD2 expression in resistant cells. To investigate the association between Sp1 and SOD2, we selected random single-cell clones that were simultaneously derived from TMZ-resistant U87MG cells. The data showed that 9 of 11 clones had increased expression of SOD2 with concurrently upregulated Sp1 (Fig. 7B), demonstrating a similar expression pattern of Sp1 and SOD2 in the majority of these resistant clones. Next, we studied U87MG xenografts and their recurrent tumors after continuous treatment with TMZ, and found increased expression of Sp1 and SOD2 in resistant tissues (Fig. 7C). Notably, areas in TMZ-resistant tumors with high Sp1 expression also exhibited increased SOD2 expression to a similar extent (upper panel, indicated by the dotted line). Subsequently, real-time PCR was performed to investigate the mRNA levels of SOD2, demonstrating that SOD2 mRNA levels were elevated in TMZ-resistant cells (Fig. 7D). Next, we used an Sp1 inhibitor to evaluate the role of Sp1 in TMZ-mediated SOD2 expression. As shown in Fig. 7E, Sp1 inhibition with MA reduced SOD2 protein expression in resistant cells. Finally, we performed a DAPA assay using a biotin-labeled Sp1 probe corresponding to the SOD2 promoter in U87MG cells. The results indicated that Sp1 bound to the SOD2 promoter, and its DNA binding was increased in resistant variants (#9 and #10, Fig. 7F). In summary, these results indicate that Sp1 plays a critical role in the upregulation of SOD2 in resistant GBM.
Fig. 7.
Sp1 activates the transcription of SOD2 in U87MG TMZ-resistant cells. (A) Schematic of the SOD2 core promoter showing several Sp1- and AP-2-binding sites. (B) U87MG cells were treated with 100 µM TMZ for one month. After treatment, eleven random single cell-derived clones from these surviving cells were analyzed by Western blotting with the indicated antibodies. (C) Control and TMZ-resistant GBM xenografts were analyzed by IHC using anti-Sp1 or anti-SOD2 antibodies. The dotted lines in the top panel outline areas with high Sp1 and SOD2 expression. The bottom panel is a magnified view of the inset red box from the top panel. (D) SOD2 mRNA levels in U87MG (Con) cells and resistant cells (TMZ-R) were quantified by real-time PCR. (E) After treatment with different doses of MA, SOD2 expression in TMZ-resistant cells was analyzed by Western blotting. (F) The level of Sp1 binding to the SOD2 promoter in TMZ-resistant clones (#9 and #10) was assessed using a DNA affinity precipitation assay (DAPA), with a biotin-labeled DAPA probe that contains two Sp1-binding sites (P: SOD2) as shown in (A). (*p < 0.05; **p < 0.01; ***p < 0.001).
3.6. Inhibition of Sp1 reinstates the TMZ treatment effect in xenograft GBM models
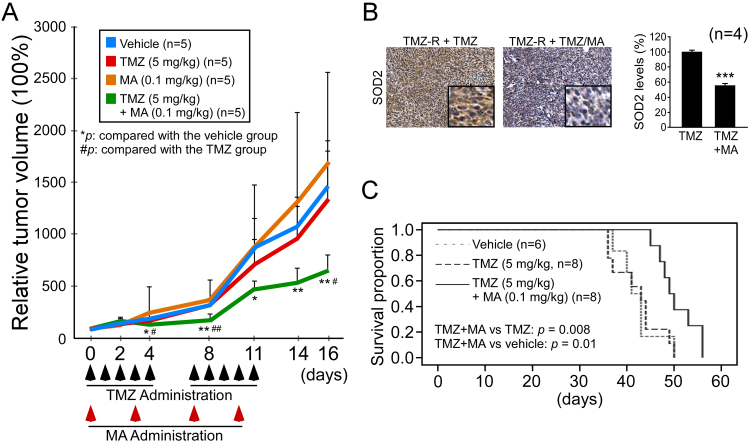
To evaluate the in vivo effect of Sp1 inhibition, a TMZ-resistant cell xenograft model was generated. Oral TMZ was given with or without intraperitoneal administration of MA (Fig. 8A). The results showed that tumor growth was significantly slower with the combined treatment compared to the other treatment groups. Notably, TMZ alone exerted only a marginal effect in resistant cells, whereas MA alone did not show any anti-tumor effect. Examination of tumor tissues following the combined treatment with TMZ/MA revealed decreased SOD2 expression (Fig. 8B), suggesting a similar mechanism to that found in the in vitro experiments.
