Abstract
Claudin-6 (CLDN6) is an integral component of the tight junction proteins in polarized epithelial and endothelial cells and plays a crucial role in maintaining cell integrity. Deregulation of CLDN6 expression and distribution in tumor tissues have been widely documented and correlated with cancer progression and metastasis. However, a complete mechanistic understanding of CLDN6 regulation and function remains to be studied. Herein, we show new potential properties of CLDN6 regulation and functions from bioinformatics analysis. Using numerous algorithms to characterize the CLDN6 gene promoter elements and the CLDN6 protein structure, physio-chemical and localization properties, and its evolutionary relationships. CLDN6 is regulated by a diverse set of transcription factors (SP1, SPR, AML-1a, CdxA, CRE-BP and CREB) and associated with the levels of methylation of CpG islands in promoters. The structural properties of CLDN6 indicate that it promotes cancer cell behavior via the ASK1-p38/JNK MAPK secretory signaling pathway. In conclusion, this information from bioinformatics analysis will help future attempts to better understand CLDN6 regulation and functions.
Keywords: CLDN6, CpG island, transcriptional factor binding sites, amino acid sequence, molecular structure, bioinformatics analysis
Introduction
Claudins (CLDNs) are critical transmembrane proteins in tight junction function primarily as a barrier against paracellular transport between epithelial cells and the CLDN family consisting of 27 members that are mostly 20–34 kDa and have four transmembrane helices with amino- and carboxyl-terminal tail extending into the cytoplasm and play a crucial role in cellular adhesion, polarity, permeability and glandular differentiation (1,2). It is reported that altered expression and mislocalization of CLDNs such as CLDN6 appear to be tissue specific in embryo epithelial development and several cancers (3–5).
CLDN6 gene is located on 16p13.3 and its expression is mainly found in mouse embryonic stem cells, epithelial lineage cells during early development and primitive germ cell tumors such as spermatocytic seminoma, embryonal carcinoma, mature teratoma and classic seminoma (6). Its expression is very weak or absent in mouse and tumor tissue (7–9). CLDN6 inhibits cancer cell growth and induces apoptosis (10–12). It is reported that CLDN6 expression is associated with ERα expression and MMP-2 and ASK1. Although some functions of CLDN6 are known, a complete understanding of CLDN6 regulation and function remains to be studied. Bioinformatic analysis to predict regulatory mechanism of the gene and protein expression greatly solves these problems.
Bioinformatics is an interdisciplinary field, which combines computer science, statistics, mathematics, and engineering to develop methods and software tools for processing and understanding biological data (13–15). In the field of genetics and genomics, it aids in sequencing and annotating genomes and their observed mutations. Sequence analysis for DNA elements helps to explain the biological meaning and functin of the gene. In addition, protein structure prediction is another important application of bioinformatics. The amino acid sequence of a protein can be easily determined from the sequence on the gene that encodes it. This primary structure uniquely determines a structure in its native environment. Knowledge of the structural information that is usually classified as one of secondary, tertiary and quaternary structure, is vital in understanding the function of the protein (16). Moreover, network analysis seeks to understand the relationships within biological networks such as metabolic or protein-protein, small molecular interaction networks. Therefore, bioinformatics tools can aid in the comparison of genetic and genomic data and more generally in the understanding of evolutionary aspects of molecular biology as well as, at a more integrative level, anlayzing and cataloguing of the biological pathways and networks that are an important part of systems biology (16).
In this study, we used bioinformatics tools to examine the CLDN6 sequence to characterize the gene TATA-box, GC-box and CAAT-box, promoter, CpG islands, potential transcriptional factors binding sites (TFBS), encoded protein structure and its structure, subcellular localization, secondary and tertiary structures, and even evolutionary relationship. These characteristics will help define the basis for CLDN6 regulation and differential expression in cancer. These various bioinformatics tools are among the common tools of molecular biology helping investigators finding leads to investigate genes/proteins.
Materials and methods
Bioinformatics databases and online software
The following were used: NCBI (http://www.ncbi.nlm.nih.gov); Neural network promoter prediction (http://www.fruitfly.org/seq_tools/promoter.html); Promoter 2.0 prediction server (http://www.cbs.dtu.dk/services/Promoter/); TFSEARCH (http://mbs.cbrc.jp/research/db/TFSEARCH.html); EMBOSS and CpG island searcher (http://www.ebi.ac.uk/Tools/emboss/); expasy (http://www.expasy.org); Protparam (http://www.expasy.org/tools/protparam.html); compute pI/mw (http://www.expasy.org/tools/pi_tool.html); ProtScale (http://www.expasy.org/tools/protscale.html); Clustalx (http://www.clustal.org/download/current/); treeview (http://www.taxonomy.zool-ogy.gla.ac.uk/rod/rod.html); GOR4 (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_gor4.html); TargetP1.1 (http://www.cbs.dtu.dk/services/TargetP/), SignalP3.0 (http://www.cbs.dtu.dk/services/SignalP/); TMHMM2.0 (http://www.cbs.dtu.dk/services/TMHMM/); Pfam24.0 (http://pfam.sanger.ac.uk/search); SOPMA (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html); Swiss-model (http://www.expasy.ch/swissmod/SWISS-MODEL.html); KEGG (http://www.genome.jp/kegg/).
Prediction methods
The following prediction methods were used for CLDN6 regulatory elements, structure and function: promoter (Neural Network Promoter Prediction), CpG island (EMBOSS and CpG Island Searcher), TFBS (TFSEARCH), the relatively molecular, amino acid sequences, protein relatively molecular quality, mass of amino acids, theoretical isoelectric point, PI, half-life, unstable factor, the total average hydrophilic (ProtParam); hydrohobicity or hydrophilicity (Prot Scale); the secondary structure (ExPASy-SOPMA and GOR4); signal lead peptide (TargetP1.1 Server) and signal peptide cutting locus (SignalP4.1Server); nuclear localization signal prediction (NLStradamus); the subcellular localization (WOLF PSORT and PSORT II); transmembrane area and across the membrane (TMpred program and TMHMM2.0); structure (SWISS-MODEL); protein structure and function (InterPro); transmembrane helices (TMpred program); evolutionary tree and homology analysis (Clustalx program and BLAST pairwise alignments); the signal pathway analysis (KEGG).
Results
Properties of the TATA-box, GC-box, CAAT-box motifs in the 5′ regulatory region of CLDN6
To determine whether there are TATA-box, GC-box, CAAT-box motifs in the 5′ regulatory region of human CLDN6 to which the transcription fators, TBP, SP1, and CBF, respectively, can bind we BLAST searched CLDN6 mRNA (NM_021195.4) and human genomic sequences between −2000 bp to 200 bp from the transcriptin start site, for the motifs TATAWAW (where W represents A or T), GGGCGG and CCAAT. We identified three GC-box fragments, but no TATA- or CAAT-boxes, suggesting that expression and transcriptional activity of CLDN6 is regulated by SP1.
