Abstract
Trefoil factor 3 (TFF3) serves an important role in intestinal mucosal damage and healing, and contributes to the pathogenesis and treatment of inflammatory bowel disease (IBD). The aim of the present study was to determine the association between TFF3 and microRNA-7-5p (miR-7-5p) in IBD. Tissue immunohistochemistry was applied to evaluate the relative expression of TFF3, and reverse transcription-quantitative polymerase chain reaction was performed to determine the expression of miR-7-5p in lesional tissue obtained from patients with IBD and healthy control tissues. A dual-luciferase reporter assay was used to investigate whether TFF3 was a target of miR-7-5p, and western blotting was performed to determine the expression of TFF3 when miR-7-5p was overexpressed or suppressed. The protein expression levels of TFF3 were decreased and miR-7-5p was overexpressed in the lesional tissue of patients with IBD compared with in healthy control tissues. TFF3 was identified as a target of miR-7-5p, and TFF3 protein expression was negatively regulated by miR-7-5p in human colonic epithelial LS174T cells. The present study demonstrated a negative association between the expression of miR-7-5p and TFF3 in IBD lesional tissues and normal tissues. In conclusion, TFF3 was identified as a novel target of miR-7-5p and miR-7-5p may serve as a promising therapeutic target for IBD.
Keywords: microRNA-7-5p, trefoil factor 3, inflammatory bowel disease, human colonic epithelial LS174T cells
Introduction
Inflammatory bowel disease (IBD) is characterized by chronic intestinal inflammation that results from an abnormal immune response to environmental factors in genetically susceptible individuals. IBD includes ulcerative colitis (UC) and Crohns disease (CD) (1). The incidence of IBD is increasing worldwide (2); however, its etiology and pathogenesis are not fully understood. Trefoil factor 3 (TFF3) belongs to the trefoil factor family and contains one trefoil domain of 59 amino acids, with a molecular weight of ~6.6 kDa (monomer) or 13 kDa (dimer). TFF3 serves an important role in intestinal mucosal damage and mucosal healing (3). TFF3 promotes epithelial restitution resulting in mucosal protection by interacting with mucin, and enhances the structural integrity of the mucosal barrier. TFF3 can reconstruct the epithelial barrier by stimulating epithelial cell migration and proliferation, and by influencing the expression of tight junction proteins. Furthermore, it maintains the integrity of the epithelial barrier by inhibiting apoptosis and regulating immune responses. The physiological functions of TFF3 have been well characterized; however, the regulatory mechanisms controlling TFF3 remain unclear.
MicroRNAs (miRNAs/miR) are small 22-nucleotide, noncoding single-stranded RNA molecules, which bind to the 3′ untranslated region (3′UTR) of their target mRNAs, interfering with RNA expression and inducing either mRNA degradation or translational repression (4). Numerous miRNA functions have been described in various autoimmune-associated conditions, including psoriasis, rheumatoid arthritis, lupus and asthma (5–7). A previous study observed differential miRNA expression in patients with active UC and CD (8). In addition, previous studies have revealed that miRNAs exert powerful gene regulatory functions, which can affect the function of Toll-like receptors (9), regulate the expression of inflammatory cytokines, enhance the permeability of tight junctions, increase apoptosis in epithelial cells and regulate the occurrence and development of IBD (10). Ye et al (11) demonstrated that tumor necrosis factor-α increased the expression of miR-122, which degraded tight junction proteins and increased the intestinal permeability of a filter-grown Caco-2 monolayer. Identification of miRNAs that regulate TFF3 expression may improve understanding regarding the pathogenesis of IBD and lead to novel approaches for the diagnosis and treatment of this disease. The present study aimed to investigate if miR-7-5p binds to the 3′UTR and regulates the expression of TFF3, and to explore the expression of miR-7-5p and TFF3 in IBD tissues.
Materials and methods
Bioinformatics prediction
Bioinformatics prediction software (Targetscan version 6.2; http://www.targetscan.org/vert_61/) was used to predict the miRNAs that would bind to the 3′UTR of TFF3.
Cell culture experiments
The human colonic epithelial cell line LS174T was obtained from American Type Culture Collection (Manassas, VA, USA). The cell line was grown to near confluence in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (both from Hyclone; GE Healthcare Life Sciences, Logan, Utah, USA), 50 U/ml penicillin and 50 mg/ml streptomycin at 37°C. The cells were subcultured following partial digestion with 0.25% trypsin and 0.9 mmol/l EDTA in Ca2+-free and Mg2+-free phosphate-buffered saline (PBS).