Fig. 8.
MA restores TMZ treatment effect in the xenograft models. (A) TMZ-resistant U87MG cells were implanted subcutaneously in SCID mice. When the inoculated TMZ-resistant tumor volume reached 200 mm3 (day 0), mice were randomly grouped and treated for two weeks. TMZ (black arrows) was administered five days/week (oral), and/or MA (red arrows) was administrated two days/week (intraperitoneal). (B) Representative images of SOD2 protein levels by IHC in the TMZ group and the TMZ plus MA group of resistant xenografts. (C) TMZ-resistant GBM inoculated orthotopic mice were randomly grouped and treated with TMZ or TMZ plus MA five days/week thereafter from day 10. Survival was plotted using a Kaplan-Meier curve. (*,#p < 0.05; **,##p < 0.01; ***,###p < 0.001).
We next used a TMZ-resistant inoculated brain orthotopic model to conduct a survival study. As shown in Fig. 8C, treatment with TMZ exhibited a similar survival curve compared to that with vehicle only (42.2 versus 42.3 days, 95% confidence intervals 38.8–45.6 versus 38.8–45.8 days). In contrast, the combination of MA with TMZ led to significantly prolonged survival, having a mean survival of 50.5 days (95% confidence interval 47.7–53.3 days).
4. Discussion
An increased understanding of the mechanism for TMZ resistance in GBM could point to potential strategies to overcome this treatment dilemma. Importantly, our study provides the first evidence that MGMT-independent TMZ resistance relies upon the defensive mechanism of Sp1-modulated ROS scavenging to mitigate TMZ-induced stress. By studying TMZ-resistant cell lines, enhanced Sp1 expression was shown to accompany enhanced SOD2 expression. Furthermore, we demonstrated that Sp1 plays a critical role in SOD2 expression through promoter binding, which could be downregulated by co-incubation with the Sp1 inhibitor MA. Cellular viability in TMZ-resistant GBM could also be decreased by Sp1 inhibition, suggesting a potential strategy to overcome the acquired TMZ resistance. These findings were further confirmed using in vivo xenograft studies. Overall, our findings indicate that the Sp1-SOD2 pathway protects GBM against TMZ toxicity, and that inhibition of this pathway could mitigate the cellular resistance to this anti-neoplastic agent.
In terms of clinical treatment of GBM, the presence of MGMT factor has been shown to contribute to the innate tumor resistance to TMZ [1]. The major treatment limitation, however, emerges from acquired resistance rather than innate resistance. Unlike findings in rodent models, to date, studies examining whether anti-cancer therapies are capable of inducing MGMT in humans have been inconclusive [23]. Our study, using MGMT-negative models, showed a lack of MGMT expression even after acquisition of TMZ resistance, thus supporting a multifactorial resistance mechanism. In response to DNA damage-mediated death signals, certain tumor cells may increase their intracellular defensive mechanisms in order to survive [24]. This theory in turn led to our interest in ROS, which has been shown to be crucial to cell death caused by various DNA-damaging cytotoxic agents [25]. With regard to TMZ, the drug itself and its analog, TMZ-perillyl alcohol conjugate, have been shown to upregulate ROS production in GBM cells and non-small cell lung cancer cells, respectively [2], [26]. However, ROS is paradoxically critical both in the promotion of cancer progression and the induction of a detrimental cytotoxic response [27]. It is therefore unsurprising that certain cells possess an enhanced inherited antioxidant ability to tightly regulate ROS levels, thus maintaining viability and avoiding oxidative stress from the anticancer therapy. For example, overexpression of ROS scavengers, including CAT and NAC, has been reported to reverse TMZ analog-induced cell death in lung cancer [26]. In this study, we identified Sp1-upregulated SOD2 expression as a critical mechanism for protecting GBM cells against TMZ-induced cytotoxicity.