Promoter, CpG island and TFBS prediction in the 5′ regulatory region of CLDN6
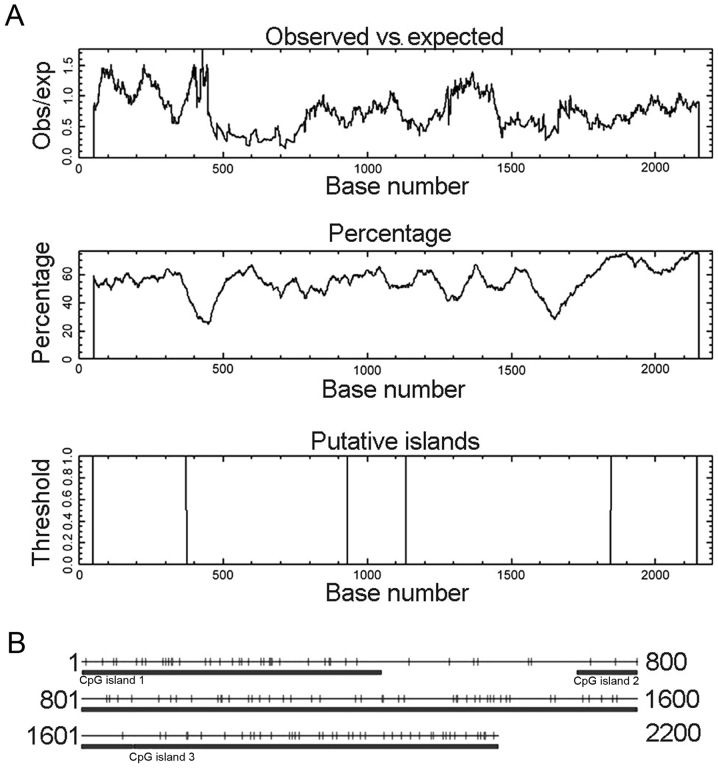
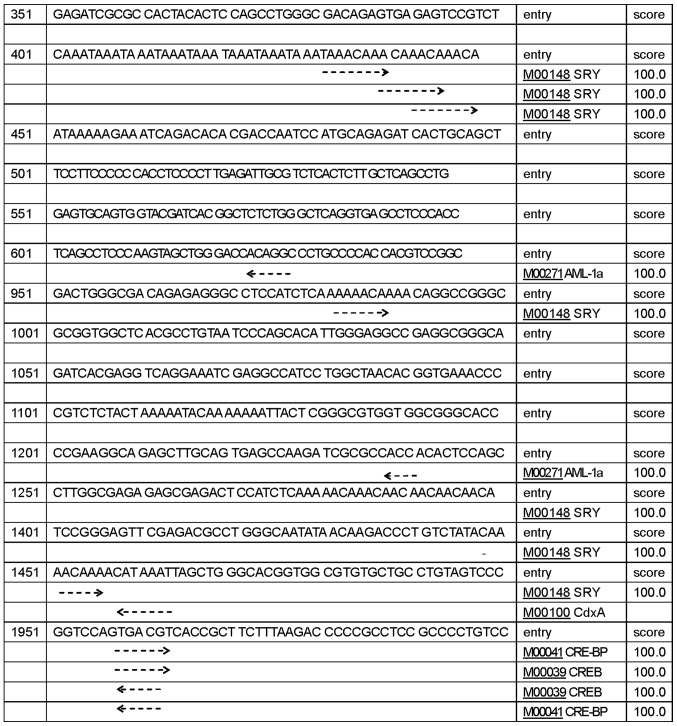
The CLDN6 promoter sequence and TFs that bind to this sequence determine the temporal and spatial expression pattern of the gene. Therefore, defining of TFBS is important in the study of gene regulation. We used online programs, such as neural network promoter prediction, EMBOSS, CpG island searcher and TFSEARCH to predict promoters, CpG islands and TFBS in 5′ regulatory region sequences of human CLDN6. We identified five promoters shown in Table I, and three CpG islands with Obs/Exp ratio >0.60, percent C + G >50% and length >200. These CpG islands have different length and location as shown in Fig. 1A. CpG Island Searcher program also identified three different CpG islands (Fig. 1B). TFSEARCH program predicted 432 potential TFBS with a score higher than 85 points, 156 potential TFBS with a score higher than 90 points, 66 potential TFBS with a score higher than 95 points, 24 potential TFBS, including SPR, AML-1a, CdxA, CRE-BP and CREB with a score over 99 points (Fig. 2). Together, these findings suggest that CLDN6 expression is associated with the levels of methylation of CpG islands in its promoters. Different transcription start sites makes CLDN6 transcription in a different way, then produce a variety of different biological functions of a transcription product.
Table I.
CLDN6 promoter site prediction using online Neural Network Promoter Prediction program.
| Software | Start sites | End sites | Score | Sequence |
|---|---|---|---|---|
| Neural network promoter prediction | 237 | 287 | 0.98 | ACTCGAAATACAAAAATTAGCCGGGCGTGGTGGCGCGCGCCTGCAATCCG |
| 975 | 1025 | 0.84 | ATCTCAAAAAACAAAACAGGCCGGGCGCGGTGGCTCACGCCTGTAATCCC | |
| 1414 | 1464 | 0.82 | GACGCCTGGGCAATATAACAAGACCCTGTCTATACAAAACAAAACATAAA | |
| 1450 | 1500 | 0.93 | AAACAAAACATAAATTAGCTGGGCACGGTGGCGTGTGCTGCCTGTAGTCC | |
| 1965 | 2015 | 0.99 | ACCGCTTCTTTAAGACCCCCGCCTCCGCCCCTGTCCCGACACTCGGCCTA |
Figure 1.
CpG island prediction for CLDN6 using two prediction programs. (A) CpG island prediction using online EMBOSS. These CpG islands were 325 bp in length (located at 49–373 bp), 204 bp (932–1135 bp) and 299 bp (1847–2145 bp). (B) CpG island prediction using CpG Island Searcher program. CpG Island Searcher program identified three different CpG islands of 432 bp (1–432 bp), 962 bp (713–1674 bp) and 526 bp (1675–2200 bp). Select lower limits: % GC=50, obsCpG/expCpG = 0.60, length = 200, distance = 100. CpG island 1 star = 1, end = 432, % GC=53.9, obsCpG/expCpG = 1.024, length = 432. CpG island 2 star = 713, end = 1674, % GC=52.8, obsCpG/expCpG = 0.735, length = 962. CpG island 3 star = 1675, end = 2200, % GC=65.4, obsCpG/expCpG = 0.711, length = 526.
Figure 2.
There are 24 potential TFBSs prediction for CLDN6, including for SPR, AML-1a, CdxA, CRE-BP and CREB with a score of 99 using TFSEARCH program prediction.