Plasmids, transfection and dual-luciferase reporter assay
miR-7-5p mimic, miR-7-5p inhibitor, mimic negative control (NC) and inhibitor NC were all purchased from Guangzhou RiboBio Co., Ltd., (Guangzhou, China). The inhibitor sequences contained nucleotides with the 2′O methyl ribose modification (m). The sequences were as follows: miR-7-5p mimic, 5′-UGGAAGACUAGUGAUUUUGUUGU-3′; miR-7-5p mimic NC, 5′-UUUGUACUACACAAAAGUACUG-3′; miR-7-5p inhibitor, 5′-mAm CmA mAm CmA mAm AmA mUm CmA mCm UmA mGm UmC mUm UmC mCmA-3′; miR-7-5p inhibitor NC, 5′-mCm AmG mUm AmC mUm UmU mUm GmU mGm UmA mGm UmA mCm AmA mA-3′. The 3′UTR of TFF3 was obtained from human genomic DNA (from LS174T cells) using reverse transcription-polymerase chain reaction (RT-PCR) (42°C 60 min, 70°C 15 min) with the following primers: Forward, 5′-CCCAAGCTTGGCACCCACGTCACAGGA-3′, and reverse, 5′-GGACTAGTAACAAAACCCAGGAATAG-3′, respectively. The TFF3 3′UTR was cloned downstream from the luciferase gene in the pMIR-Reporter luciferase vector (Promega Corporation, Madison, WI, USA). Mutant vectors with altered predicted miR-7-5p binding sites were constructed using a site-directed mutagenesis kit (Beyotime Institute of Biotechnology, Shanghai, China) and the following primer: 5′-GCAAGCCAATAAAACTGCTGTCCAAAGTGGTCCTTTA-3′. The TFF3 gene with wild-type 3′UTR (TFF3-WtUTR) or mutant 3′UTR (TFF3-MutUTR) was inserted into the pMIR-Report luciferase vector. HEK293 cells (acquired from Shanghai Institute for Biological Sciences, Chinese Academy of Sciences, Shanghai, China) were seeded at a density of 2×105 cells/well in 24-well plate and cotransfected, respectively with 200 ng pMIR-TFF3-WtUTR and pMIR-TFF3-MutUTR together with 100 ng pRL-TK-Renilla-luciferase plasmid (Promega Corporation) and 100 nM miR-7-5p mimic or miRNA negative control using Lipofectamine® 3000 (Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's protocol. Cells were harvested 48 h post transfection, and luciferase activity was measured using a dual-luciferase reporter system (Promega Corporation). The pRL-TK-Renilla-luciferase plasmid was cotransfected as the internal Renilla control. Each transfection was performed in triplicate.
In vitro transfection of miR-7-5p mimic and inhibitor
The effects of miR-7-5p on TFF3 were determined by western blotting, post-transfection of miR-7-5p into LS174T cells. The transfection solution was prepared in two microcentrifuge tubes, one containing DMEM (25 µl) and Lipofectamine® 3000 (5 µl), the other containing miR-7-5p mimic, miR-7-5p inhibitor, miR-7-5p mimic NC or miR-7-5p inhibitor NC (10 µl; 100 nM), DMEM (25 µl) and P3000 (5 µl; Invitrogen; Thermo Fisher Scientific, Inc.). The contents of the two tubes were mixed together, incubated for 5 min at room temperature, then added to the LS174T cells, which had been grown to 60–80% confluence in 6-well plates. Following 48 h culture at 37°C, the cells were harvested and prepared for the next experiment.
Western blot analysis
LS174T cells were seeded in 6-well plates and grown to 60–80% confluence. The cells were transfected with miR-7-5p mimic, miR-7-5p inhibitor, miR-7-5p mimic NC or miR-7-5p inhibitor NC for 48 h. Following incubation, the cells were scraped from the culture plates and lysed with radioimmunoprecipitation assay lysis buffer [50 mM Tris (pH 7.4), 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, inhibitors including sodium orthovanadate, sodium fluoride, EDTA and leupeptin, and proteinase/phosphatase inhibitors] (Beyotime Institute of Biotechnology, Shanghai, China). Equal quantities of protein (40 mg) were separated by 15% SDS-PAGE and the proteins were blotted onto a polyvinylidene fluoride membrane (EMD Millipore, Billerica, MA, USA). The membrane was blocked in 5% non-fat milk, followed by incubation at 4°C overnight with primary antibodies against TFF3 (rabbit monoclonal antibody; dilution, 1:1,000; ab108599; Abcam, Cambridge, MA, USA) and GAPDH (rabbit monoclonal antibody; dilution, 1:5,000; ab9485; Abcam). After washing in TBS-Tween-20 (0.1%), membranes were incubated with the secondary antibody [horseradish peroxidase-labeled goat anti-rabbit immunoglobulin G (H+L); dilution, 1:2,000; A0208; Beyotime Institute of Biotechnology] at room temperature for 2 h. Bands were visualized using the enhanced chemiluminescence method (Pierce™ ECL Western Blotting Substrate; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Semi-quantification was performed using ImageJ v1.48u software (National Institutes of Health, Bethesda, Maryland, USA).
Immunohistochemistry and hematoxylin and eosin (H&E) staining
Ulcerative colonic tissue specimens (0.2-cm3) were obtained from children with IBD (5 CD patients and 3 UC patients) by colonoscopy, and normal colonic tissues were obtained from patients who had undergone intestinal surgery for other diseases. Patients were recruited from October-December 2015 in the pediatric digestive internal medicine ward and the pediatric surgical ward of Shengjing Hospital of China Medical University (Shenyang, China). Normal control colon tissues were collected from 8 patients. The inclusion criteria was as follows: i) Diagnosis with intestinal polyps or congenital megacolon; ii) pathology revealed normal intestinal tissue, with no inflammation and ulcers. The exclusion criteria was as follows: i) Diagnosis with intestinal perforation, necrosis, or destruction of the mucosal structure; ii) diagnosis with a tumor, due to the impact on miRNA levels in these patients. The clinical parameters of the patients were as follows: IBD patients, age 3.5–14 years; male: Female=7:1; UC: CD=3:5. The age of the control patients ranged 33 months-10 years; male: Female=5:3; 7 patients were diagnosed with colonic polyps, 1 patient was diagnosed with congenital megacolon. The present study was approved by the ethics committee of the Affiliated Shengjing Hospital of China Medical University (approval no. 2015PS281K; Shenyang, China). Written informed consent was obtained from the relatives of the children.