Sp1 has been recognized as critical in regulating multiple genes and influencing cellular functions in many cancer cell types such as breast, gastric, cervical, and pancreatic cancers [7], [28]. In glioma, the expression of Sp1 is associated with disease grade and clinical outcome [29]. Although previous studies have shown that Sp1 inhibition by MA enhances the chemotherapeutic effect of TMZ [30] and reduces cancer stemness [31], [32] in GBM, it remains unclear whether environmental stress induced by chemotherapy affects Sp1 and how Sp1 affects genes protective against the therapeutic stress. In this study, we showed that Sp1 was overexpressed in TMZ-resistant GBM and that it modulated SOD2 expression, which enabled cells to survive TMZ by eliminating ROS. Supported by studies showing that oxidative stress enhances DNA binding affinity, transactivation capacity, cofactor recruitment, and protein levels of Sp1 [21], [33], we propose that this protein plays a key role in acquired TMZ drug resistance. Importantly, co-treatment of resistant cells with an Sp1 inhibitor reversed TMZ resistance. In summary, our study indicates an indispensable role for Sp1 in GBM TMZ resistance and proposes a possible therapeutic strategy involving Sp1 and its downstream target SOD2 in the prevention of cancer resistance to chemotherapy.
Our current study is focused on understanding the intracellular response to TMZ and therefore has a limitation with respect to microenvironmental factors such as the inflammatory response [34] and hypoxia that could also contribute to chemo-resistance [35]. Multiple environmental stress-related transcription factors including Nrf2 and HIF-1 have been shown to be responsible for the induction of detoxifying enzyme gene expression [36]. Additionally, FoxO3a, a transcription factor produced in response to oxidative stress, has been shown to block hypoxia-mediated ROS generation [37]. These factors are, however, not related to increased SOD2 gene expression [37], [38], [39]. Although Sp1 is a stress-induced transcription factor [9], [21] and our study indicated that Sp1 indeed plays an essential role in the transcriptional regulation of SOD2 gene, the inhibition of Sp1 by MA, which led to a decrease in SOD2, only diminished tumor growth and prolonged survival, but did not eliminate the tumor in these animal models. An important limitation of this study that needs to be recognized is that we only focused on the Sp1/SOD2 pathway and disregarded other transcription factors and ROS scavenging proteins, whose expression levels were also found to be increased in the clinical database, as shown in Fig. 5A. Thus, the role of microenvironment factor-mediated cellular protection resulting in chemotherapy resistance in GBM still needs to be further examined.
In conclusion, our study describes a mechanism by which TMZ resistance may be acquired, and suggests a treatment strategy to eliminate resistance by modulating oxidative pathways. This mechanism can be summarized as follows: firstly, Sp1 is upregulated in MGMT-deficient cells. Secondly, SOD2 accumulation regulated by Sp1 protects the GBM, resulting in TMZ tolerance. Finally, Sp1 and SOD2 antagonists allow TMZ susceptibility to be restored. Inhibition of this pathway may be of benefit in the clinical treatment with TMZ and represents a potential therapeutic strategy for the treatment of GBM.
Conflicts of interest
The authors declare no conflicts of interest in this work.
Acknowledgments
This work was supported by the Ministry of Science and Technology, Taiwan (103-2320-B-038-046-MY3, 103-2314-B-400-011-MY3, 104-2923-B-038-002-MY3, 106-2320-B-038-002, and 106-2320-B-038-028), the Ministry of Health and Welfare (MOHW106-TDU-B-212-144001), and National Health Research Institutes (CA-106-PP-08). The ethical use of human samples was under regulation of National Health Research Institutes IRB (EC1021209-E) and Taipei Medical University IRB (201006011).
