Abstract
To investigate the role of centromere protein U (CENPU) in human bladder cancer (BCa), CENPU gene expression was evaluated in human BCa tissues. We used real-time quantitative PCR (qPCR) and found that CENPU gene expression in human BCa tissues was higher compared to that observed in cancer-adjacent normal tissues. High CENPU expression was found to be strongly correlated with tumor size and TNM stage. Kaplan-Meier survival analysis indicated that high CENPU levels were associated with reduced survival. We used a lentivirus to silence endogenous CENPU gene expression in the BCa T24 cell line. CENPU knockdown was confirmed by qPCR. Cellomic imaging and BrdU assays showed that cell proliferation was significantly reduced in the CENPU-silenced cells compared to that noted in the control cells. Flow cytometry revealed that in the CENPU-silenced cells the cell cycle was arrested at the G1 phase relative to that in the control cells. In addition, apoptosis was significantly increased in the CENPU-silenced cells. Giemsa staining showed that CENPU-silenced cells, compared to control cells, displayed a significantly lower number of cell colonies. The genome-wide effect of CENPU knockdown showed that a total of 1,274 differentially expressed genes was found, including 809 downregulated genes and 465 upregulated genes. Network analysis by Ingenuity Pathway Analysis (IPA) resulted in 25 distinct signaling pathways, including the top-ranked network: ‘Cellular compromise, organismal injury and abnormalities, skeletal and muscular disorders’. In-depth IPA analysis revealed that CENPU was associated with the HMGB1 signaling pathway. qPCR and western blot analysis demonstrated that in the HMGB1 signaling pathway, CENPU knockdown downregulated expression levels of ILB, CXCL8, RAC1 and IL1A. In conclusion, our data may provide a potential pathway signature for therapeutic targets with which to treat BCa.
Keywords: bladder cancer, CENPU, HMGB1 signaling pathway, Ingenuity Pathway Analysis
Introduction
Bladder cancer (BCa) is the fourth most common type of cancer in the United States (1,2). Approximately 75% of BCa patients are diagnosed with non-muscle invasive bladder cancer (NMIBC) (3). Among the majority of NMIBC cases that recur, roughly 15–20% progress to MIBC and lead to local invasion and distant metastasis, representing the main cause of death (4,5). Due to the high mortality rate associated with BCa, novel treatment methods to fight the disease are clearly warranted (6). Currently, chemotherapy combined with gemcitabine and cisplatin treatment is the most common treatment strategy for BCa, as well as other carcinomas (7,8). However, in approximately 60–70% of patients with metastatic BCa, relapse occurs in the first year and drug resistance is becoming a major concern. The underlying mechanisms of BCa remain unclear. Thus, it is of utmost importance to gain a better understanding of the underlying mechanisms of BCa.
Centromere protein U (CENPU) (9,10), also known as KLIP1, MLF1IP (11,12), CENP-50/PBIP1 and Plk1 (13,14) is required for maintenance of sister chromatid adhesion during recovery from spindle damage in chicken cells (10,15). In human cells, loss of CENPU can cause mitotic defects in chromosome attachment (16). As a phosphorylation substrate of Polo-like kinase 1 (Plk1), a member of the serine/threonine kinase family and regulator of a variety of functions during the cell cycle, Plk1 recruitment to interphase and mitosis kinetochore requires a phophorylation-dependent CENPU-Plk1 interaction (10,15). Moreover, CENPU plays a key role in kinetochore-microtubule attachment by interacting with Hec1 (9).
Accumulating evidence has demonstrated that CENPU is associated with tumorigenesis. One study showed that CENPU (MLF1IP) was increased in various glioblastoma cell lines and plays an important role in erythroleukemias (12). Moreover, CENPU overexpression showed a positive correlation with progression and prognosis in luminal breast cancer (17). Although CENPU significantly promotes prostate cancer cell proliferation, a difference in CENPU mRNA expression between CENPU upregulation and downregulation was not found (18) and to date the functional role of CENPU in human BCa has not yet been established.
In this study, we evaluated the role of CENPU to better determine its role in human bladder carcinogenesis.
Materials and methods
Human BCa specimens
In the present study, 100 patients with BCa who were diagnosed at the First Affiliated Hospital of Bengbu Medical College between January 2011 and December 2013 were enrolled. From each patient, a tissue biopsy was obtained via surgical excision. All samples were obtained after informed consent was provided and used with approval from the Review Board of the First Affiliated Hospital of Bengbu Medical College. At the time of diagnosis, the ages of the patients ranged from 18 to 95 years, the median age being 56 years. From all samples, 5-µm-thick formalin-fixed paraffin-embedded slides were prepared in accordance with the protocol of the Department of Pathology at the First Affiliated Hospital of Bengbu Medical College. From all patients, data were obtained including age at diagnosis, sex, tumor size, histological grade, TNM stage and vital status of patients relative to disease-specific survival at the 3-year follow-up appointment.
Immunohistochemistry
Immunohistochemical staining for CENPU was conducted on 5-µm-thick formalin-fixed paraffin-embedded tissues. First, the tissue was heated at 60°C, deparaffinized, and hydrated by sequential washes in xylene, graded alcohol followed by phosphate-buffered saline (PBS). The CENPU antigen was retrieved by incubation with 0.1 mol/l citrate buffer (pH 6.0) for 30 min at 95°C. To quench endogenous peroxidase activity, tissues were washed with PBS and incubated with 3% (v/v) H2O2 for 10 min at room temperature. Following three additional washes with PBS, non-specific binding sites were blocked by incubation with 10% (w/v) normal goat serum diluted in 1 part (w) bovine serum albumin (BSA) and 99 parts (v) PBS for 30 min at room temperature. Tissues were incubated overnight with 1 part anti-CENPU monoclonal antibody (Abcam, Cambridge, MA, USA) and 999 parts blocking solution at 4°C. Tissues were then washed three times with PBS, incubated in blocking solution for 10 min at room temperature followed by incubation with a biotinylated antibody and horseradish peroxidase-labeled streptavidin for 1 h at room temperature. The enzymatic reaction was developed with 3,3′-diaminobenzidine (DAB; Sangon Biotechnology, Shanghai, China) for 5 min at room temperature. Tissues were counterstained with hematoxylin (Sigma, St. Louis, MO, USA) for 30 sec, and washed with PBS for 5 min.
CENPU immunostaining was scored independently by two pathologists. Scoring of CENPU expression was as previously described (19) and based on two variables: i) the percentage of positively stained tumor cells (0 to <5% positively stained cells = 0, 5–25% positively stained cells = 1, or >50% positively stained cells =2) and ii) the staining intensity (absent or low staining =0, moderate staining =1, or high staining =2). Both scores were multiplied to obtain an overall score for each specimen and was dichotomized into low (scores of 0–1) or high (scores >2) classifications.