Immunohistochemical staining was performed using a two-step immunohistochemistry kit (ZSGB-BIO, Beijing, China). The samples were fixed in 4.0% formaldehyde at room temperature for 24 h, embedded in paraffin and sectioned at 5.0-µm. The sections were dewaxed (xylene for 10 min, xylene for 20 min), rehydrated [graded ethanol (100, 95, 85, 70%) for 5 min, respectively], and treated with 3.0% H2O2 to block endogenous peroxidases. Antigens were recovered in microwave-heated citrate acid, and the sections were incubated with primary TFF3 antibody (dilution, 1:200; ab108599; Abcam) overnight at 4°C. The sections were recovered at 37°C for 20 min, then incubated with the secondary antibody (biotin labeled goat anti-rabbit IgG; dilution, 1:50; SP-9001; ZSGB-BIO, Beijing, China) at room temperature for 30 min. The color was developed using 5% diaminobenzidine for 5 min at room temperature and nuclei were stained with hematoxylin (Beyotime Institute of Biotechnology) for 10 min at room temperature. Staining was observed with an Eclipse Ci Plus optical microscope (Nikon Corporation, Tokyo, Japan). PBS was substituted for the primary antibody as negative control both in the IBD group and the NC group.
For H&E staining, samples were washed in ice-cold PBS, fixed in 4% paraformaldehyde for 24 h at room temperature and embedded in paraffin. Consecutive colon sections (4-µm) were stained with 0.1–0.5% H&E at room temperature for 4 min for pathomorphological examination by optical microscope.
RNA extraction and quantitative (q)-PCR
Ulcerative and normal colonic tissues were obtained as aforementioned. Total miRNA was extracted using a mirVana™ miRNA Isolation kit (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Complementary DNA was synthesized from 10 ng total RNA, using a TaqMan® miRNA reverse transcription kit (4366596; Thermo Fisher Scientific, Inc.). The reaction mixture contained the following: 100 nM dNTPs with dTTP, 0.15 µl; MutiScribe™ Reverse Transcription 50 U/µl, 1 µl; 10X reverse buffer, 1.5 µl; RNase inhibitor 20 U/µl, 0.19 µl; nuclease-free water, 4.16 µl; total RNA (1–10 ng), 5 µl; 5X RT primer, 3 µl. The thermocycling conditions were: 16°C 30 min, 42°C 30 min, 85°C 5 min, hold 4°C. The expression levels of miR-7-5p were quantified using a TaqMan® small RNA assay kit (PN4427975; Thermo Fisher Scientific, Inc.). The qPCR reaction mixture contained the following; TaqMan Universal PCR Master MixII noUNG+ (4440049; Thermo Fisher Scientific, Inc.) 10 µl; nuclease-free water, 7.67 µl; Taqman Small RNA assay (20X), 1.0 µl; product from RT reaction, 1.33 µl. The following thermocycling conditions were employed: Optimal AmpErase UNG activity 50°C 2 min; enzyme activation 95°C 10 min; 40 cycles of, denaturation at 95°C for 15 sec and extension at 60°C for 60 sec. Quantification was performed using the 2−ΔΔCq method (12), with U6 snRNA as an endogenous control.
Statistical analysis
All values were expressed as the mean ± standard error of the mean. Statistical analyses were performed using SPSS 13.0 (SPSS, Inc., Chicago, IL, USA). Differences between groups were analyzed by one-way analysis of the variance followed by Bonferroni post hoc analyses as appropriate. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-7-5p may regulate TFF3 protein expression
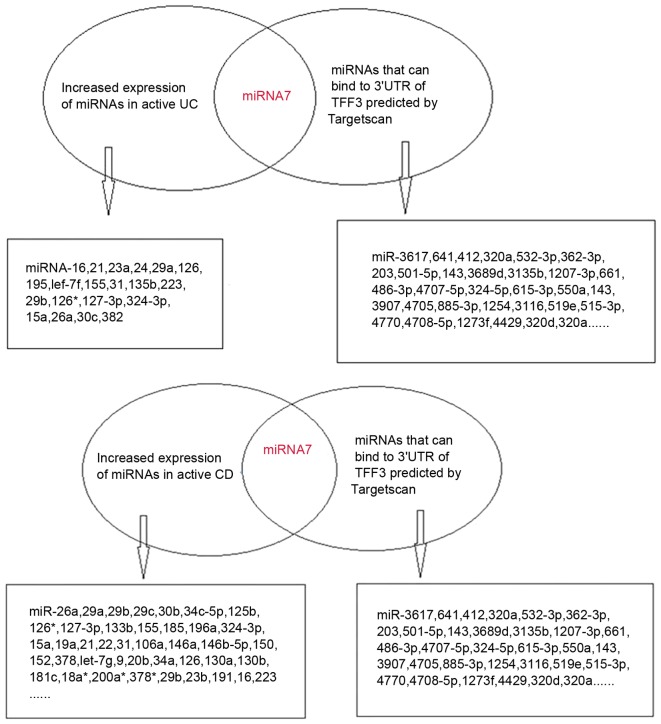
The bioinformatics prediction software Targetscan v6.2 identified three high-likelihood miRNAs and 123 miRNAs with relatively lower probabilities, which were able to bind to the 3′UTR of TFF3. Based on previous literature, there were eight miRNAs that were associated with IBD, including miR-7, miR-320a, miR-375, miR-532-3p, miR-362-3p, miR-203, miR-501-5p and miR-143 (8,13–16). Expression of miR-320a was decreased in IBD (8); miR-203, miR-501-5p and miR-143 were increased in intestinal dysplasia tissues (13–15); and miR-532-3p and miR-362-3p expression was elevated in the peripheral blood of patients with CD (16). In addition, miR-7 exhibited increased expression in active UC and active CD tissues (8).