Contributor Information
Tzu-Jen Kao, Email: geokao@tmu.edu.tw.
Jian-Ying Chuang, Email: chuangcy@tmu.edu.tw.
References
- 1.Stupp R., Hegi M.E., Mason W.P., van den Bent M.J., Taphoorn M.J., Janzer R.C., Ludwin S.K., Allgeier A., Fisher B., Belanger K., Hau P., Brandes A.A., Gijtenbeek J., Marosi C., Vecht C.J., Mokhtari K., Wesseling P., Villa S., Eisenhauer E., Gorlia T., Weller M., Lacombe D., Cairncross J.G., Mirimanoff R.O., R. European Organisation for, T. Treatment of Cancer Brain, G. Radiation Oncology, G. National Cancer Institute of Canada Clinical Trials Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 2.Zhang W.B., Wang Z., Shu F., Jin Y.H., Liu H.Y., Wang Q.J., Yang Y. Activation of amp-activated protein kinase by temozolomide contributes to apoptosis in glioblastoma cells via p53 activation and mTORC1 inhibition. J. Biol. Chem. 2010;285:40461–40471. doi: 10.1074/jbc.M110.164046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wick W., Weller M., van den Bent M., Sanson M., Weiler M., von Deimling A., Plass C., Hegi M., Platten M., Reifenberger G. MGMT testing–the challenges for biomarker-based glioma treatment. Nat. Rev. Neurol. 2014;10:372–385. doi: 10.1038/nrneurol.2014.100. [DOI] [PubMed] [Google Scholar]
- 4.Oliva C.R., Nozell S.E., Diers A., McClugage S.G., 3rd, Sarkaria J.N., Markert J.M., Darley-Usmar V.M., Bailey S.M., Gillespie G.Y., Landar A., Griguer C.E. Acquisition of temozolomide chemoresistance in gliomas leads to remodeling of mitochondrial electron transport chain. J. Biol. Chem. 2010;285:39759–39767. doi: 10.1074/jbc.M110.147504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma L., Liu J., Zhang X., Qi J., Yu W., Gu Y. p38 MAPK-dependent Nrf2 induction enhances the resistance of glioma cells against TMZ. Med. Oncol. 2015;32:69. doi: 10.1007/s12032-015-0517-y. [DOI] [PubMed] [Google Scholar]
- 6.Wang L., Wei D., Huang S., Peng Z., Le X., Wu T.T., Yao J., Ajani J., Xie K. Transcription factor Sp1 expression is a significant predictor of survival in human gastric cancer. Clin. Cancer Res. 2003;9:6371–6380. [PubMed] [Google Scholar]
- 7.Chuang J.Y., Wang Y.T., Yeh S.H., Liu Y.W., Chang W.C., Hung J.J. Phosphorylation by c-Jun NH2-terminal kinase 1 regulates the stability of transcription factor Sp1 during mitosis. Mol. Biol. Cell. 2008;19:1139–1151. doi: 10.1091/mbc.E07-09-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chuang J.Y., Wang S.A., Yang W.B., Yang H.C., Hung C.Y., Su T.P., Chang W.C., Hung J.J. Sp1 phosphorylation by cyclin-dependent kinase 1/cyclin B1 represses its DNA-binding activity during mitosis in cancer cells. Oncogene. 2012;31:4946–4959. doi: 10.1038/onc.2011.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chuang J.Y., Kao T.J., Lin S.H., Wu A.C., Lee P.T., Su T.P., Yeh S.H., Lee Y.C., Wu C.C., Chang W.C. Specificity protein 1-zinc finger protein 179 pathway is involved in the attenuation of oxidative stress following brain injury. Redox Biol. 2017;11:135–143. doi: 10.1016/j.redox.2016.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yeh S.H., Yang W.B., Gean P.W., Hsu C.Y., Tseng J.T., Su T.P., Chang W.C., Hung J.J. Translational and transcriptional control of Sp1 against ischaemia through a hydrogen peroxide-activated internal ribosomal entry site pathway. Nucleic Acids Res. 2011;39:5412–5423. doi: 10.1093/nar/gkr161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thews O., Lambert C., Kelleher D.K., Biesalski H.K., Vaupel P., Frank J. Impact of therapeutically induced reactive oxygen species and radical scavenging by alpha-tocopherol on tumor cell adhesion. Oncol. Rep. 2007;18:965–971. [PubMed] [Google Scholar]