Cell lines
Human bladder cancer cell lines T24 and 5637 were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA) and cultured in Dulbecco's modified Eagles medium (DMEM) supplemented with 10% fetal calf serum (both from Gibco, Shanghai, China), 100 µg/ml streptomycin and 100 U/ml penicillin (both from Sangon Biotech, Shanghai, China) in 5% CO2 at 37°C. Media were replaced every other day and passaged upon reaching 70–80% confluency.
RNA isolation and quantitative real-time PCR
Total RNA was extracted using a TRIzol kit (Invitrogen, Shanghai, China) according to the manufacturer's instructions. RNA quality was assessed using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Shanghai, China). To synthesize cDNA from total RNA, reverse transcription was performed using the M-MLV-RTase kit (Promega, Shanghai, China). For quantitative real-time PCR (qPCR; Applied Biosystems, Foster City, CA, USA), SYBR-Green PCR Master Mix was used according to the manufacturer's instructions. For each sample, 2.0 µg of total RNA was used as a template for qPCR. The primer sequences are listed in Table I. Results were normalized to GAPDH (20) and data were analyzed using the Pfaffl's method (21).
Table I.
Primers used in this study.
| Gene | Accession no. | Primer sequences | Size (bp) |
|---|---|---|---|
| CENPU | NM_024629 | Forward: 5′-ATGAACTGCTTCGGTTAGAGC-3′ | 246 |
| Reverse: 5′-TATTTCGCAGATGGCTTTCGG-3′ | |||
| CXCL8 | NM_000584 | Forward: 5′-TGGCAGCCTTCCTGATTT-3′ | 236 |
| Reverse: 5′-AACCCTCTGCACCCAGTT-3′ | |||
| RAC1 | NM_018890 | Forward: 5′-GTAGCAGCTCAGCTCTTTGGA-3′ | 228 |
| Reverse: 5′-TACCCGTGACACTTTCATTCC-3′ | |||
| CCND3 | NM_001287427 | Forward: 5′-ACCTGGCTGCTGTGATTGC-3′ | 158 |
| Reverse: 5′-GATCATGGATGGCGGGTAC-3′ | |||
| IL1A | NM_000575 | Forward: 5′-AGATGCCTGAGATACCCAAAACC-3′ | 147 |
| Reverse: 5′-CCAAGCACACCCAGTAGTCT-3′ | |||
| IL6 | NM_000600 | Forward: 5′-CAAATTCGGTACATCCTCG-3′ | 259 |
| Reverse: 5′-CTCTGGCTTGTTCCTCACTA-3′ | |||
| IL1B | NM_000576 | Forward: 5′-TCCTGTTGTCTACACCAATGCCCA-3′ | 190 |
| Reverse: 5′-GAACCAAATGTGGCCGTGGTTTCT-3′ | |||
| TNFRSF11B | NM_002546 | Forward: 5′-CCTTGCCCTGACCACTAC-3′ | 207 |
| Reverse: 5′-TTGCACCACTCCAAATCC-3′ | |||
| PTGS2 | NM_000963 | Forward: 5′-CTCCTGTGCCTGATGATTGC-3′ | 215 |
| Reverse: 5′-CAGCCCGTTGGTGAAAGC-3′ | |||
| MAD2L1 | NM_002358 | Forward: 5′-GAGTCGGGACCACAGTTTAT-3′ | 97 |
| Reverse: 5′-TTTTGTAGGCCACCATGCTA-3′ | |||
| FN1 | NM_212482 | Forward: 5′-GAGAATAAGCTGTACCATCGCAA-3′ | 200 |
| Reverse: 5′-CGACCACATAGGAAGTCCCAG-3′ | |||
| GAPDH | Forward: 5′-TGACTTCAACAGCGACACCCA-3′ | 121 | |
| Reverse: 5′-CACCCTGTTGCTGTAGCCAAA-3′ |
Lentiviral transfection of T24 cells
Based on the gene expression profile in the human BCa cell lines T24 and 5637, the BCa T24 cell line was selected for the remaining experiments. When in the logarithmic phase, T24 cells were digested with trypsin (Sangon Biotech) and resuspended in DMEM. Cells were seeded in 6-well plates (5×104 cells/well) and incubated in 5% CO2 at 37°C until ~30% confluency was reached. Next, two experimental groups were constructed: i) an shCENPU group in which cells were transfected with CENPU-siRNA GFP lentivirus and ii) a control (shCtrl) group in which cells were transfected with a scramble sequence GFP lentivirus (Genechem, Shanghai, China). An appropriate amount of lentivirus was added according to the multiplicity of infection (MOI). When, after 12 h, no cytotoxic effects were observed, transfection was continued. After 24 h, the standard medium was replaced. GFP fluorescence was used to evaluate transfection efficiency 3 days post-transfection. When the transfection efficiency was >80%, the cells were divided into two groups: one group was transferred to a 12-well plate for future RNA extraction, the other group was transferred to a 6-well plate for future protein extraction.
Cell proliferation assays
ShCENPU and shCtrl cells were trypsinized and after a logarithmic proliferation phase was reached, they were resuspended in DMEM. Cells were plated in 96-wells at equal cell density (2,000 cells/100 µl/well) followed by incubation at 37°C with 5% CO2. After 24 h, a Cellomics ArrayScan VTI imager (Thermo Fisher Scientific, Waltham, MA, USA) was used to continuously measure GFP expression in each well over a 5-day time period. The data were used for performing statistical data mapping and construction of cell proliferation curves.
A BrdU assay for cell proliferation was conducted according to the manufacturer's instructions (CST, Danvers, MA, USA). Briefly, shCENPU and shCtrl cells were trypsinized and after a logarithmic proliferation phase was reached, they were resuspended in standard medium. The cells were plated in a 96-well plate at an equal density (2,500 cell/100 µl/well) and incubated at 37°C in 5% CO2 for continuous detection over a 3-day time period. Subsequently, the cells were collected, fixing/denaturing solution (CST) was applied and the cells were incubated for 30 min at room temperature. Detection antibody solution was added (100 µl/well) and the cells were incubated for 1 h at room temperature. After removal of the medium, the cells were washed 3 times with washing buffer. A HRP-conjugated secondary antibody (100 µl/well) was added and the cells were incubated for 1 h at room temperature. After washing with washing buffer, 100 µl TMB substrate was added to each well and incubated for 30 min at 25°C. One hundred microliters of STOP solution was added to each well to terminate the reaction. The plates were oscillated for 10 min at room temperature. Cell proliferation was measured using a microplate reader (Elx800; BioTek Instruments, Inc., Winooski, VT, USA) at a wavelength of 450 nm.