TargetScan-predicted and literature-identified miRNAs are presented in Fig. 1. From these findings, miR-7-5p was hypothesized to be a modulatory miRNA of the TFF3 protein. Fig. 2A demonstrates the binding site of miR-7-5p on the 3′UTR of TFF3.
Figure 1.
(Left) miRNAs that were upregulated in the active IBD tissues were identified in the literature. miR-7-5p expression was increased in active UC and CD tissues. (Right) miRNAs that may bind to the 3′UTR of TFF3 were predicted by TargetScan. CD, Crohn's disease; miR, microRNA; TFF3, trefoil factor 3; UC, ulcerative colitis.
Figure 2.
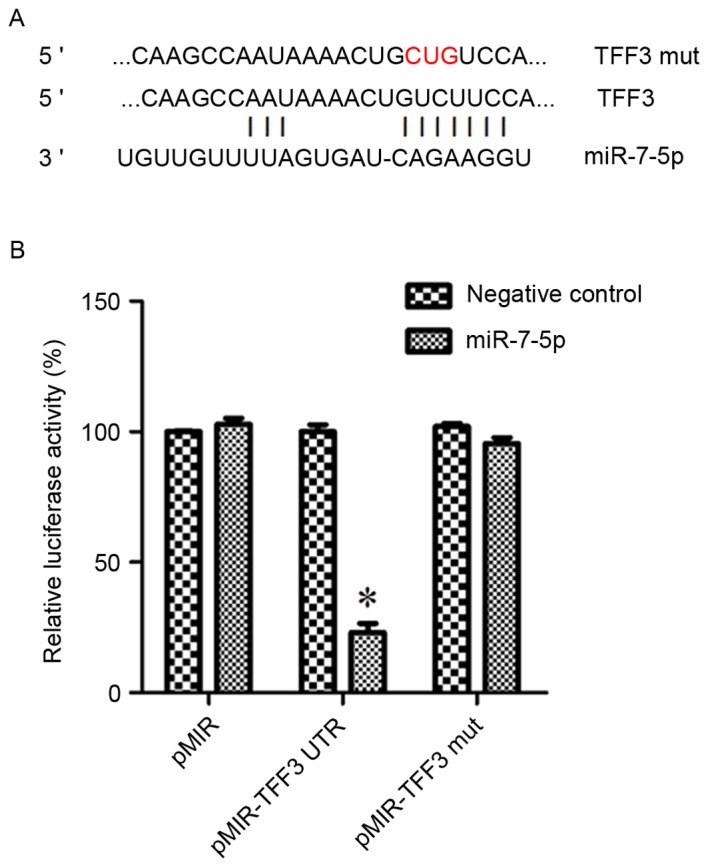
(A) Targetscan software predicted that miR-7-5p binds to the 3′UTR of TFF3. (B) Luciferase reporter assay of miR-7-5p in HEK293 cells. Normal and mutated 3′UTRs from TFF3 were subcloned into the pMIR vector. miR-7-5p was cotransfected with pMIR-TFF3-wild-type 3′UTR or pMIR-TFF3-Mut 3′UTR plasmids into HEK293 cells. The percentage of relative luciferase activity was recorded. *P<0.05 vs. pMIR and pMIR-TFF3 mut. miR, microRNA; mut, mutant; TFF3, trefoil factor 3; 3′UTR, 3′untranslated region.
TFF3 is a target of miR-7-5p in HEK293 cells
A dual-luciferase reporter assay was performed to determine whether TFF3 was a direct target of miR-7-5p in HEK293 cells. The results of the dual-luciferase reporter assays demonstrated that post-transfection with pMIR-TFF3-WtUTR in miR-7-5p-transfected cells, there was a significant decrease in Renilla/firefly luciferase activity; however, there was no difference among the other groups (Fig. 2B). These findings indicated that TFF3 may be a target of miR-7-5p.
TFF3 protein expression is negatively regulated by miR-7-5p in LS174T cells
The protein expression levels of TFF3 were evaluated in LS174T cells where miR-7-5p was overexpressed or suppressed (Fig. 3). As presented in Fig. 3B, overexpression of miR-7-5p significantly inhibited the protein expression levels of TFF3. Conversely, suppression of miR-7-5p expression, via transfection with the miR-7-5p inhibitor, significantly increased the protein expression levels of TFF3 in LS174T cells. The relative expression of miR-7-5p is depicted in Fig. 3A. These findings indicated that miR-7-5p may negatively regulate TFF3 protein expression at the post-transcriptional level.