The amino acid sequence and physicochemical properties of CLDN6 protein
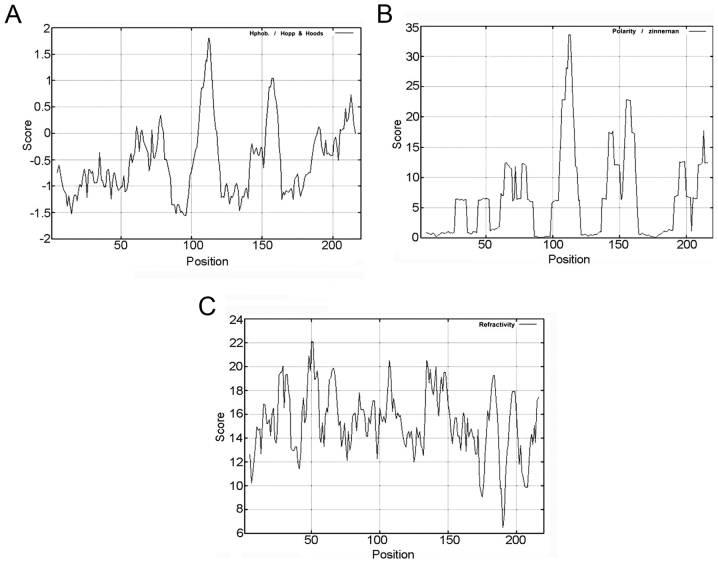
To predict the protein structure of CLDN6, we used ProtParam online software (http://au.expasy.org/tools/) to analyze amino acid composition, molecular formula, molecular weight and isoelectric point. CLDN6 consists of 220 amino acids with 20 different amino acids, including alanine (10.90%), glycine (9.50%), leucine (14.50%), serine (8.20%) and valine (10.90%) (Table IIA). CLDN6 has the following properties: protein formula C1054H1682N268O291S16, molecular weight 23,2775 kDa, theoretical PI 8.32, estimated half-life in vitro 30 h, instability index 42.03 and the hydrophilic residue ratio is lower than that of the hydrophobic residues. The threshold value of CLDN6 hydrophilicity is highest at: 105–118 bp, 153–161 and 205–216 bp (Fig. 3A and Table IIB), and the threshold value of CLDN6 polarity is highest at: 106–116, 142–145 and 154–161 bp (Fig. 3B and Table IIB), the threshold value of turn-back coefficient is highest at 14–22, 26–34, 43–55, 60–71, 80–89, 92–97, 100–115, 133–151, 154–156, 179–186, 195–200 and 213–216 bp (Fig. 3C and Table IIB). The overlapping range of these three parameters is shown in Table IIB at 106–115 and 154–156 bp. Taken together, these data suggest that CLDN6 mainly contains hydrophilic amino acid and polar amino acids, functional overlapping structure ranges from 106 to 156 bp, and that CLDN6 protein maybe a hydrophobic and unstable protein.
Table II.
The basic properties of CLDN6 analyzed by using ProtParam online software.
| A, Amino acid composition of CLDN6. | |||
|---|---|---|---|
| Amino acid | Abbreviations | Number | Composition (%) |
| Alanine | Ala(A) | 24 | 10.90 |
| Arginine | Arg(R) | 6 | 2.70 |
| Asparagine | Asn(N) | 4 | 1.80 |
| Aspartate | Asp(D) | 4 | 1.80 |
| Cystine | Cys(C) | 10 | 4.50 |
| Glutamine | Gln(Q) | 9 | 4.10 |
| Glutamate | Glu(E) | 6 | 2.70 |
| Glycine | Gly(G) | 21 | 9.50 |
| Histidine | His(H) | 2 | 0.90 |
| Isoleucine | Ile(I) | 9 | 4.10 |
| Leucine | Leu(L) | 32 | 14.50 |
| Lysine | Lys(K) | 7 | 3.20 |
| Methionine | Met(M) | 6 | 2.70 |
| Phenylalanine | Phe(F) | 4 | 1.80 |
| Proline | Pro(P) | 9 | 4.10 |
| Serine | Ser(S) | 18 | 8.20 |
| Threonine | Thr(T) | 11 | 5.00 |
| Tryptophan | Trp(W) | 6 | 2.70 |
| Tyrosine | Tyr(Y) | 8 | 3.60 |
| Valine | Val(V) | 24 | 10.90 |
| B, Molecular formula, molecular weight, isoelectric point and other basic properties of CLDN6. | |||
| Parameters | Prediction results | ||
| Formula | C1054H1682N268O291S16 | ||
| Molecular weight | 23277.5 | ||
| Theoretical pI | 8.32 | ||
| Total number of atoms | 3311 | ||
| Number of amino acids | 220 | ||
| Total number of negatively charged residues | 10 | ||
| Total number of positively charged residues | 13 | ||
| Estimated half-life (mammalian reticulocytes, in vitro) | 30 h | ||
| Instability index | 42.03 | ||
| Hydrophilic | 105–118, 153–161, 205–216 | ||
| Polarity | 106–116, 142–145, 154–161 | ||
| Turn-back coefficient | 14–22, 26–34, 43–55, 60–71, 80–89, 92–97, 100–115, 133–151, 154–156, 179–186, 195–200, 213–216 | ||
| The predicted results of the overlapping area | 106–115, 154–156 | ||
Figure 3.
Nucleotide and deduced amino acid sequences of human CLDN6 analyzed using Proparam. (A) It encodes a 220 amino acid polypeptide with a predicted molecular mass of 23,2775 kD. (B) Hydrophilicity plot analysis of CLDN6. The plot records the average hydrophilicity along the sequence over a window of 10 residues. Hydrophilic and hydrophobic residues are in the lower and upper part of the frame, respectively. The axis is numbered in amino acid residues. (C) Polarity analysis of CLDN6.