Cell cycle assay
shCENPU and shCtrl cells were grown to ~80% confluency, the supernatant was aspirated and after a single wash with washing buffer, the cells were trypsinized. The cells were washed with PBS and counted using a hemocytometer to ensure a sufficient number of cells (>1×106/well, 3 wells per experimental group). Cells were transferred to 5-ml centrifuge tubes and centrifuged at 1,500 rpm for 5 min at 4°C. The supernatant was discarded and the cell pellet was washed once with ice cold PBS (pH 7.2–7.4). Cells were centrifuged for 5 min at 1,500 rpm at 4°C and fixed in ice cold 70% ethanol for 1 h. Subsequently, the cells were centrifuged for 5 min at 1,500 rpm, washed once with ice-cold PBS, and then centrifuged for 5 min at 1,500 rpm at 4°C. Based on the amount of cells, 1–1.5 ml cell-staining solution [40X PI liquor (2 mg/ml, P4170; Sigma), 100X RNase mother liquor (10 mg/ml, EN0531; Fermentas, USA) and 1X PBS (Gibco, USA)] was used to resuspend the fixed cells. The cell suspension was filtered through a 300-mesh nylon mesh prior to flow cytometry on a FACSCalibur instrument (Becton-Dickinson, USA) at a flow rate of 200–350 cells/sec.
Apoptosis assay
ShCENPU and shCtrl cells were trypsinized and resuspended in standard medium after the logarithmic proliferation phase was reached. After the cells were washed, Annexin V-APC apoptosis detection kit and propidium iodide (PI, cat. no. 88-8007; eBioscience, San Diego, CA, USA) were applied to determine apoptotic cells according to the manufacturer's instructions. Briefly, the cells were resuspended in 1X binding buffer at a concentration of 1×106 cells/ml. Next, 100 µl of the cell suspension was added to each of the following tubes: i) an empty tube, ii) a tube containing Annexin V-FITC (5 µl); iii) a tube containing PI (10 µl) and (iv) a tube containing both Annexin V-FITC (5 µl) and PI (10 µl). After the tubes were gently mixed in the dark for 15 min at room temperature, 1X binding buffer (400 µl) was added to each tube and flow cytometry was conducted within 1 h.
Colony formation assay
ShCENPU and shcontrol cells were trypsinized and resuspended in standard medium after the logarithmic proliferation phase was reached. Cells were counted using a hemocytometer and seeded at a density of 800 cells/well into a 6-well plate (3 wells per experimental group). The cells were incubated at 37°C in 5% CO2 and maintained for 14 days. Every three days, half of the medium was replaced with fresh medium.
Images of cell colonies were captured under a fluorescence microscope (MicroPublisher 3.3RTV; Olympus, Japan). In brief, cells were washed with PBS and fixed with 4% paraformaldehyde (1 ml/well; Shanghai Sangon, China) for 30–60 min at room temperature. Next, the cells were washed with PBS and stained with 500 µl Giemsa (Sigma-Aldrich, Shanghai, China) for 20 min at room temperature. Then, cells were washed with ddH2O three times and left to dry. Under light microscopy, a digital camera was used to capture images of each slide and colony counts were obtained.
CENPU gene detection in human bladder tissue
Gene expression of CENPU was determined in both 10 BCa tissue samples and cancer-adjacent normal tissues. The Ethics Committee of the First Affiliated Hospital of Bengbu Medical College approved this clinically oriented experiment. Prior to sampling, written informed consent was obtained from all donors. The protocol for performing qPCR was as described above.
Microarray analysis
The genome-wide effects of CENPU knockdown were assessed by a GeneChip® PrimeView™ Human Gene Expression array (Affymetrix, Santa Clara, CA, USA), which represents 20,000 human transcripts. Three biological replicates of T24 cells transfected with the shCENPU lentivirus (for 72 h) and shCtrl lentivirus were included in the array experiment. RNA was isolated via the TRIzol method (Invitrogen) and RNA quality was determined by NanoDrop using a NanoDrop 2000 spectrophotometer and Agilent Bioanalyzer 2100. For gene expression profiling purposes, individual microarrays were used per sample. Briefly, 500 ng of total RNA was reverse transcribed and labeled with biotin using the GeneChip® 3 IVT labeling kit (Affymetrix) according to the manufacturer's instructions. The labeled amplified RNA was hybridized overnight to GeneChip® PrimeView™ Human Gene Expression array (Affymetrix) at 60°C. Arrays were performed with GeneChip® Hybridization Wash and Stain kit (Affymetrix) using GeneChip® Fluidics Station 450 (Affymetrix), according to the manufacturer's instructions. Data were normalized using the GeneSpring normalization algorithms according to the manufacturer's instructions. The normalized data were used to generate lists of genes that were differentially expressed by at least ±1.5-fold. Moreover, a differential score p-value <0.05 among the test samples was considered a differentially expressed gene.
Ingenuity Pathway Analysis
Datasets derived from microarray analysis and representing differentially expressed genes were imported into the Ingenuity Pathway Analysis (IPA) tool (Ingenuity® Systems, Redwood City, CA, USA; http://www.ingenuity.com). Differentially expressed genes were mapped in the Ingenuity database to the genetic networks available and then ranked by score according to the manufacturer's instructions.
The IPA tool allows for the identification of biological networks from a particular dataset, as well as its global functions and functional pathways. In this context, a network is a graphical representation of the relationships between molecules. Molecules are represented as nodes and the biological relationship between two nodes is represented as an edge (line). All edges are supported by at least 1 reference, a textbook, or from canonical information stored in the Ingenuity Pathways Knowledge Base. The intensity of the node color indicates the degree of up (red) or down (green) regulation. Nodes are displayed using various shapes that represent the functional class of the gene product. The IPA tool presents a significant value of genes; it shows interaction with and how gene products act on each other, directly or indirectly. This includes genes that are not part of the microarray analysis. The networks created are ranked by the number of significantly expressed genes and diseases that are most significant.
