Abstract
Low-grade gliomas (LGGs) are associated with neurological disability. The present study used microRNA (miRNA) expression profiles to identify risk miRNAs for potential prognosis of cerebral LGGs. miRNA expression profiles and clinical data from 408 patients with cerebral LGGs were obtained from the Cancer Genome Atlas database. Risk miRNAs were identified by plotting Kaplan-Meier curves and Cox proportional hazard regression analysis with the survival and KMsurv packages in R. A regulatory network of miRNA-targets was constructed, followed by gene ontology (GO) function and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis using the Database for Annotation, Visualization and Integrated Discovery. A protein-protein interaction (PPI) network of miRNA targets was built using Search Tool for the Retrieval of Interacting Genes software, and sub-pathway identification was performed using the iSubpathwayMiner package in R. In total, 39 miRNAs had significant effect on survival curves. Following the Cox analysis and construction of miRNA-targets regulatory network, hsa-miRNA (miR)-326 was identified to regulate 397 target genes. Additionally, targets of miR-326 were primarily enriched in the GO terms of cell proliferation, epithelial growth factor receptor and nerve growth factor signaling pathways. Additionally, son of sevenless homolog 1 (SOS1), neuroblastoma RAS viral oncogene homolog (NRAS), vitamin D receptor (VDR) and mothers against decapentaplegic family member 3 (SMAD3) were most enriched in the PPI network. Targets of miR-326 were primarily enriched in sub-pathways including sphingolipid metabolism and arachidonic acid metabolism, in which sphingomyelin synthase 1 (SGMS1) and hematopoietic prostaglandin D synthase (HPGDS) were screened out. Hsa-miR-326 was identified as a risk miRNA for prognosis and may improve the outcome prediction of patients with cerebral LGG. This miRNA may regulate cancer cell proliferation by targeting SOS1, NRAS, VDR, SMAD3, SGMS1 and HPGDS.
Keywords: cerebral low-grade gliomas, microRNA, regulatory network, sub-pathway
Introduction
Cerebral low-grade gliomas (LGGs) are associated with neurological disability and present a challenge to neurosurgeons and neuro-oncologists (1). Although they are a relatively slow-growing brain tumor, LGGs have complex clinical manifestations (1). LGG frequently occur in Caucasians, particularly males and typically affect young patients. Therefore, scientific and clinical advances are required. However, predictive markers for diagnosis and prognosis of cerebral LGG are rare and mortality is high.
Clinical outcomes may be improved by identification of potential molecular biomarkers of LGG. Candidate therapeutic biomarkers have been identified by high-throughput technologies. The identification of microRNAs (miRNAs) has revealed novel insights into diagnosis and prognosis of cancers (2,3). miRNAs are a class of small, non-coding RNAs which have roles in cell apoptosis, differentiation, proliferation and stress responses (4,5). Previous studies have determined that alteration of miRNA expression level is associated with development and prognosis of human cancers, including pancreatic, breast, non-small cell lung and ovarian cancer (6–9). For example, miRNA-26a is overexpressed in high-grade glioma and directly targets phosphatase and tensin homolog which suppresses protein kinase B (Akt) signaling (10). miRNA (miR)-221 has been identified to be upregulated in glioblastoma and directly targets the tumor suppressor p27 (11). Conversely, downregulated miR-7 has been identified to reduce proliferation and invasiveness in cultured glioma cells by targeting the epidermal growth factor receptor (12). Additionally, miR-205 has been identified as a potential prognostic indicator for human glioma (13). A previous study has demonstrated that miR-221/222 may be a predictive marker for increased cell invasion and poor prognosis in glioma (14). However, miRNA biomarkers for tumorigenesis and prognosis of LGG have not been determined and candidate therapeutic targets remain to be identified.
The present study aimed to gain further insight into the clinical outcome of LGG by miRNA profile analysis. miRNA expression and clinical data of LGGs from the Cancer Genome Atlas (TCGA) database was downloaded, followed by identification of risk miRNAs using survival and Cox proportional hazard regression analysis. Functional annotation and protein-protein interaction (PPI) network construction of targets of miRNAs were performed. Additionally, sub-pathways were mined for further investigation of the function of risk miRNAs.
Materials and methods
Data collection
Clinical data and miRNA expression profiles were obtained from the TCGA database (http://cancergenome.nih.gov/) based on the platform of BCGSC_IlluminaHiSeq_miRNASeq. On the 11 August, 2014, there were 529 miRNA expression profiles and 411 clinical data from patients with cerebral LGGs, 408 of which contained associated miRNA expression profiles.
Survival analysis
The reads per kilobase of exon model per million mapped (RPKM) value which estimated the expression value for each gene was calculated to detect present miRNAs. In order to analyze the association between a queried miRNA and survival, the patients were grouped according to the median expression of the selected miRNA (or upper or lower quartile). In order to identify the genomic factors associated with survival, patient survival Kaplan-Meier (KM) curves were plotted using the survival (15) and KMsurv (16) packages in R and differences between curves were evaluated by two-sided log-rank test.