Figure 3.
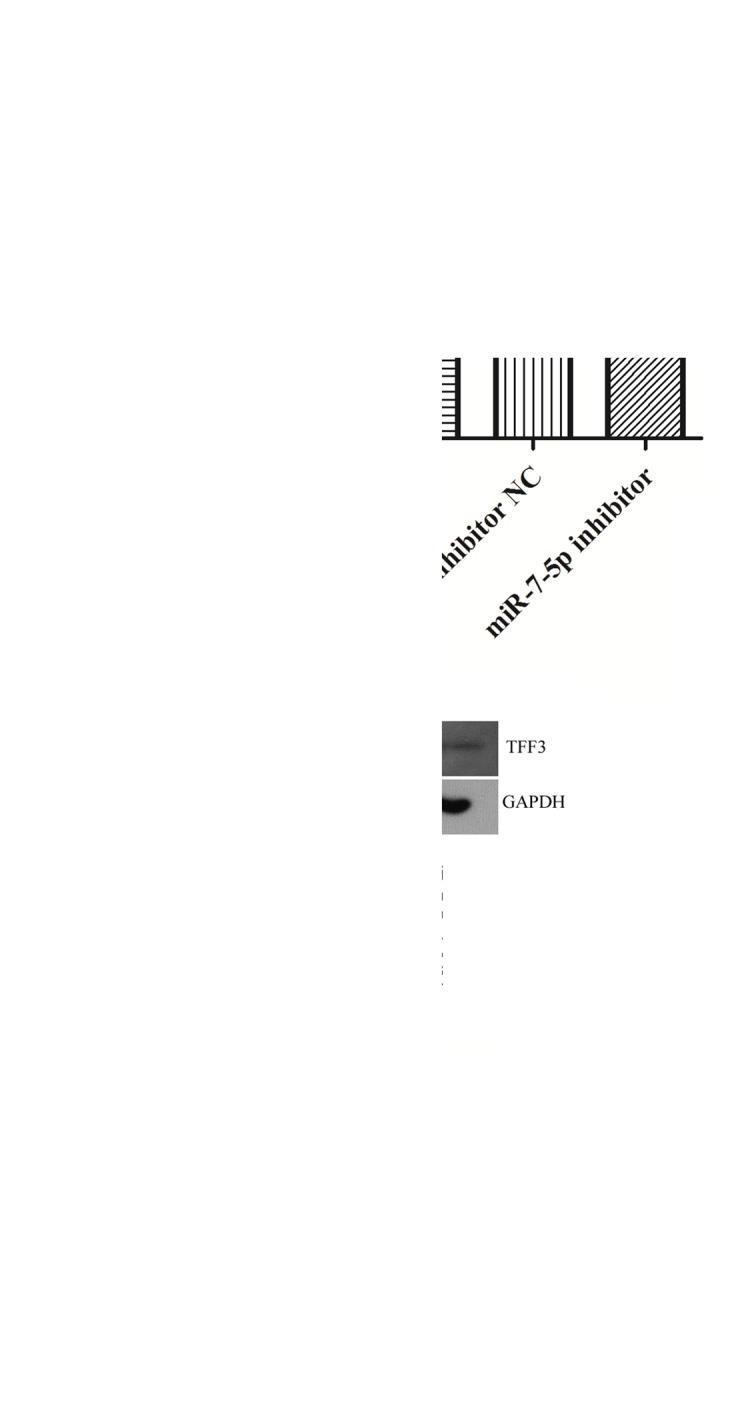
miR-7-5p negatively regulated TFF3 protein expression. (A) Quantitative-polymerase chain reaction was performed to determine the expression of miR-7-5p post-transfection of LS174T cells with miR-7-5p mimic or inhibitor. (B) Western blotting was performed to determine the expression of TFF3 protein post-transfection of LS174T cells with miR-7-5p mimic or inhibitor. Results were normalized to GAPDH. *P<0.05 vs. NC cells. NC, cells without transfection; miR-7-5p mimic NC, cells transfected with miR-7-5p mimic NC miRNA; miR-7-5p mimic, cells transfected with miR-7-5p mimics; miR-7-5p inhibitor NC, cells transfected with miR-7-5p inhibitor NC miRNA; miR-7-5p inhibitor, cells transfected with miR-7-5p inhibitor. miR, microRNA; NC, negative control; TFF3, Trefoil factor 3.
TFF3 expression is decreased in tissues from children with IBD
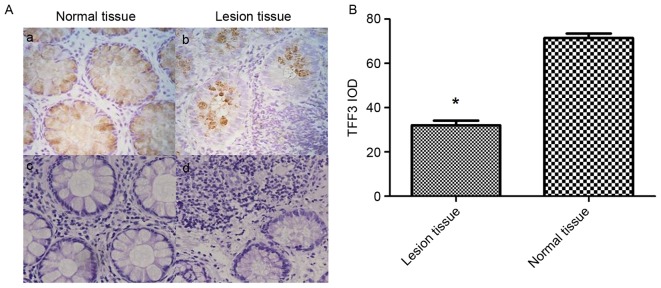
TFF3 expression in IBD tissues from children was examined by immunohistochemistry (Fig. 4). As presented in Fig. 4Aa and Ab, when compared to normal colonic tissues, the relative expression of TFF3 was decreased in IBD lesional tissues. The integrated optical density of TFF3 staining in normal colonic tissues was significantly higher compared with in the IBD lesional tissues (71.5226±4.34 vs. 32.0481±4.76; P<0.05; Fig. 4B). Normal bowel structures were observed in the H&E stain of normal colon tissue (Fig. 4Ac). In contrast, a large number of inflammatory cells, irregular intestinal shape and unevenly distributed goblet cells were present in the H&E stain of IBD lesional tissue (Fig. 4Ad).