Western blotting
At the protein level, the expression of target genes was determined by immunostaining using specific antibodies. Forty-eight hours post lentiviral transfection, the cells were lysed using ice-cold RIPA lysis buffer (cat. no. P0013C; Beyotime, Beijing, China). Lysates were centrifuged at 12,000 × g for 10 min at 4°C, supernatants were collected and the protein concentration was determined using a BCA protein assay kit (cat no. P0010S; Beyotime). Per treatment, 20 µg of protein was separated by 10% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene difluoride (PVDF) membrane. Membranes were blocked in Tris-based buffered saline with 0.5% Tween-20 (TBST), containing 5% skimmed milk for 1 h at room temperature. Membranes were then incubated overnight at 4°C with the following antibodies: mouse monoclonal anti-Flag® M2 antibody (cat no. F1804, 1:1,000 dilution; Sigma), rabbit polyclonal anti-IL1B antibody (cat no. ab9722, 0.2 µg/ml), mouse monoclonal anti-CXCL8 antibody (cat no. ab18672, 0.1 µg/ml), rabbit polyclonal anti-RAC1 antibody (cat no. ab97568, 1:1,000 dilution), rabbit monoclonal anti-TNFRSF11B antibody (cat no. ab73400, 1 µg/ml), and rabbit monoclonal anti-IL1A antibody (cat no. ab9614, 0.2 µg/ml) (all from Abcam) and mouse monoclonal anti-GAPDH antibody (cat no. sc-32233, 1:2,000 dilution; Santa Cruz Biotechnology). After incubation with primary antibodies, membranes were incubated with secondary antibodies: HRP-conjugated goat anti-mouse (cat no. sc-2005, 1:5,000 dilution) or HRP-conjugated goat anti-rabbit IgG (cat no. sc-2004, 1:5,000 dilution) (both from Santa Cruz Biotechnology) for 1 h at 37°C. Signals were visualized using the ECL-Plus kit (cat no. M3121; Thermo Fisher Scientific) according to the manufacturer's instructions. GAPDH was used as the loading control.
Statistical analysis
Statistical analyses were performed using SPSS software version 20.0 (IBM, Armonk, NY, USA). Data are represented as mean ± SD. Categorical data between groups were compared by Chi-square test. Comparisons of data between groups were performed using Student's t-test. Kaplan-Meier analysis was used for survival analysis. A p-value of <0.05 was considered significant.
Results
CENPU expression and its association with clinicopathological characteristics of non-muscle invasive BCa tissues
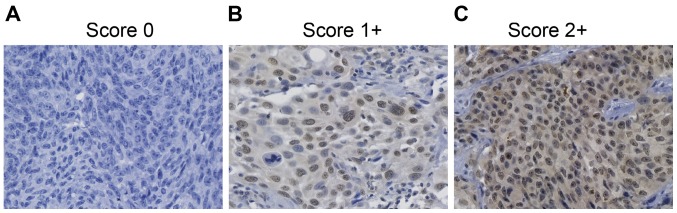
Using immunohistochemical analysis, CENPU expression was evaluated in 100 NMIBC tissues (Fig. 1). Sixty-seven patients showed high levels of CENPU expression, whereas 33 patients showed low expression of CENPU.
Figure 1.
CENPU expression in NMIBC tissues. Representative images are shown (original magnification, ×400). (A and B) Low expression; (C) high expression. CENPU, centromere protein U. NMIBC, non-muscle invasive bladder cancer.
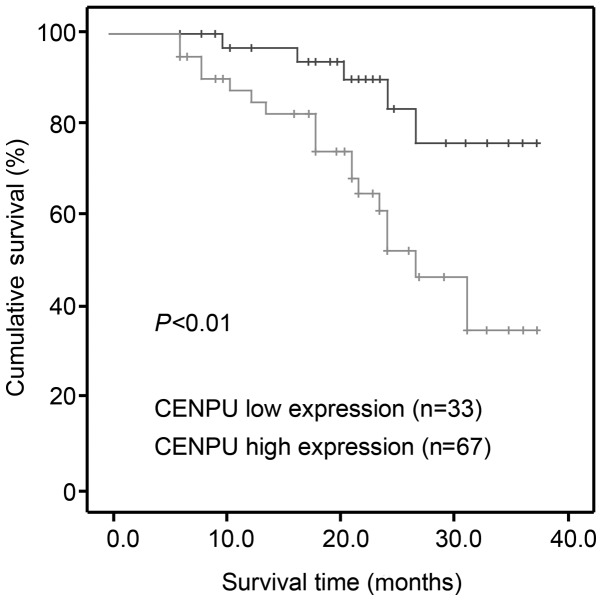
Table II shows the association between CENPU expression and clinicopathological characteristics. The results indicated that no significant correlation was observed between CENPU expression and sex (P=0.356), age (P=0.424), or histological grade (P=0.211). Instead, high CENPU expression was associated with larger tumor size (P=0.032) and advanced TNM stage (P=0.029). Kaplan-Meier survival analysis indicated that high CENPU expression was correlated with a worse outcome compared to low CENPU expression (P<0.001; Fig. 2).
Table II.
Association between CENPU expression and clinicopathological characteristics of the bladder cancer cases (n=100).
| CENPU expression | ||||
|---|---|---|---|---|
| Variables | Total no. n (%) | Low n (%) | High n (%) | P-value |
| Sex | ||||
| Male | 58 (58.0) | 17 (51.5) | 41 (61.2) | 0.356 |
| Female | 42 (42.0) | 16 (48.5) | 26 (38.8) | |
| Age (years) | ||||
| ≤60 | 34 (34.0) | 13 (39.4) | 21 (31.3) | 0.424 |
| >60 | 66 (66.0) | 20 (60.6) | 46 (68.7) | |
| Tumor size (cm) | ||||
| ≤3 | 29 (29.0) | 5 (15.2) | 24 (35.8) | 0.032 |
| >3 | 71 (71.0) | 28 (84.8) | 43 (64.2) | |
| Histological grade | ||||
| Urothelial | 61 (61.0) | 23 (69.7) | 38 (56.7) | 0.211 |
| Other | 39 (39.0) | 10 (30.3) | 29 (43.3) | |
| TNM stage | ||||
| 0/I | 18 (18.0) | 2 (93.9) | 16 (23.9) | 0.029 |
| II/III | 82 (82.0) | 31 (6.1) | 51 (76.1) | |
Figure 2.
Association between CENPU expression and clinicopathologic characteristics of the patients with NMIBC. Patients with high CENPU expression demonstrated significantly lower survival rate compared to those with low CENPU expression (P<0.01). CENPU, centromere protein U; NMIBC, non-muscle invasive bladder cancer.