Secondary structure prediction for CLDN6 protein
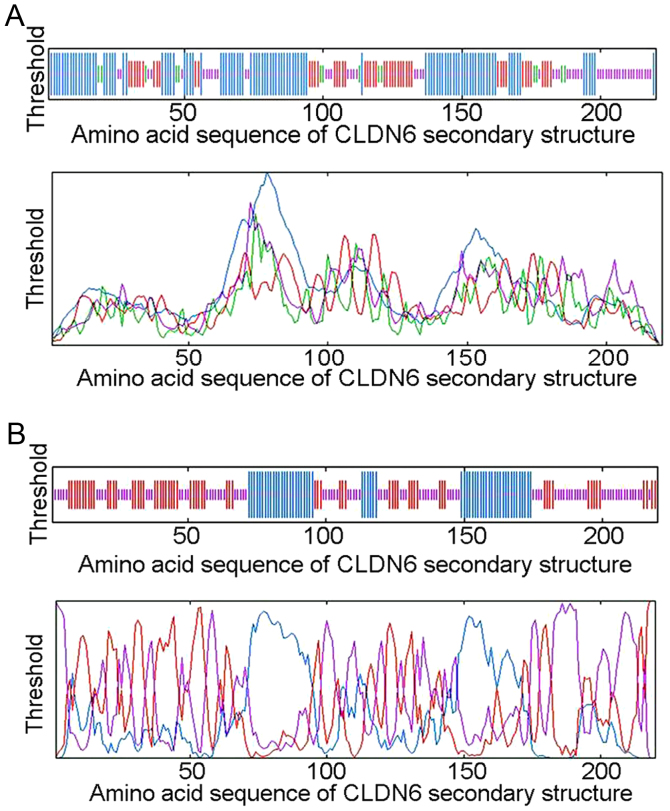
The arrangement of atoms in space of the main polypeptide chain (α helix, extended strand, β turn and random coil) determines basic protein secondary structure. Determination of this arrangement can help predict functions, and protein modifications. We used ExPASy-SOPMA and GOR4 secondary structure prediction module to calculate CLDN6 secondary structure, and to draw the structure model. Module output prediction results can be shown as a peak figure or the diagram can be simplified to show as the random of coiled and folded areas. The SOPMA method identified 104 (47.27%) α helixs, 48 (21.82%) extended strands, 14 (6.36%) β turns and 54 (24.55%) random coils and irregular coiled and folded structures located mainly at 1–70, 96–114, 120–150 and 176–220 bp between the peak figure and simplified diagram (Fig. 4A and Table III). The GOR4 method identified 56 (25.45%) α helixs, 67 (30.45%) extended strands, and 97 (44.09%) random coils and irregular coiled and folded structures located mainly at 26–42, 102–112, 131–139, 147–156, 173–194 and 196–220 bp (Fig. 4B and Table III). The comparison of two secondary structure prediction results is shown in Table III. CLDN6 secondary structure mainly consists of the irregular curl overlapping areas at 26–42, 102–112, 131–139, 147–150, 222–238, 176–194 and 196–220 bp, suggesting that these areas are mainly composed of α helix, extended strand and random coil structure. Taken together, the functional domain of CLDN6 protein is likely to be limited to these overlapped areas.
Figure 4.
Secondary structure analysis of CLDN6 protein using SOPMA (A, up: schematic illustration; down: the peak figure) and GOR4 (B, up: schematic illustration; down: the peak figure) prediction software. Helix (blue), spiral structure; Sheet (red), folding; Turn (green), corner structure; Coil (purple), irregular curly structure.
Table III.
Comparison of CLDN6 secondary structure prediction results between SOPMA and GOR4 methods.
| Secondary structure prediction methods | Prediction results |
|---|---|
| SOPMA | 1–70, 96–114, 120–150, 176–220 |
| GOR4 | 26–42, 102–112, 131–139, 147–156, 173–194, 196–220 |
| The predicted results of the overlapping area | 26–42, 102–112, 131–139, 147–150, 222–238, 176–194, 196–220 |
Analysis of signal peptide cleavage site, subcellular location, transmembrane domains in the CLDN6 protein
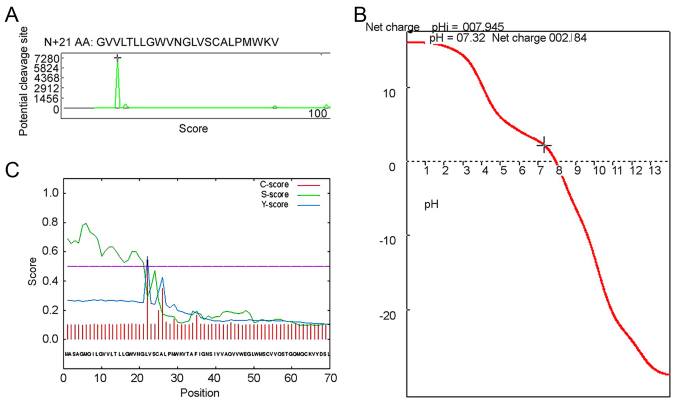
Signal peptides direct protein localization in cells and usually consists of 15–30 N-terminal amino acid residues. To analyze the CLDN6 signal peptide, we first used the Anthprot signal peptide cutting locus analysis module and the result showed CLDN6 has a short signal peptide composed of 21 amino acid residues, and its sequence is shown in Figs. 5A and 6. Consistent with previous results, the CLDN6 isoelectric point is approximately near 8.0, and the physiological state of CLDN6 molecules is closest to pH 7.3, with a positive charge of 2.184, as shown in Fig. 5B.
Figure 5.
Signal peptide cleavage site and titration curve of CLDN6 using Anthprot and SignalP analysis. (A) Anthprot signal peptide cleavage site, subcellular localization prediction using TargetP1.1 and PSORT II Prediction. (B) Titration curve of CLDN6 using Anthprot analysis. (C) Curve of CLDN6 protein signal peptide using SignalP-4.1 euk predictions.
Figure 6.
A short peptide composed of 21 amino acid residues.
We also used SignalP4.1 to predict the signal peptide and its cleavage site. As shown in Fig. 5C and Table IV the cleavage site is between amino acids 21 and 22: VNG-LV. We also used NLStradamus, a simple Hidden Markov Model for nuclear localization signal prediction, to show that there were no nuclear localization signal sequences, suggesting that CLDN6 does not enter the nucleus. TargetP1.1 predicts CLDN6 to be located in the secretory pathway (98.7%) (Table V), while WoLF PSORT predicts CLDN6 to be a multi-pass membrane protein with subcellular localization to tight junction or the cell membrane. However, PSORT II Prediction indicated that CLDN6 may locate to the endoplasmic reticulum (ER, 66.7%) and mitochondria (33.3%) (Table VI). The review by Koval on differential pathways of claudin oligomerization and integration into tight junctions described the three potential models for claudin oligomerization (cis interactions) occurring in the endoplasmic reticulum (ER) (17). A requirement for CLDN quality control early in the secretory pathway and mutant CLDNs which are misfolded accumulate in the ER (18,19). CLDN oligomerization is more likely to happen in the TGN or another late secretory pathway and formation of stable claudin-claudin intermediates, however, given the structural diversity of different claudin family members, it is unlikely that all claudins will oligomerize via the same pathway. There is some evidence that CLDN16, CLDN3 and CLDN4 tagged with an ER retentin signal, His-Lys-Lys-Ser-Leu (HKKSL) are retained in the ER (20,21), while there is no reported for CLDN6 retaining in ER. We considered that CLDN6, as common protein functional maturation steps, oligomerizes in ER and is transported via the secretory pathway and integrates into tight junction.
Table IV.
Results of CLDN6 protein signal peptide using SignalP-4.1 euk predictions.
| # Measure | Position | Value | Cut off | Signal peptide |
|---|---|---|---|---|
| Max. C | 22 | 0.545 | No | |
| Max. Y | 22 | 0.570 | No | |
| Max. S | 6 | 0.795 | No | |
| Mean S | 1–21 | 0.635 | No | |
| D | 1–21 | 0.596 | 0.500 | Yes |
Table V.