- 12.Trachootham D., Alexandre J., Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat. Rev. Drug Discov. 2009;8:579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- 13.Josson S., Xu Y., Fang F., Dhar S.K., Clair D.K., St, Clair W.H. St. RelB regulates manganese superoxide dismutase gene and resistance to ionizing radiation of prostate cancer cells. Oncogene. 2006;25:1554–1559. doi: 10.1038/sj.onc.1209186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takada Y., Hachiya M., Park S.H., Osawa Y., Ozawa T., Akashi M. Role of reactive oxygen species in cells overexpressing manganese superoxide dismutase: mechanism for induction of radioresistance. Mol. Cancer Res. 2002;1:137–146. [PubMed] [Google Scholar]
- 15.Glaser R., Zhang H.Y., Yao K.T., Zhu H.C., Wang F.X., Li G.Y., Wen D.S., Li Y.P. Two epithelial tumor cell lines (HNE-1 and HONE-1) latently infected with Epstein-Barr virus that were derived from nasopharyngeal carcinomas. Proc. Natl. Acad. Sci. USA. 1989;86:9524–9528. doi: 10.1073/pnas.86.23.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rhodes D.R., Yu J., Shanker K., Deshpande N., Varambally R., Ghosh D., Barrette T., Pandey A., Chinnaiyan A.M. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su T.C., Lin S.H., Lee P.T., Yeh S.H., Hsieh T.H., Chou S.Y., Su T.P., Hung J.J., Chang W.C., Lee Y.C., Chuang J.Y. The sigma-1 receptor-zinc finger protein 179 pathway protects against hydrogen peroxide-induced cell injury. Neuropharmacology. 2016;105:1–9. doi: 10.1016/j.neuropharm.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsu C.C., Chang W.C., Hsu T.I., Liu J.J., Yeh S.H., Wang J.Y., Liou J.P., Ko C.Y., Chang K.Y., Chuang J.Y. Suberoylanilide hydroxamic acid represses glioma stem-like cells. J. Biomed. Sci. 2016;23:81. doi: 10.1186/s12929-016-0296-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsai S.Y., Chuang J.Y., Tsai M.S., Wang X.F., Xi Z.X., Hung J.J., Chang W.C., Bonci A., Su T.P. Sigma-1 receptor mediates cocaine-induced transcriptional regulation by recruiting chromatin-remodeling factors at the nuclear envelope. Proc. Natl. Acad. Sci. USA. 2015;112:E6562–E6570. doi: 10.1073/pnas.1518894112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rocha C.R., Kajitani G.S., Quinet A., Fortunato R.S., Menck C.F. NRF2 and glutathione are key resistance mediators to temozolomide in glioma and melanoma cells. Oncotarget. 2016;7:48081–48092. doi: 10.18632/oncotarget.10129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryu H., Lee J., Zaman K., Kubilis J., Ferrante R.J., Ross B.D., Neve R., Ratan R.R. Sp1 and Sp3 are oxidative stress-inducible, antideath transcription factors in cortical neurons. J. Neurosci. 2003;23:3597–3606. doi: 10.1523/JNEUROSCI.23-09-03597.