CENPU gene expression in tissues and cell lines
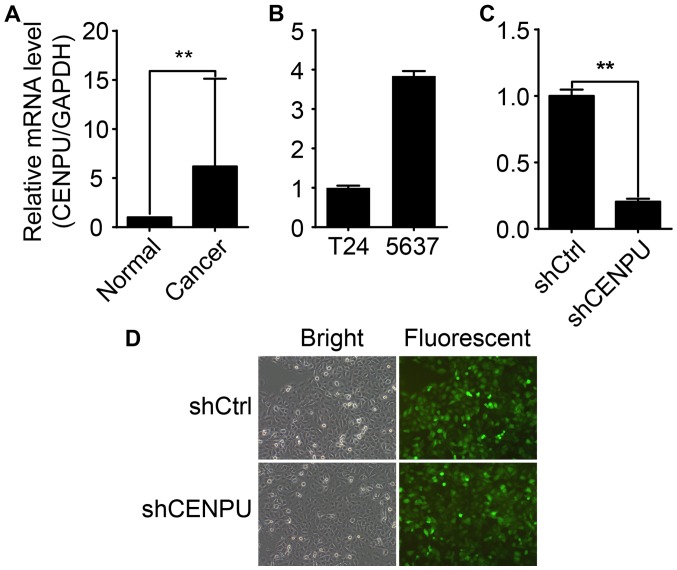
In this study, qPCR was performed to analyze the CENPU gene expression profile in 10 BCa tissue samples and cancer-adjacent normal tissue samples. Compared to CENPU mRNA levels in normal tissues, the expression of CENPU in BCa tissues was increased >6 fold (Fig. 3A). CENPU mRNA expression varied between the T24 and 5637 cell lines (Fig. 3B). T24 displayed the highest endogenous CENPU expression and was therefore selected for further investigation. Post-lentiviral transfection significantly inhibited CENPU mRNA expression in the T24 shCENPU cells compared to that noted in the control cells (Fig. 3D), indicating successful transfection and CENPU gene expression knockdown (Fig. 3C).
Figure 3.
CENPU mRNA expression in bladder cancer tissues and cell lines. (A) CENPU mRNA levels in bladder cancer tissues and adjacent normal tissues. (B) CENPU mRNA expression varied between the T24 and 5637 cell lines. GAPDH was used as an internal control. (C and D) Post-lentiviral transfection, relative CENPU mRNA expression was significantly inhibited in the T24 CENPU-siRNA knockdown cells as compared to the control cells. GAPDH was used as an internal control. **P<0.01. CENPU, centromere protein U.
CENPU knockdown inhibits T24 cell proliferation
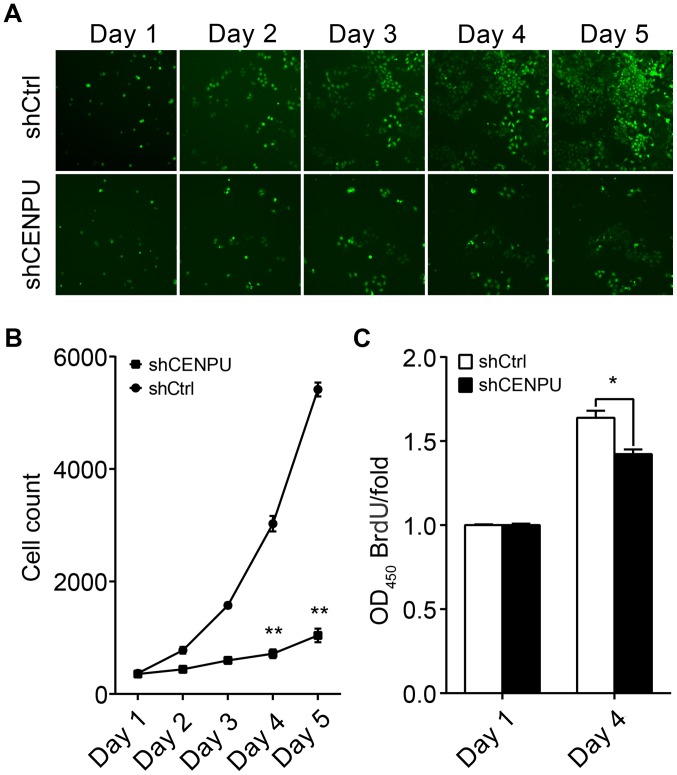
To evaluate the effect of CENPU expression on cell proliferation, T24 cells transfected with either shCENPU or shCtrl were plated into 96-well plates and analyzed for 5 days by Cellomics. As illustrated in Fig. 4A and confirmed by quantification in Fig. 2B, the number of cells in the shCtrl group significantly increased over the duration of 4 days; however the number of cells in the shCENPU group did not change (Fig. 4B). In addition, BrdU assay demonstrated that the BrdU ratio was significantly reduced in the shCENPU-transfected cells as compared to that noted in the shCtrl-transfected cells (Fig. 4C).
Figure 4.
Cell proliferation analysis by GFP-based imaging and BrdU assay. (A) Representative image of GFP-based Cellomics ArrayScan VTI imaging after transfection of T24 cells with shCENPU or shCtrl. (B) After lentiviral transfection of T24 cells, cell proliferation was significantly inhibited in the shCENPU cells when compared to that noted in the shCtrl cells. (C) The BrdU ratio was significantly reduced in the shCENPU cells when compared to the control cells. *P<0.05, **P<0.01. CENPU, centromere protein U.
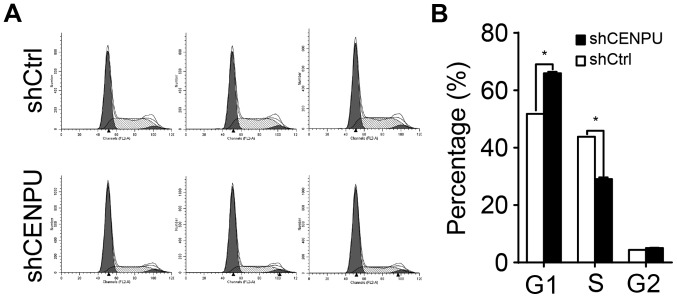
CENPU knockdown leads to cell cycle arrest
To determine whether CENPU is required for cell cycle progression, cell cycle distribution of the T24 cells was evaluated by flow cytometry (Fig. 5A). As shown in Fig. 5B, shCENPU-knockdown cells demonstrated a significant increase in the percentage of cells in the G1 phase when compared to the percentage in the control cells (P<0.05) and a decrease in the percentage of cells in the S phase (P<0.05). Taken together, these data suggest that CENPU is involved in cell proliferation regulation and blocks cell cycle progression in the G1 phase.
Figure 5.
CENPU knockdown leads to cell cycle arrest in the G1 phase. (A) The cell cycle of T24 cells was analyzed in triplicate by flow cytometry. (B) Cell cycle phases as determined by flow cytometry. Compared with the shCtrl group, shCENPU cells showed a significant increase in the proportion of cells in the G1 phase. *P<0.05. CENPU, centromere protein U.