Subcellular localization prediction of CLDN6 using TargetP1.1.
| Name | Len | mTP | SP | Other | Loc | RC |
|---|---|---|---|---|---|---|
| Sequence | 220 | 0.022 | 0.987 | 0.023 | S | 1 |
| Cut off | 0.000 | 0.000 | 0.000 |
The location assignment is based on the predicted presence of any CLDN6 N-terminal presequences: mitochondrial targeting peptide (mTP) or secretory pathway signal peptide; other, other localization within cells. Number of query sequences: 1, cleavage site predictions are not included using Non-Plant networks.
Table VI.
CLDN6 protein may locate in the endoplasmic reticulum (ER, 66.7%) and mitochondria (33.3%) using PSORT II Prediction.
| K=9/23 |
|---|
| 66.7%: Endoplasmic reticulum |
| 33.3%: Mitochondrial |
| >> Prediction for QUERY is end (k=9) |
Prediction of CLDN6 protein domain and function site
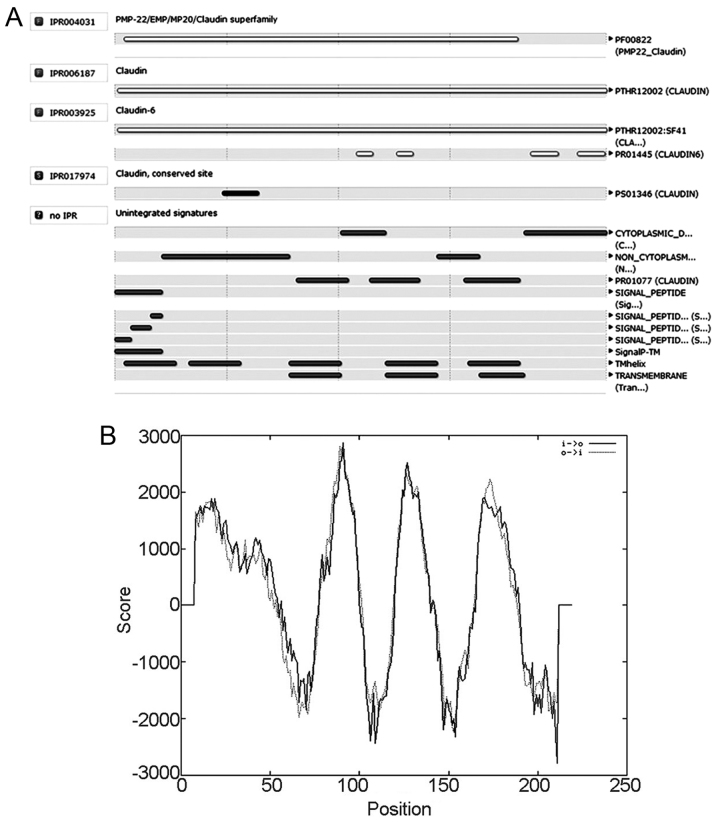
To predict CLDN6 structural domain and important functional sites, we used InterPro software. The results show that CLDN6 belongs to the PMP-22/EMP/CLDN superfamily (IPR004031) and CLDN (IPR006187) as shown in Fig. 7A. Its function is associated with structural molecular activity (GO: 0005198) and that the action contributes to cellular component in bicellular tight junction (GO: 0005923) and the structure integrity of a complex or assembly within or outside a cell (GO: 0016021).
Figure 7.
Transmembrane structural domain and function of CLDN6. (A) CLDN6 belongs to the PMP-22/EMP/CLDN superfaminly (IPR004031) and CLDN (IPR006187) according to the InterPro software. (B) CLDN6 possibly has four strong transmembrane helices in strongly preferred model.
CLDN6 has a four-element fingerprint that provides a signature for CLDNs. The fingerprint was derived from an initial alignment of two sequences. The motifs were drawn from conserved regions within the C-terminal half of the alignment, focusing on those sections that characterize CLDN6 and distinguish it from other family members: motif 1 lies in the intracellular loop; motif 2 resides within TM domain 3; and motifs 3 and 4 lie within the cytoplasmic C-terminal region. A single iteration on SPTR39_14f was required to reach convergence, no further sequences were identified beyond the starting set. CLDN6 possibly has four strong transmembrane helices according to the strongly preferred model (Fig. 7B). Taken together, CLDN6 is four-transmembrane tight junction protein.
Advanced structure of human CLDN6 protein
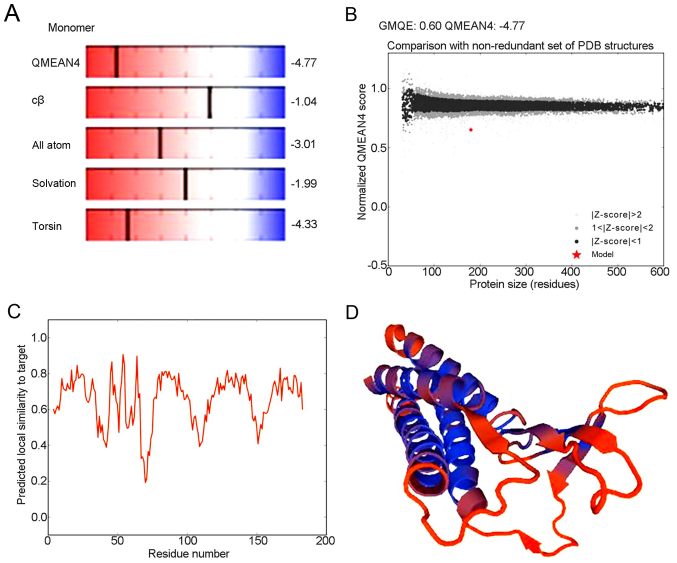
Analysis of the detailed structures of CLDN6 will further our understanding of its biological role. To obtain a high level protein structure simulation map, the amino acid sequence was analyzed using Swissmodel server. CLDN6 protein and its structure database template 3×29.1.A, has 42.62% amino acid sequence, which is derived from the template CLDN19 that GMQE is 0.60 and QMEAN4 is −4.77, which 3D structure as shown in Fig. 8A-D.
Figure 8.
Predicted three-dimensional structure of CLDN6 with Swissmodel server. (A) CLDN6 protein and its structure database template 3×29.1.A. (B) CLDN6 protein has 42.62% amino acid sequence. (C) GMQE is 0.60 and QMEAN4 is −4.77. (D) The predicted 3D structure of CLDN6.