Figure 4.
TFF3 expression in lesional tissue from patients with IBD and normal control tissues. (A) Immunohistochemical staining was performed to determine the relative protein expression of TFF3 in IBD lesion colon tissues and normal control tissues (n=8; magnification, ×400). Immunohistochemical staining of (a) normal colonic tissue and (b) lesional colonic tissue; H&E staining of (c) normal colonic tissue and (d) lesional colonic tissue. (B) IOD of TFF3 staining. *P<0.05 vs. normal tissue. H&E, hematoxylin and eosin staining; IBD, inflammatory bowel disease; IOD, integrated optical density; TFF3, trefoil factor 3.
miR-7-5p expression is increased in tissues from children with IBD
The expression levels of miR-7-5p were detected in healthy control tissues and IBD lesional tissues (Fig. 5). The expression levels of miR-7-5p were significantly increased in IBD lesional tissues compared with in normal colonic tissues (P<0.05). These results indicated that miR-7-5p expression in IBD lesional tissues was negatively associated with TFF3 protein expression.
Figure 5.
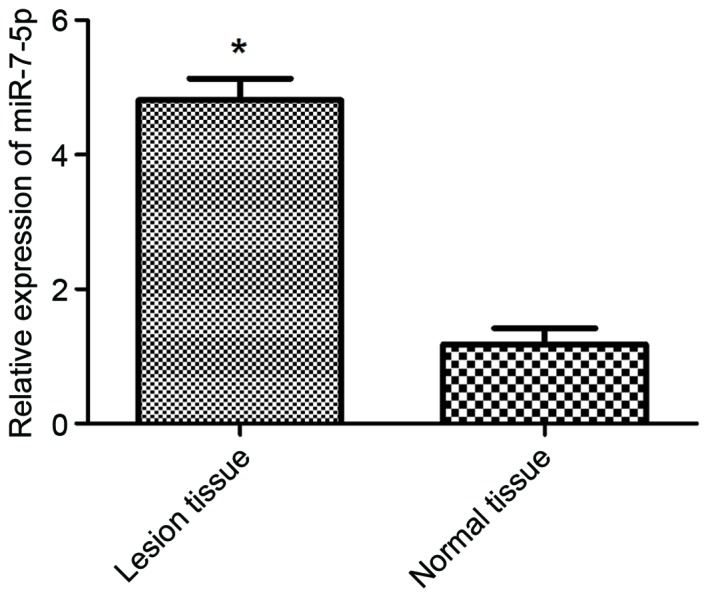
miR-7-5p expression was determined using quantitative polymerase chain reaction in normal and lesional colonic tissue. miR-7-5p was highly expressed in lesional tissue compared with in normal tissue. *P<0.05 vs. normal tissue. miR, microRNA.
Discussion
The incidence of childhood IBD has been reported to increase annually. A Scottish study, which was conducted over 40 years, demonstrated that the incidence of IBD in pediatric patients (<16 years old) increased from 4.45/100,000 between 1990 and 1995, to 7.82/100,000 between 2003 and 2008 (17). In Shanghai, a 10-year study in patients with IBD reported that the incidence of IBD increased from 0.5 per million in 2001 to 6.0 per million in 2010 (18). Younger patients have a particularly difficult time dealing with IBD; therefore, it is of great importance to understand the molecular regulatory mechanisms contributing to the pathogenesis and treatment of IBD.
TFF3 is a small peptide secreted by goblet cells that is highly expressed in intestinal mucosa. It can enhance restitution in intestinal epithelial cells and sustain mucosal integrity (19,20). Mice overexpressing intestinal TFF3 display increased resistance to intestinal damage and ulceration (21), whereas mice with TFF3 gene deletion have a higher susceptibility to gastrointestinal injury (22). TFF3, in conjunction with mucin glycoproteins, can protect gastrointestinal mucosa from various insults (20). Our previous studies have demonstrated that TFF3 protects the intestinal epithelium against platelet activating factor-induced disruption, by restricting the rearrangement of the F-actin cytoskeleton and tight junctions, thereby decreasing mucosal permeability (23,24). Given the potential of TFF3 therapy for treating IBD, the regulatory mechanisms controlling TFF3 expression are of great interest.