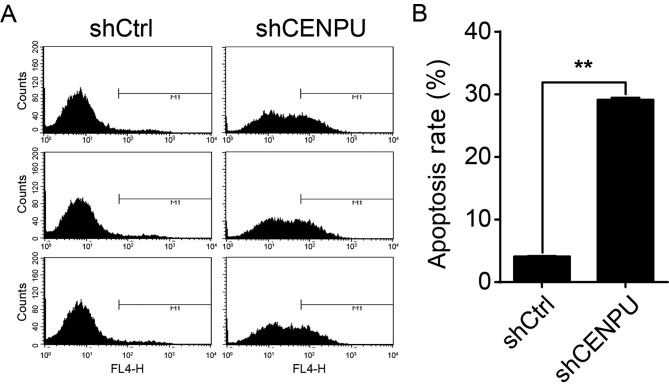
CENPU knockdown in T24 cells augments cell apoptosis
To test whether or not CENPU expression affects T24 cell apoptosis, CENPU was knocked down in T24 cells and the presence of apoptotic cells was determined by flow cytometry (Fig. 6A). As shown in Fig. 6B, apoptosis was significantly increased in shCENPU cells compared to that noted in the shCtrl group (P<0.01). These results suggest that, in T24 cells, CENPU expression is a regulator of apoptosis.
Figure 6.
CENPU knockdown augments apoptosis in the T24 cells. (A) Cell death was determined by Annexin V staining and flow cytometry. (B) Quantification of results shown in A. **P<0.01. CENPU, centromere protein U.
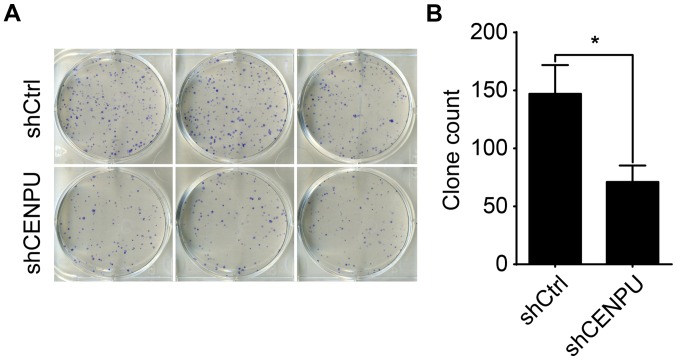
Knockdown of CENPU suppressed cell colony formation
Next, we studied the colony-formation capacity of T24 cells treated by the shCENPU lentivirus. Two groups of T24 cells (shCENPU and control cells) were grown for 14 days and allowed to form colonies. As shown in Fig. 7, CENPU knockdown resulted in a nearly 0.5-fold decrease in the number of colonies as compared with the shCtrl group (P<0.05).
Figure 7.
CENPU silencing suppresses bladder cancer cell colony formation. (A) Photomicrographs of Giemsa-stained colonies of T24 cells 14 days after transfection. (B) The number of cells present in each T24 cell colony was counted. The cell number in the shCENPU group was significantly reduced compared to the shCtrl group. *P<0.05. CENPU, centromere protein U.
Genome-wide effects of CENPU knockdown in T24 cells
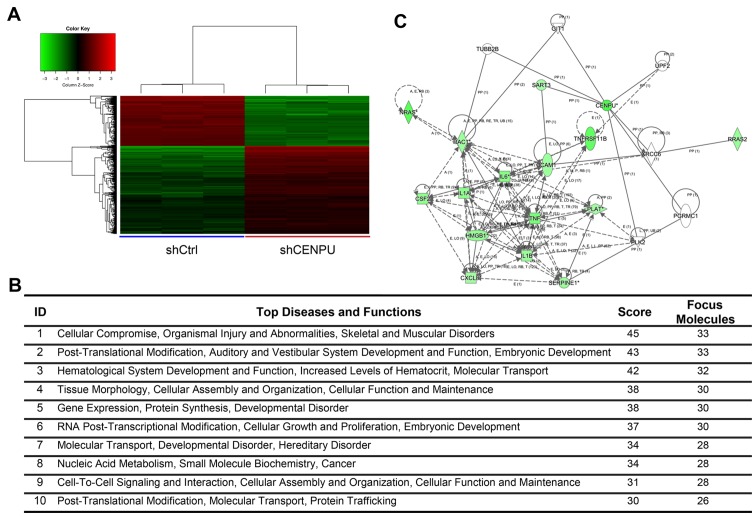
The gene expression profiles of T24 cells knocked down for CENPU or shCtrl cells were determined by GeneChip® PrimeView™ Human Gene Expression array. Three biological replicates were used and raw data were analyzed using GeneSpring v11 software. Data were average normalized and filtered by detection of a p-value <0.05 and differential p-value <0.05. The gene expression array identified 1,274 differentially expressed genes (809 genes were downregulated and 465 upregulated) (Fig. 8A).
Figure 8.
Ingenuity Pathway Analysis (IPA) summary. (A) Clustering of differentially expressed gene sets. (B) List of top 10 networks with their respective scores obtained from IPA. (C) Most highly rated network in IPA analysis influenced by CENPU knockdown. CENPU, centromere protein U.
Elucidation of pathways and interactions among differentially expressed genes
To determine possible biological interactions of the differently regulated genes, datasets representing genes with an altered expression profile derived from microarray analyses were imported in the IPA tool.
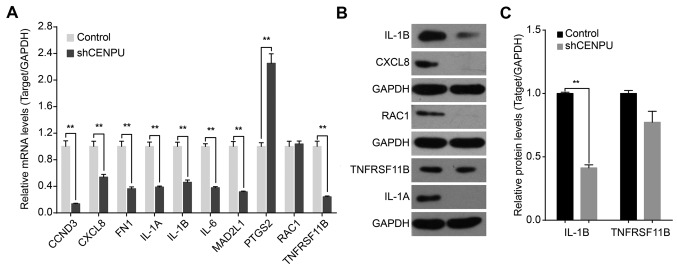
IPA analysis identified 25 significant networks (data not shown). Fig. 8B represents the list of the top 10 networks identified by IPA. Of these networks, ‘Cellular compromise, organismal injury and abnormalities, skeletal and muscular disorders’ was the highest rated network with 25 focus molecules and had a significance score of 45 and implicated 33 genes (Fig. 8B). Additional IPA analysis showed that CENPU knockdown was involved in the regulation of numerous genes that are involved in the HMGB1 signaling pathway. These include CXCL8, IL1A, NRAS, ICAM1, RAC1, IL6, HMGB1, RRAS2, IL1B, CSF2, SERPINE1, TNF, TNFRSF11B, and PLAT. The Ingenuity Pathway of CENPU and the HMGB1 signaling pathway is shown in Fig. 8C. Results of qPCR analysis demonstrated that CENPU knockdown significantly downregulated the expression of CCND3, CXCL8, FN1, IL1A, IL1B, IL6, MAD2L1, RAC1, and TNFRSF11B. In addition, CENPU knockdown upregulated PTGS2 expression (Fig. 9A). Western blot analysis further confirmed that CENPU knockdown affected the downregulation of IL1B, CXCL8, RAC1 and IL1A expression (Fig. 9B and C).
Figure 9.