The evolutionary tree of CLDN family amino acid sequence rendering system and homology analysis of human CLDN6 protein sequences
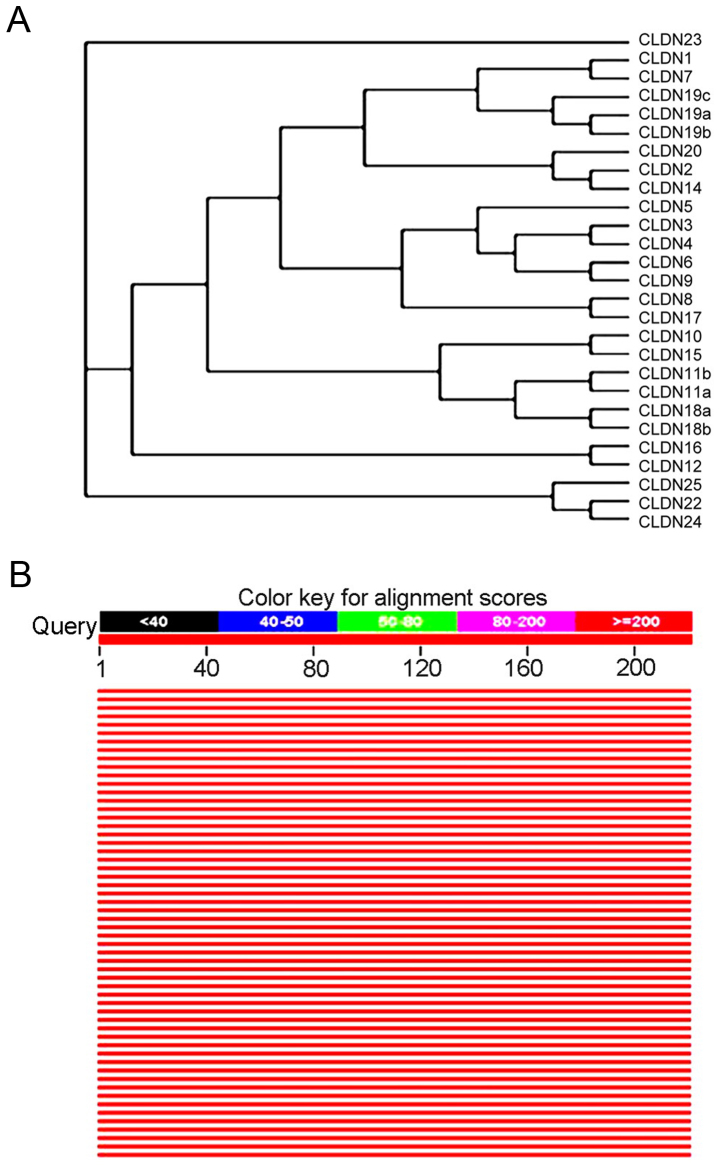
CLDN6 was identified by searching expressed sequence tag (EST) databases for sequences similar to CLDN1 and CLDN2 (22). It was subsequently cloned and expressed in cells, where it was shown to concentrate at tight junctions (22) and human and mouse isoforms have been identified. With Clustalx program construct phylogenetic tree, and with the Treeview software on the system of evolutionary tree edit and comparison, the family system evolutionary tree of CLDNs protein amino acid sequence was drawn. All members of CLDNs are clustered closely except CLDN23, and except CLDN5, CLDN19c, CLDN20 and CLDN25, 22 members of the others exist in pairs and CLDN6 in particular is paired with CLDN9, suggesting they are the closest (Fig. 9A). Comparison of the amino acid seuqences showed that CLDN6 shares 25–70% overall similarity with other CLDNs at the amino acid level with highest similarity to CLDN9. Through the relevant data in the library collection download sequence similarity information encoding protein >50% of the species to build the system tree, as shown in Fig. 9B, the CLDN6 protein with human and rodent coding product has high homology of other animals in the phylogenetic tree perimeter, they may play the same biological functions.
Figure 9.
Evolutionary tree of the CLDN family. Drosophila Tap protein amino acid sequence with other species Phylogenetic tree between the two systems. (A) The family system evolutionary tree of CLDNs protein amino acid sequence was drawn (B) through the relevant data in the library collection download sequence similarity information encoding protein >50% of the species to build the system tree.
CLDN6 related signaling pathway analysis
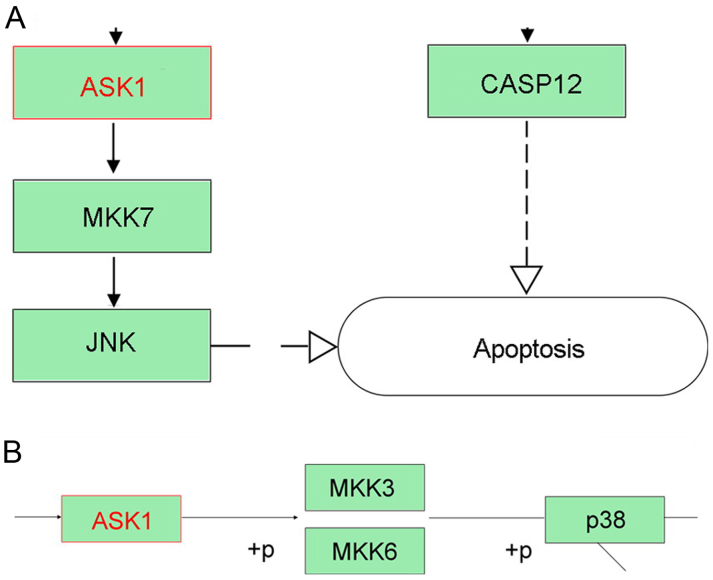
To identify CLDN6-related signaling pathway, We focused on CLDN6-related molecular network information in KEGG pathway maps. CLDN6 is related to four signaling pathways including hsa04514; hsa04530; hsa04670; hsa05160 and has been identified as exosomal proteins of cancer cells such as ovarian cancer, and colorectal cancer, are closely related to CLDN6. CLDNs activated by miR-122 can activate RIP1, TRAF6, p38 and JNK in hsa05160, and apoptosis signal-regulating kinase 1 (ASK1) can activate downstream p38 and JNK-induced apoptosis (Fig. 10A and B). However, the other signaling pathways are not involved in apoptosis. ASK1, a member of the mitogen-activated protein kinase (MAPK) kinase family has been proved to positively correlate with the level of CLDN6 and associated with its pro-apoptosis effect in cervical carcinoma and breast cancer (12,23,24). Taken together, CLDN6 may induce cancer cell apoptosis via ASK1-p38/JNK MAPK pathway.
Figure 10.
CLDN6-related signaling pathways, identified using KEGG pathway searches. (A) Cell apoptosis is induced by ASK1-MKK7-JNK signaling and casp12. (B) The effects of ASK1 on p38 expression is shown through downstream MKK3 and MKK6 molecules.
Discussion
CLDN6 on chromosome 16p13.3 encodes a 23 kDa four-transmembrane protein. CLDN6 has been shown to be expressed differentially in various tumor tissues and cells (25–27), and to a certain degree its alteration can inhibit or pomote tumor cell growth, apoptosis, invastion, migration and EMT in vitro, and methylation of the gene may be involved in tumorigenesis (7,26,28). Although we have clarified some functions of CLDN6, more questions remain to be answered, e.g. why it is differentially expressed in different cells, which signal pathway relates to it and how it is regulated, suggesting alternative ways are needed to answer the questions. Bioinformics may be a way complementary to experimental biology in understanding CLDN6 regulation and function, which necessarily must be validated by experiments in vitro or in vivo.