Bioinformatics analysis by Targetscan demonstrated that miR-7-5p can bind to the 3′UTR of TFF3 mRNA. miR-7 is a well-studied miRNA, and numerous studies have revealed that miR-7 influences cancer cell metastasis, proliferation and apoptosis by targeting various mRNAs (25–27). Fang et al (26) demonstrated that miR-7 inhibited tumor growth and cell metastasis by targeting the phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma, and Xu et al (27) reported that miR-7 inhibited colorectal cancer cell proliferation and induced apoptosis by targeting X-ray repair cross complementing 2. In addition, Nguyen et al (28) revealed that miR-7 modulated cluster of differentiation 98 expression during intestinal epithelial cell differentiation. To the best of our knowledge, the target of miR-7 in IBD has not been previously reported. Therefore, the present study evaluated the expression levels of miR-7-5p in lesional tissues from patients with IBD and matched normal colonic tissues. The results demonstrated that miR-7-5p expression was markedly increased in patients with IBD compared with in control patients. This result was consistent with a previous study by Pekow et al (29). In addition, Fasseu et al (8) demonstrated that miR-7 was overexpressed in inflamed colonic tissues from patients with UC or CD. The present study also demonstrated that TFF3 expression was decreased in lesional tissues from patients with IBD compared with in normal colonic tissue, and was negatively associated with miR-7-5p expression. The present study identified TFF3 as a target of miR-7-5p in HEK293 cells, and further demonstrated that miR-7-5p negatively regulated the expression of TFF3 at a post-transcriptional level in LS174T cells, indicating that the effects of TFF3 on IBD may be mediated by miR-7-5p. The functional implications of the modulation of TFF3 by miR-7-5p require further study.
In conclusion, the present study demonstrated differential expression of TFF3 and miR-7-5p in patients with IBD, and revealed that TFF3 is a novel target of miR-7-5p. In LS174T cells, miR-7-5p modulated protein expression of TFF3 at a post-transcriptional level. Further functional studies are required to fully understand the role of TFF3 in IBD and its regulation by miR-7-5p.
Acknowledgements
The present study was supported by grants from the National Natural Science Foundation of China Youth Foundation (grant no. 81400585) and the Natural Science Foundation of Liaoning Province, China (grant no. 2014021042).
References
- 1.Madsen JR, Laursen LS, Lauritsen K. Chronic inflammatory bowel disease-current status. Ugeskr Laeger. 1992;154:2243–2250. (In Danish) [PubMed] [Google Scholar]
- 2.Ng SC, Zeng Z, Niewiadomski O, Tang W, Bell S, Kamm MA, Hu P, de Silva HJ, Niriella MA, Udara WS, et al. Early course of inflammatory bowel disease in a population-based inception cohort study from 8 countries in Asia and Australia. Gastroenterology. 2016;150:82–95. e3; quiz e13-e14. doi: 10.1053/j.gastro.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 3.Kjellev S. The trefoil factor family-small peptides with multiple functionalities. Cell Mol Life Sci. 2009;66:1350–1369. doi: 10.1007/s00018-008-8646-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Stanczyk J, Pedrioli DM, Brentano F, Sanchez-Pernaute O, Kolling C, Gay RE, Detmar M, Gay S, Kyburz D. Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 2008;58:1001–1009. doi: 10.1002/art.23386. [DOI] [PubMed] [Google Scholar]
- 6.Tang Y, Luo X, Cui H, Ni X, Yuan M, Guo Y, Huang X, Zhou H, de Vries N, Tak PP, et al. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 2009;60:1065–1075. doi: 10.1002/art.24436. [DOI] [PubMed] [Google Scholar]
- 7.Tan Z, Randall G, Fan J, Camoretti-Mercado B, Brockman-Schneider R, Pan L, Solway J, Gern JE, Lemanske RF, Nicolae D, Ober C. Allele-specific targeting of microRNAs to HLA-G and risk of asthma. Am J Hum Genet. 2007;81:829–834. doi: 10.1086/521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fasseu M, Tréton X, Guichard C, Pedruzzi E, Cazals-Hatem D, Richard C, Aparicio T, Daniel F, Soulé JC, Moreau R, et al. Identification of restricted subsets of mature microRNA abnormally expressed in inactive colonic mucosa of patients with inflammatory bowel disease. PLoS One. 2010;5:pii: e13160. doi: 10.1371/journal.pone.0013160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheedy FJ, Palsson-McDermott E, Hennessy EJ, Martin C, O'Leary JJ, Ruan Q, Johnson DS, Chen Y, O'Neill LA. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol. 2010;11:141–147. doi: 10.1038/ni.1828. [DOI] [PubMed] [Google Scholar]