CENPU knockdown affects the expression levels of downstream genes. (A) Quantitative PCR confirmation of mRNA expression of CCND3, CXCL8, FN1, IL-1A, IL-1B, IL-6, MAD2L1, PTGS2, RAC1 and TNFRSF11B in T24 cells transfected with either a control or shCENPU lentivirus. Data represent mean ± SD of relative mRNA quantity normalized to GAPDH mRNA expression. (B) Western blot analysis of protein expression of IL-1B, CXCL8, RAC1, TNFRSF11B and IL-1A. (C) Densitometric analysis of target proteins in T24 cells transfected with control or shCENPU lentivirus. **P<0.01. CENPU, centromere protein U.
Discussion
To the best of our knowledge, our study is the first to demonstrate the expression, clinical value, and the potential functional mechanism of action of CENPU in BCa. As expected, our qPCR data demonstrated that CENPU was abundantly expressed in cancer-related tissues, whereas in cancer-adjacent normal tissues CENPU expression was weak. Immunohistochemical studies showed that, in BCa, high CENPU expression was significantly correlated with unfavorable clinicopathological characteristics such as tumor size and TNM stage. After CENPU expression in normal and BCa tissue was confirmed, we used BCa cell lines for further experiments to better establish the mechanistic role of CENPU in human bladder carcinogenesis.
Our in vitro studies indicated a variable CENPU expression in two BCa cell lines that represent different molecular features. The 5637 cell line is an in vitro model for high-risk NMIBC (22,23), whereas the T24 cell line is an invasive BC cell line (24). The finding that the T24 cell line which has a higher invasive potential displayed higher CENPU expression as determined by qPCR is consistent with the immunohistochemical analysis. These results confirmed CENPU expression according to the invasive potential of the T24 cells. We also found that T24 cell proliferation and cell colony formation were significantly reduced in the shCENPU-transfected cells. Apoptosis was significantly increased in the CENPU-silenced BCa cells. CENPU-silenced T24 cells showed significant cell cycle arrest at the G1 phase. To date, only one study has demonstrated that CENPU knockdown in prostate cancer cell line PC-3 inhibited cell proliferation, colony formation, increased apoptosis. However, in the study, the cell cycle did not appear to be affected (18).
The CENPU gene (also known as MLF1IP, Cenp-50/PBIP1 and KLIP1) encodes a 46-kDa nuclear-localizing transcription suppressor protein that has been associated with malignancy in previous research (25). In addition to its transcription suppressor activity, CENPU is required for stable kinetochore-microtubule attachment, proper chromosome segregation and recovery from spindle damage during mitosis (9,10,14). CENPU was initially identified in 2004 by Hanissian et al, who implied a possible role for CENPU deregulation in the pathogenesis of erythroleukemias (11). Subsequently, the same group found that CENPU upregulation was associated with enhanced neuropoiesis and glioblastoma tumor development in both humans and rodents (12). More recently, CENPU upregulation has been identified in human breast cancer (17) and familial colorectal cancer (Lynch syndrome) patients (26). However, no obvious alterations of CENPU expression were observed in human prostate cancer tissue (18).
Little is known concerning the role of CENPU in human BCa. Therefore, we evaluated the gene profiling in shCENPU-transfected T24 cells to identify the mechanism of action of CENPU knockdown. A total of 1,274 differentially expressed genes were identified, including 809 downregulated genes and 465 upregulated genes. In this study, IPA was used to visualize the co-deregulated genes affected by CENPU knockdown. Network analysis identified 25 distinct signaling pathways, including the top-ranked network ‘Cellular Compromise, organismal injury and abnormalities, skeletal and muscular disorders’ (Fig. 8B). In-depth IPA analysis revealed that CENPU plays a role in HMGB1 signaling (Fig. 8C). Overexpression of HMGB1 is associated with progression and poor prognosis in human BCa (27,28), however downregulation of HMGB1 is known to inhibit the bioactivity of BCa cell lines (29). The qPCR results obtained in this study demonstrated that CENPU knockdown downregulated the expression of members involved in HMGB1 signaling, including CCND3, CXCL8, FN1, IL1A, IL1B, IL6, MAD2L1, RAC1, and TNFRSF11B, and upregulated PTGS2 expression. Western blot analysis of key players in the HMGB1 pathway, including IL1B, CXCL8, RAC1, and IL1A, demonstrated that all of these molecules were significantly downregulated. Taken together, knockdown of CENPU in T24 cells deregulated cell proliferation, colony formation, cell cycle arrest and apoptosis via the HMGB1 signaling pathway and therefore, high expression of CENPU causes an increased risk for BCa tumorigenesis.
In conclusion, in T24 cells, CENPU knockdown significantly inhibited cell proliferation, colony formation and cell cycle arrest at the G1 stage and significantly promoted apoptosis. The mechanism underlying the effect of CENPU knockdown in BCa cells may involve the HMGB1 signaling pathway.
Acknowledgements
This study was supported by Key Projects of Science Research for Colleges and Universities in Anhui Province (KJ2015A280).