We studied the 5′ regulatory region of CLDN6 using bioinformatics tools and found that it contained three GC-boxes, three CpG islands and in the 5′ regulatory region sequence 432 and 24 potential TFBS were predicted with a score of 85–99 and diverse TFs such as SP1 are predicted to bind to these TFBS. These results are supported by published literatures, e.g. Anelli and Sitia documented that the associations between CLDN6 expression and mRNA levels of SP1 in ovarian carcinoma effusions (18), as well as transcription factors such as CRE-BP and CREB (17), the expression of CLDN6 is associated with methylating CpG islands; treatment with TSA and/or 5-aza or DMSO induced marked decreases in the levels of methylation of CpG islands in the promoters of CLDN6. The above are also supported by our experiments, which shows that DNA methyltransferase (DNMT1) inhibits the expression of CLDN6 to affect the function of tight junction in breast cancer MCF-7 cells in our unpublished work. In addition, it is reported that some TFBS, including those for AP-1, c-Jun, ATF-2, HNF-4α and COUP-TF have been shown to contribute to CLDN6 regulation (29–34).
The amino acid sequence of CLDN6 was also analyzed by bioinformatics methods. The results showed that the isoelectric point was 8.32; leucine, valine, glycine, serine and alanine were the most abundant amino acids; CLDN6 was an unstable protein because of hydrophobic amino acids. Further, the investigation also found that CLDN6 was a trans-membrane protein with leader peptides and a signal peptide at the N-terminus that was hydrophilic. CLDN conformation in the plane of the membrane shows the four transmembrane α-helical domain as a tightly packed complex, classes of claudin-claudin interactions within a tight junction strand and they can interact via head-to-head binding in the extracellular environment between adjacent cells and within the plane of the plasma membrane in the same cell (21). The main predicted secondary structures of CLDN6 are α helices, extended strands and random coils in our results. CLDN6 encodes four-transmembrane domain protein components of tight junction strands (22). Our CLDN evolutionary tree shows CLDN6 and CLDN9 are most closely related, which is similar to a report by Lal-Nag et al (35) and it is reported that both CLDN6 and CLDN9 function as additional coreceptors for hepatitis C virus (25), suggesting they have common functions. CLDN6 related signaling pathway analysis results obtained from these tools are used to support CLDN6-induced apoptosis via regulating ASK1-p38/JNK signaling in breast cancer MCF-7 cells (36).
In summary, these results reveal that CLDN6 gene may have diverse transcription start sites and its transcription is regulated by DNA methylation and transcription factors such as SP1. Additionally, CLDN6 may be oligomerized in ER and transported to cell membrane via secretory pathway and finally integrated into tight junctions.
Acknowledgements
This study was supported by National Natural Science Foundation of China (code: 81172499) and Jilin Province Science and Technology Development Projects (20140414036GH) This study is funded by Key Laboratory of Natural Resources of Changbai Mountain & Functional Molecules (Yanbian University), Ministry of Education.
Glossary
Abbreviations
- CLDN
claudin
- GC
cytosine and guanine
- CpG
cytosine-phosphoric acid-guanine
- ASK1
Apoptosis signal regulating kinase 1
- JNK
c-Jun NH2-terminal kinase
- ERα
estrogen receptor α
- MMP-2
matrix metalloproteinase-2
- TFBS
transcriptional factor binding sites
- TBP
TATA binding protein
- SP1
specificity protein 1
- CBF
C-repeat binding factor
- AML-1a
acute myeloid leukemia 1a
- CREB
cAMP-response element bingding protein
- CRE-BP
CREB-binding protein
- ER
endoplasmic reticulum
- EST
expressed sequence tag
References
- 1.Günzel D, Yu AS. Claudins and the modulation of tight junction permeability. Physiol Rev. 2013;93:525–569. doi: 10.1152/physrev.00019.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu S, Singh K, Mangray S, Tavares R, Noble L, Resnick MB, Yakirevich E. Claudin expression in high-grade invasive ductal carcinoma of the breast: Correlation with the molecular subtype. Mod Pathol. 2013;26:485–495. doi: 10.1038/modpathol.2012.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mineta K, Yamamoto Y, Yamazaki Y, Tanaka H, Tada Y, Saito K, Tamura A, Igarashi M, Endo T, Takeuchi K, et al. Predicted expansion of the claudin multigene family. FEBS Lett. 2011;585:606–612. doi: 10.1016/j.febslet.2011.01.028. [DOI] [PubMed] [Google Scholar]
- 4.Kwon MJ. Emerging roles of claudins in human cancer. Int J Mol Sci. 2013;14:18148–18180. doi: 10.3390/ijms140918148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li X, Li Y, Qiu H, Wang Y. Downregulation of claudin 7 potentiates cellular proliferation and invasion in endometrial cancer. Oncol Lett. 2013;6:101–105. doi: 10.3892/ol.2013.1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rendon-Huerta EP, Torres-Martínez AC, Montaño L. CLDN6 (claudin 6) Atlas Genet Cytogenet Oncol Haematol. 2013;17:396–399. [Google Scholar]
- 7.Zavala-Zendejas VE, Torres-Martinez AC, Salas-Morales B, Fortoul TI, Montaño LF, Rendon-Huerta EP. Claudin-6, 7, or 9 overexpression in the human gastric adenocarcinoma cell line AGS increases its invasiveness, migration, and proliferation rate. Cancer Invest. 2011;29:1–11. doi: 10.3109/07357907.2010.512594. [DOI] [PubMed] [Google Scholar]
- 8.Wang L, Xue Y, Shen Y, Li W, Cheng Y, Yan X, Shi W, Wang J, Gong Z, Yang G, et al. Claudin 6: A novel surface marker for characterizing mouse pluripotent stem cells. Cell Res. 2012;22:1082–1085. doi: 10.1038/cr.2012.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Q, Zhang Y, Zhang T, Han ZG, Shan L. Low claudin-6 expression correlates with poor prognosis in patients with non-small cell lung cancer. Onco Targets Ther. 2015;8:1971–1977. doi: 10.2147/OTT.S85478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu X, Jin H, Liu Y, Liu L, Wu Q, Guo Y, Yu L, Liu Z, Zhang T, Zhang X, et al. The expression patterns and correlations of claudin-6, methy-CpG binding protein 2, DNA methyltransferase 1, histone deacetylase 1, acetyl-histone H3 and acetyl-histone H4 and their clinicopathological significance in breast invasive ductal carcinomas. Diagn Pathol. 2012;7:33. doi: 10.1186/1746-1596-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ren Y, Wu Q, Liu Y, Xu X, Quan C. Gene silencing of claudin-6 enhances cell proliferation and migration accompanied with increased MMP-2 activity via p38 MAPK signaling pathway in human breast epithelium cell line HBL-100. Mol Med Rep. 2013;8:1505–1510. doi: 10.3892/mmr.2013.1675. [DOI] [PubMed] [Google Scholar]