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Y., Porntadavity S., Clair D.K., St Transcriptional regulation of the human manganese superoxide dismutase gene: the role of specificity protein 1 (Sp1) and activating protein-2 (AP-2) Biochem. J. 2002;362:401–412. doi: 10.1042/0264-6021:3620401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christmann M., Kaina B. Transcriptional regulation of human DNA repair genes following genotoxic stress: trigger mechanisms, inducible responses and genotoxic adaptation. Nucleic Acids Res. 2013;41:8403–8420. doi: 10.1093/nar/gkt635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bouwman P., Jonkers J. The effects of deregulated DNA damage signalling on cancer chemotherapy response and resistance. Nat. Rev. Cancer. 2012;12:587–598. doi: 10.1038/nrc3342. [DOI] [PubMed] [Google Scholar]
- 25.Kang M.A., So E.Y., Simons A.L., Spitz D.R., Ouchi T. DNA damage induces reactive oxygen species generation through the H2AX-Nox1/Rac1 pathway. Cell Death Dis. 2012;3:e249. doi: 10.1038/cddis.2011.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song X., Xie L., Wang X., Zeng Q., Chen T.C., Wang W., Song X. Temozolomide-perillyl alcohol conjugate induced reactive oxygen species accumulation contributes to its cytotoxicity against non-small cell lung cancer. Sci. Rep. 2016;6:22762. doi: 10.1038/srep22762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gorrini C., Harris I.S., Mak T.W. Modulation of oxidative stress as an anticancer strategy. Nat. Rev. Drug Discov. 2013;12:931–947. doi: 10.1038/nrd4002. [DOI] [PubMed] [Google Scholar]
- 28.Yang H.C., Chuang J.Y., Jeng W.Y., Liu C.I., Wang A.H., Lu P.J., Chang W.C., Hung J.J. Pin1-mediated Sp1 phosphorylation by CDK1 increases Sp1 stability and decreases its DNA-binding activity during mitosis. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guan H., Cai J., Zhang N., Wu J., Yuan J., Li J., Li M. Sp1 is upregulated in human glioma, promotes MMP-2-mediated cell invasion and predicts poor clinical outcome. Int J. Cancer. 2012;130:593–601. doi: 10.1002/ijc.26049. [DOI] [PubMed] [Google Scholar]
- 30.Seznec J., Silkenstedt B., Naumann U. Therapeutic effects of the Sp1 inhibitor mithramycin A in glioblastoma. J. Neurooncol. 2011;101:365–377. doi: 10.1007/s11060-010-0266-x. [DOI] [PubMed] [Google Scholar]
- 31.Singh D.K., Kollipara R.K., Vemireddy V., Yang X.L., Sun Y., Regmi N., Klingler S., Hatanpaa K.J., Raisanen J., Cho S.K., Sirasanagandla S., Nannepaga S., Piccirillo S., Mashimo T., Wang S., Humphries C.G., Mickey B., Maher E.A., Zheng H., Kim R.S., Kittler R., Bachoo R.M. Oncogenes activate an autonomous transcriptional regulatory circuit that drives glioblastoma. Cell Rep. 2017;18:961–976. doi: 10.1016/j.celrep.2016.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gopisetty G., Xu J., Sampath D., Colman H., Puduvalli V.K. Epigenetic regulation of CD133/PROM1 expression in glioma stem cells by Sp1/myc and promoter methylation. Oncogene. 2013;32:3119–3129. doi: 10.1038/onc.2012.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin M., Ande A., Kumar A., Kumar S. Regulation of cytochrome P450 2e1 expression by ethanol: role of oxidative stress-mediated pkc/jnk/sp1 pathway. Cell Death Dis. 2013;4:e554. doi: 10.1038/cddis.2013.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yeung Y.T., McDonald K.L., Grewal T., Munoz L. Interleukins in glioblastoma pathophysiology: implications for therapy. Br. J. Pharmacol. 2013;168:591–606. doi: 10.1111/bph.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsieh C.H., Lin Y.J., Wu C.P., Lee H.T., Shyu W.C., Wang C.C. Livin contributes to tumor hypoxia-induced resistance to cytotoxic therapies in glioblastoma multiforme. Clin. Cancer Res. 2015;21:460–470. doi: 10.1158/1078-0432.CCR-14-0618. [DOI] [PubMed] [Google Scholar]