- 10.McKenna LB, Schug J, Vourekas A, McKenna JB, Bramswig NC, Friedman JR, Kaestner KH. MicroRNAs control intestinal epithelial differentiation, architecture, and barrier function. Gastroenterology. 2010;139:1654–1664, e1. doi: 10.1053/j.gastro.2010.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ye D, Guo S, Al-Sadi R, Ma TY. MicroRNA regulation of intestinal epithelial tight junction permeability. Gastroenterology. 2011;141:1323–1333. doi: 10.1053/j.gastro.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Bansal A, Lee IH, Hong X, Anand V, Mathur SC, Gaddam S, Rastogi A, Wani SB, Gupta N, Visvanathan M, et al. Feasibility of microRNAs as biomarkers for Barrett's Esophagus progression: A pilot cross-sectional, phase 2 biomarker study. Am J Gastroenterol. 2011;106:1055–1063. doi: 10.1038/ajg.2011.37. [DOI] [PubMed] [Google Scholar]
- 14.Olaru AV, Selaru FM, Mori Y, Vazquez C, David S, Paun B, Cheng Y, Jin Z, Yang J, Agarwal R, et al. Dynamic changes in the expression of MicroRNA-31 during inflammatory bowel disease-associated neoplastic transformation. Inflamm Bowel Dis. 2011;17:221–231. doi: 10.1002/ibd.21359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanaan Z, Rai SN, Eichenberger MR, Barnes C, Dworkin AM, Weller C, Cohen E, Roberts H, Keskey B, Petras RE, et al. Differential microRNA expression tracks neoplastic progression in inflammatory bowel disease-associated colorectal cancer. Hum Mutat. 2012;33:551–560. doi: 10.1002/humu.22063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu F, Guo NJ, Tian H, Marohn M, Gearhart S, Bayless TM, Brant SR, Kwon JH. Peripheral blood microRNAs distinguish active ulcerative colitis and Crohn's disease. Inflamm Bowel Dis. 2011;1:241–250. doi: 10.1002/ibd.21450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henderson P, Wilson DC. The rising incidence of paediatric-onset inflammatory bowel disease. Arch Dis Child. 2012;97:585–586. doi: 10.1136/archdischild-2012-302018. [DOI] [PubMed] [Google Scholar]
- 18.Wang XQ, Zhang Y, Xu CD, Jiang LR, Huang Y, Du HM, Wang XJ. Inflammatory bowel disease in Chinese children: A multicenter analysis over a decade from Shanghai. Inflamm bowel Dis. 2013;19:423–428. doi: 10.1097/MIB.0b013e318286f9f2. [DOI] [PubMed] [Google Scholar]
- 19.Sun Z, Liu H, Yang Z, Shao D, Zhang W, Ren Y, Sun B, Lin J, Xu M, Nie S. Intestinal trefoil factor activates the PI3K/Akt signaling pathway to protect gastric mucosal epithelium from damage. Int J Oncol. 2014;45:1123–1132. doi: 10.3892/ijo.2014.2527. [DOI] [PubMed] [Google Scholar]
- 20.Kindon H, Pothoulakis C, Thim L, Lynch-Devaney K, Podolsky DK. Trefoil peptide protection of intestinal epithelial barrier function: Cooperative interaction with mucin glycoprotein. Gastroenterology. 1995;109:516–523. doi: 10.1016/0016-5085(95)90340-2. [DOI] [PubMed] [Google Scholar]
- 21.Marchbank T, Cox HM, Goodlad RA, Giraud AS, Moss SF, Poulsom R, Wright NA, Jankowski J, Playford RJ. Effect of ectopic expression of rat trefoil factor family 3 (intestinal trefoil factor) in the jejunum of transgenic mice. J Biol Chem. 2001;276:24088–24096. doi: 10.1074/jbc.M101363200. [DOI] [PubMed] [Google Scholar]
- 22.Mashimo H, Wu DC, Podolsky DK, Fishman MC. Impaired defense of intestinal mucosa in mice lacking intestinal trefoil factor. Science. 1996;274:262–265. doi: 10.1126/science.274.5285.262. [DOI] [PubMed] [Google Scholar]
- 23.Xu LF, Teng X, Guo J, Sun M. Protective effect of intestinal trefoil factor on injury of intestinal epithelial tight junction induced by platelet activating factor. Inflammation. 2012;35:308–315. doi: 10.1007/s10753-011-9320-x. [DOI] [PubMed] [Google Scholar]
- 24.Xu LF, Xu C, Mao ZQ, Teng X, Ma L, Sun M. Disruption of the F-actin cytoskeleton and monolayer barrier integrity induced by PAF and the protective effect of ITF on intestinal epithelium. Arch Pharm Res. 2011;34:245–251. doi: 10.1007/s12272-011-0210-4. [DOI] [PubMed] [Google Scholar]
- 25.Ma J, Fang B, Zeng F, Pang H, Zhang J, Shi Y, Wu X, Cheng L, Ma C, Xia J, Wang Z. Curcumin inhibits cell growth and invasion through up-regulation of miR-7 in pancreatic cancer cells. Toxicol Lett. 2014;231:82–91. doi: 10.1016/j.toxlet.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 26.Fang Y, Xue JL, Shen Q, Chen J, Tian L. MicroRNA-7 inhibits tumor growth and metastasis by targeting the phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma. Hepatology. 2012;55:1852–1862. doi: 10.1002/hep.25576. [DOI] [PubMed] [Google Scholar]
- 27.Xu K, Chen Z, Qin C, Song X. miR-7 inhibits colorectal cancer cell proliferation and induces apoptosis by targeting XRCC2. Onco Targets Ther. 2014;7:325–332. doi: 10.2147/OTT.S59364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen HT, Dalmasso G, Yan Y, Laroui H, Dahan S, Mayer L, Sitaraman SV, Merlin D. MicroRNA-7 modulates CD98 expression during intestinal epithelial cell differentiation. J Biol Chem. 2010;285:1479–1489. doi: 10.1074/jbc.M109.057141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pekow JR, Kwon JH. MicroRNAs in inflammatory bowel disease. Inflamm Bowel Dis. 2012;18:187–193. doi: 10.1002/ibd.21691. [DOI] [PMC free article] [PubMed] [Google Scholar]