- 12.Zhang X, Ruan Y, Li Y, Lin D, Liu Z, Quan C. Expression of apoptosis signal-regulating kinase 1 is associated with tight junction protein claudin-6 in cervical carcinoma. Int J Clin Exp Pathol. 2015;8:5535. [PMC free article] [PubMed] [Google Scholar]
- 13.Luscombe NM, Greenbaum D, Gerstein M. What is bioinformatics? A proposed definition and overview of the field. Methods Inf Med. 2001;40:346–358. [PubMed] [Google Scholar]
- 14.Ilzins OA, Isea R, Hoebeke J., III Can bioinformatics be considered as an experimental biological science? Open Sci J Biosci Bioeng. 2015;2:60–62. [Google Scholar]
- 15.Isea R. The present-day meaning of the word bioinformatics. Glob J Adv Res. 2015;2:70–73. [Google Scholar]
- 16.Eck RV, Dayhoff MO. Evolution of the structure of ferredoxin based on living relics of primitive amino acid sequences. Science. 1966;152:363–366. doi: 10.1126/science.152.3720.363. [DOI] [PubMed] [Google Scholar]
- 17.Koval M. Differential pathways of claudin oligomerization and integration into tight junctions. Tissue Barriers. 2013;1:e24518. doi: 10.4161/tisb.24518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anelli T, Sitia R. Protein quality control in the early secretory pathway. EMBO J. 2008;27:315–327. doi: 10.1038/sj.emboj.7601974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piontek J, Winkler L, Wolburg H, Müller SL, Zuleger N, Piehl C, Wiesner B, Krause G, Blasig IE. Formation of tight junction: Determinants of homophilic interaction between classic claudins. FASEB J. 2008;22:146–158. doi: 10.1096/fj.07-8319com. [DOI] [PubMed] [Google Scholar]
- 20.Hou J, Renigunta A, Konrad M, Gomes AS, Schneeberger EE, Paul DL, Waldegger S, Goodenough DA. Claudin-16 and claudin-19 interact and form a cation-selective tight junction complex. J Clin Invest. 2008;118:619–628. doi: 10.1172/JCI33970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Overgaard CE, Daugherty BL, Mitchell LA, Koval M. Claudins: control of barrier function and regulation in response to oxidant stress. Antioxid Redox Signal. 2011;15:1179–1193. doi: 10.1089/ars.2011.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morita K, Furuse M, Fujimoto K, Tsukita S. Claudin multigene family encoding four-transmembrane domain protein components of tight junction strands. Proc Natl Acad Sci USA. 1999;96:511–516. doi: 10.1073/pnas.96.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo Y, Xu X, Liu Z, Zhang T, Zhang X, Wang L, Wang M, Liu Y, Lu Y, Liu YA, et al. Apoptosis signal-regulating kinase 1 is associated with the effect of claudin-6 in breast cancer. Diagn Pathol. 2012;7:111. doi: 10.1186/1746-1596-7-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, Ruan Y, Li Y, Lin D, Quan C. Tight junction protein claudin-6 inhibits growth and induces the apoptosis of cervical carcinoma cells in vitro and in vivo. Med Oncol. 2015;32:148. doi: 10.1007/s12032-015-0600-4. [DOI] [PubMed] [Google Scholar]
- 25.Zheng A, Yuan F, Li Y, Zhu F, Hou P, Li J, Song X, Ding M, Deng H. Claudin-6 and claudin-9 function as additional coreceptors for hepatitis C virus. J Virol. 2007;81:12465–12471. doi: 10.1128/JVI.01457-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu Q, Wu X-Y, Zhang H-Y, Liu Y-F, Ren Y, Qu S-S, Quan C-S, Li Y-L. Expression of tight junctions protein claudin-6 in breast cancer tissues and cell lines and its relationship with metastasis of breast cancer. J Jilin Univ Med Edit. 2008;34:274–279. (In Chinese) [Google Scholar]
- 27.Lin Z, Zhang X, Liu Z, Liu Q, Wang L, Lu Y, Liu Y, Wang M, Yang M, Jin X, et al. The distinct expression patterns of claudin-2, −6, and −11 between human gastric neoplasms and adjacent non-neoplastic tissues. Diagn Pathol. 2013;8:133. doi: 10.1186/1746-1596-8-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu Q, Liu Y, Ren Y, Xu X, Yu L, Li Y, Quan C. Tight junction protein, claudin-6, downregulates the malignant phenotype of breast carcinoma. Eur J Cancer Prev. 2010;19:186–194. doi: 10.1097/CEJ.0b013e328337210e. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y, Jin X, Li Y, Ruan Y, Lu Y, Yang M, Lin D, Song P, Guo Y, Zhao S, et al. DNA methylation of claudin-6 promotes breast cancer cell migration and invasion by recruiting MeCP2 and deacetylating H3Ac and H4AcJ. J Exp Clin Cancer Res. 2016;35:120. doi: 10.1186/s13046-016-0396-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohazama A, Sharpe PT. Expression of claudins in murine tooth development. Dev Dyn. 2007;236:290–294. doi: 10.1002/dvdy.21001. [DOI] [PubMed] [Google Scholar]
- 31.Osanai M, Murata M, Chiba H, Kojima T, Sawada N. Epigenetic silencing of claudin-6 promotes anchorage-independent growth of breast carcinoma cells. Cancer Sci. 2007;98:1557–1562. doi: 10.1111/j.1349-7006.2007.00569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ribeiro A, Archer A, Le Beyec J, Cattin AL, Saint-Just S, Pinçon-Raymond M, Chambaz J, Lacasa M, Cardot P. Hepatic nuclear factor-4, a key transcription factor at the crossroads between architecture and function of epithelia. Recent Pat Endocr Metab Immune Drug Discov. 2007;1:166–175. doi: 10.2174/187221407780832000. [DOI] [Google Scholar]
- 33.Hui PJH. QSPACE. Kingston, Ontario, Canada: Queen's University; 2008. Small proline rich protein-2 expression and regulation in the Caco-2 model of intestinal epithelial differentiation along the crypt-villus axis. [Google Scholar]
- 34.Nishikiori N, Sawada N, Ohguro H. Prevention of murine experimental corneal trauma by epigenetic events regulating claudin 6 and claudin 9. Jpn J Ophthalmol. 2008;52:195–203. doi: 10.1007/s10384-008-0524-z. [DOI] [PubMed] [Google Scholar]
- 35.Lal-Nag M, Battis M, Santin A, Morin P. Claudin-6: A novel receptor for CPE-mediated cytotoxicity in ovarian cancer. Oncogenesis. 2012;1:e33. doi: 10.1038/oncsis.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo Y, Lin D, Zhang M, Zhang X, Li Y, Yang R, Lu Y, Jin X, Yang M, Wang M, et al. CLDN6-induced apoptosis via regulating ASK1-p38/JNK signaling in breast cancer MCF-7 cells. Int J Oncol. 2016;48:2435–2444. doi: 10.3892/ijo.2016.3469. [DOI] [PubMed] [Google Scholar]