- 36.Espinosa-Diez C., Miguel V., Mennerich D., Kietzmann T., Sanchez-Perez P., Cadenas S., Lamas S. Antioxidant responses and cellular adjustments to oxidative stress. Redox Biol. 2015;6:183–197. doi: 10.1016/j.redox.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferber E.C., Peck B., Delpuech O., Bell G.P., East P., Schulze A. FOXO3a regulates reactive oxygen metabolism by inhibiting mitochondrial gene expression. Cell Death Differ. 2012;19:968–979. doi: 10.1038/cdd.2011.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao Y.H., Li C.X., Shen S.M., Li H., Chen G.Q., Wei Q., Wang L.S. Hypoxia-inducible factor 1alpha mediates the down-regulation of superoxide dismutase 2 in von Hippel-Lindau deficient renal clear cell carcinoma. Biochem. Biophys. Res. Commun. 2013;435:46–51. doi: 10.1016/j.bbrc.2013.04.034. [DOI] [PubMed] [Google Scholar]
- 39.Miller C.J., Gounder S.S., Kannan S., Goutam K., Muthusamy V.R., Firpo M.A., Symons J.D., Paine R., 3rd, Hoidal J.R., Rajasekaran N.S. Disruption of Nrf2/ARE signaling impairs antioxidant mechanisms and promotes cell degradation pathways in aged skeletal muscle. Biochim. Biophys. Acta. 2012;1822:1038–1050. doi: 10.1016/j.bbadis.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 40.Sun L., Hui A.M., Su Q., Vortmeyer A., Kotliarov Y., Pastorino S., Passaniti A., Menon J., Walling J., Bailey R., Rosenblum M., Mikkelsen T., Fine H.A. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9:287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 41.Bredel M., Bredel C., Juric D., Harsh G.R., Vogel H., Recht L.D., Sikic B.I. Functional network analysis reveals extended gliomagenesis pathway maps and three novel MYC-interacting genes in human gliomas. Cancer Res. 2005;65:8679–8689. doi: 10.1158/0008-5472.CAN-05-1204. [DOI] [PubMed] [Google Scholar]
- 42.Murat A., Migliavacca E., Gorlia T., Lambiv W.L., Shay T., Hamou M.F., de Tribolet N., Regli L., Wick W., Kouwenhoven M.C., Hainfellner J.A., Heppner F.L., Dietrich P.Y., Zimmer Y., Cairncross J.G., Janzer R.C., Domany E., Delorenzi M., Stupp R., Hegi M.E. Stem cell-related "self-renewal" signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J. Clin. Oncol. 2008;26:3015–3024. doi: 10.1200/JCO.2007.15.7164. [DOI] [PubMed] [Google Scholar]
- 43.Liang Y., Diehn M., Watson N., Bollen A.W., Aldape K.D., Nicholas M.K., Lamborn K.R., Berger M.S., Botstein D., Brown P.O., Israel M.A. Gene expression profiling reveals molecularly and clinically distinct subtypes of glioblastoma multiforme. Proc. Natl. Acad. Sci. USA. 2005;102:5814–5819. doi: 10.1073/pnas.0402870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shai R., Shi T., Kremen T.J., Horvath S., Liau L.M., Cloughesy T.F., Mischel P.S., Nelson S.F. Gene expression profiling identifies molecular subtypes of gliomas. Oncogene. 2003;22:4918–4923. doi: 10.1038/sj.onc.1206753. [DOI] [PubMed] [Google Scholar]
- 45.Lee J., Kotliarova S., Kotliarov Y., Li A., Su Q., Donin N.M., Pastorino S., Purow B.W., Christopher N., Zhang W., Park J.K., Fine H.A. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]