References
- 1.Gandaglia G, Popa I, Abdollah F, Schiffmann J, Shariat SF, Briganti A, Montorsi F, Trinh QD, Karakiewicz PI, Sun M. The effect of neoadjuvant chemotherapy on perioperative outcomes in patients who have bladder cancer treated with radical cystectomy: A population-based study. Eur Urol. 2014;66:561–568. doi: 10.1016/j.eururo.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 2.Yu HJ, Chang YH, Pan CC. Prognostic significance of heat shock proteins in urothelial carcinoma of the urinary bladder. Histopathology. 2013;62:788–798. doi: 10.1111/his.12087. [DOI] [PubMed] [Google Scholar]
- 3.Sun L, Lu J, Niu Z, Ding K, Bi D, Liu S, Li J, Wu F, Zhang H, Zhao Z, et al. A potent chemotherapeutic strategy with Eg5 inhibitor against gemcitabine resistant bladder cancer. PLoS One. 2015;10:e0144484. doi: 10.1371/journal.pone.0144484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kang HW, Yoon HY, Ha YS, Kim WT, Kim YJ, Yun SJ, Lee SC, Kim WJ. FAM70B as a novel prognostic marker for cancer progression and cancer-specific death in muscle-invasive bladder cancer. Korean J Urol. 2012;53:598–606. doi: 10.4111/kju.2012.53.9.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allory Y, Beukers W, Sagrera A, Flández M, Marqués M, Márquez M, van der Keur KA, Dyrskjot L, Lurkin I, Vermeij M, et al. Telomerase reverse transcriptase promoter mutations in bladder cancer: High frequency across stages, detection in urine, and lack of association with outcome. Eur Urol. 2014;65:360–366. doi: 10.1016/j.eururo.2013.08.052. [DOI] [PubMed] [Google Scholar]
- 6.Rosenberg JE. Current status of neoadjuvant and adjuvant chemotherapy for muscle-invasive bladder cancer. Expert Rev Anticancer Ther. 2007;7:1729–1736. doi: 10.1586/14737140.7.12.1729. [DOI] [PubMed] [Google Scholar]
- 7.Hodge LS, Taub ME, Tracy TS. Effect of its deaminated metabolite, 2,2-difluorodeoxyuridine, on the transport and toxicity of gemcitabine in HeLa cells. Biochem Pharmacol. 2011;81:950–956. doi: 10.1016/j.bcp.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 8.Leijen S, Veltkamp SA, Huitema AD, van Werkhoven E, Beijnen JH, Schellens JH. Phase I dose-escalation study and population pharmacokinetic analysis of fixed dose rate gemcitabine plus carboplatin as second-line therapy in patients with ovarian cancer. Gynecol Oncol. 2013;130:511–517. doi: 10.1016/j.ygyno.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 9.Hua S, Wang Z, Jiang K, Huang Y, Ward T, Zhao L, Dou Z, Yao X. CENP-U cooperates with Hec1 to orchestrate kinetochore-microtubule attachment. J Biol Chem. 2011;286:1627–1638. doi: 10.1074/jbc.M110.174946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Minoshima Y, Hori T, Okada M, Kimura H, Haraguchi T, Hiraoka Y, Bao YC, Kawashima T, Kitamura T, Fukagawa T. The constitutive centromere component CENP-50 is required for recovery from spindle damage. Mol Cell Biol. 2005;25:10315–10328. doi: 10.1128/MCB.25.23.10315-10328.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanissian SH, Akbar U, Teng B, Janjetovic Z, Hoffmann A, Hitzler JK, Iscove N, Hamre K, Du X, Tong Y, et al. cDNA cloning and characterization of a novel gene encoding the MLF1-interacting protein MLF1IP. Oncogene. 2004;23:3700–3707. doi: 10.1038/sj.onc.1207448. [DOI] [PubMed] [Google Scholar]
- 12.Hanissian SH, Teng B, Akbar U, Janjetovic Z, Zhou Q, Duntsch C, Robertson JH. Regulation of myeloid leukemia factor-1 interacting protein (MLF1IP) expression in glioblastoma. Brain Res. 2005;1047:56–64. doi: 10.1016/j.brainres.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 13.Dai W, Wang X. Grabbing Plk1 by the PBD. Mol Cell. 2006;24:489–490. doi: 10.1016/j.molcel.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 14.Lee KS, Oh DY, Kang YH, Park JE. Self-regulated mechanism of Plk1 localization to kinetochores: Lessons from the Plk1-PBIP1 interaction. Cell Div. 2008;3:4. doi: 10.1186/1747-1028-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hori T, Okada M, Maenaka K, Fukagawa T. CENP-O class proteins form a stable complex and are required for proper kinetochore function. Mol Biol Cell. 2008;19:843–854. doi: 10.1091/mbc.E07-06-0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR, III, Cleveland DW. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 17.Huang DP, Luo RC. MLF1IP is correlated with progression and prognosis in luminal breast cancer. Biochem Biophys Res Commun. 2016;477:923–926. doi: 10.1016/j.bbrc.2016.06.159. [DOI] [PubMed] [Google Scholar]
- 18.Zhang L, Ji G, Shao Y, Qiao S, Jing Y, Qin R, Sun H, Shao C. MLF1 interacting protein: A potential gene therapy target for human prostate cancer? Med Oncol. 2015;32:454. doi: 10.1007/s12032-014-0454-1. [DOI] [PubMed] [Google Scholar]
- 19.Radhika K, Prayaga AK. Estrogen and progesterone hormone receptor status in breast carcinoma: Comparison of immunocytochemistry and immunohistochemistry. Indian J Cancer. 2010;47:148–150. doi: 10.4103/0019-509X.63006. [DOI] [PubMed] [Google Scholar]
- 20.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vasconcelos-Nóbrega C, Pinto-Leite R, Arantes-Rodrigues R, Ferreira R, Brochado P, Cardoso ML, Palmeira C, Salvador A, Guedes-Teixeira CI, Colaço A, et al. In vivo and in vitro effects of RAD001 on bladder cancer. Urol Oncol. 2013;31:1212–1221. doi: 10.1016/j.urolonc.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 23.Gazzaniga P, Silvestri I, Gradilone A, Scarpa S, Morrone S, Gandini O, Gianni W, Frati L, Aglianò AM. Gemcitabine-induced apoptosis in 5637 cell line: An in-vitro model for high-risk superficial bladder cancer. Anticancer Drugs. 2007;18:179–185. doi: 10.1097/CAD.0b013e328010ef47. [DOI] [PubMed] [Google Scholar]
- 24.Lee YG, Macoska JA, Korenchuk S, Pienta KJ. MIM, a potential metastasis suppressor gene in bladder cancer. Neoplasia. 2002;4:291–294. doi: 10.1038/sj.neo.7900231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki H, Arakawa Y, Ito M, Saito S, Takeda N, Yamada H, Horiguchi-Yamada J. MLF1-interacting protein is mainly localized in nucleolus through N-terminal bipartite nuclear localization signal. Anticancer Res. 2007;27:1423–1430. [PubMed] [Google Scholar]
- 26.Dominguez-Valentin M, Therkildsen C, Veerla S, Jönsson M, Bernstein I, Borg A, Nilbert M. Distinct gene expression signatures in lynch syndrome and familial colorectal cancer type x. PLoS One. 2013;8:e71755. doi: 10.1371/journal.pone.0071755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang W, Jiang H, Zhu H, Zhang H, Gong J, Zhang L, Ding Q. Overexpression of high mobility group box 1 and 2 is associated with the progression and angiogenesis of human bladder carcinoma. Oncol Lett. 2013;5:884–888. doi: 10.3892/ol.2012.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang GL, Zhang LH, Bo JJ, Huo XJ, Chen HG, Cao M, Liu DM, Huang YR. Increased expression of HMGB1 is associated with poor prognosis in human bladder cancer. J Surg Oncol. 2012;106:57–61. doi: 10.1002/jso.23040. [DOI] [PubMed] [Google Scholar]
- 29.Huang Z, Zhong Z, Zhang L, Wang X, Xu R, Zhu L, Wang Z, Hu S, Zhao X. Down-regulation of HMGB1 expression by shRNA constructs inhibits the bioactivity of urothelial carcinoma cell lines via the NF-κB pathway. Sci Rep. 2015;5:12807. doi: 10.1038/srep12807. [DOI] [PMC free article] [PubMed] [Google Scholar]