Cox proportional hazard regression analysis
To identify prognosis-associated miRNAs, the joint effect of variables with a significant P-value were examined using the Cox proportional hazard regression model which was built with the aforementioned two packages. P<0.05 was considered to indicate a statistically significant difference.
Regulatory network construction of miRNA-targets
In order to predict target genes of risk miRNA, which were selected from the miRNecords (17) and MiRWalk (18) databases, or recorded in at least 3 databases of the following databases: miRanda (19), MirTarget2 (20), PicTar (21), PITA (22) and TargetScan (23). A regulatory network of miRNA targets was constructed using the combined databases and visualized using Cytoscape software version 2.8 (24).
Functional annotation of miRNA target genes
In order to annotate functions of miRNA targets, GO (Gene Ontology) (25) function in biological process, cellular components, molecular function and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway (26) enrichment analysis were performed using Database for Annotation, Visualization, and Integrated Discovery (DAVID) (27). P<0.05 was considered to indicate a statistically significant difference.
PPI network construction for miRNA targets
In order to determine the interaction of miRNA targets, a PPI network was built using STRING software version 9.1 (28) and visualized with Cytoscape for protein-protein pairs, where the combined score was >0.4.
Sub-pathway analysis for miRNA targets
In order to identify the risk disease-associated sub-pathway of miRNA targets, the present study used a k-clique concept from the SubpathwayMiner package in R (29). P was calculated using a hypergeometric distribution and P<0.05 was considered to indicate a statistically significant difference.
Results
Results of survival analysis
The median miRNA intensity value of the 408 patients was used as the cut-off point in the KM curve analysis. A total of 39 miRNAs were obtained, which significantly affected survival in the KM curve (data not shown). From the survival analysis, patients with high expression of these 39 miRNAs had reduced survival compared with patients with low expression.
Risk miRNA for cerebral LGG prognosis
The cerebral LGGs risks were estimated as a hazard ratio (HR) and 95% confidence intervals (CI) using the Cox proportional hazard regression model. In the Cox proportional hazard regression analysis, 3 miRNAs including has-miR-1287, has-miR-326 and has-miR-1275 were considered as risk miRNAs for cerebral LGGs prognosis (Table I).
Table I.
Identification of risk microRNAs in patients with cerebral low-grade glioma using Cox proportional hazard regression analysis.
| Name | β | HR | P | Lower CI | Upper CI |
|---|---|---|---|---|---|
| hsa-miR-1287 | 0.016169188 | 1.016300616 | 1.69446×10−7 | 1.010161075 | 1.022477472 |
| hsa-miR-326 | −0.008757427 | 0.991280808 | 0.005923248 | 0.985117424 | 0.997482752 |
| hsa-miR-1275 | −0.193926634 | 0.823718335 | 0.035076194 | 0.687784787 | 0.986517742 |
β, coefficient in Cox regression model; HR, hazard ratio; P adjusted by likelihood ratio test; CI, 95% confidence intervals; miR, miRNA.
Regulatory network of risk miRNA-targets
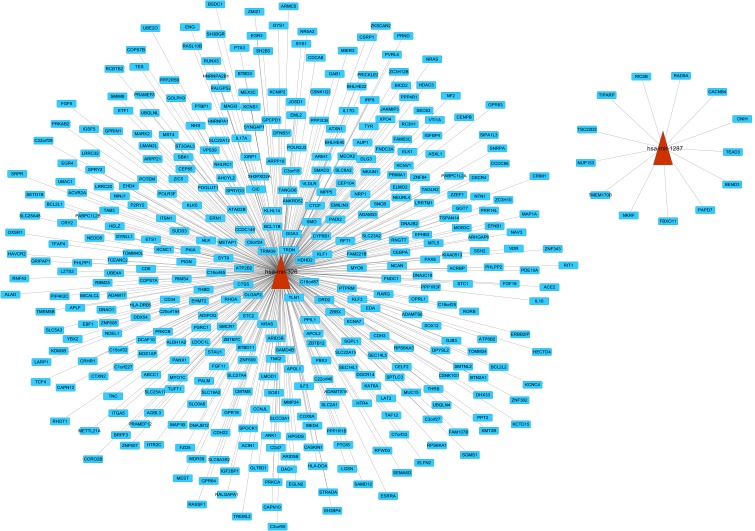
In order to investigate regulatory function of risk miRNAs, a regulatory network for miRNA-target genes was constructed (Fig. 1). However, no predicted target genes were identified for has-miR-1275. The present study predicted 13 and 397 targets regulated by hsa-miR-1287 and hsa-miR-326, respectively. In the network, there were 410 links and 412 nodes. In the network, son of sevenless homolog 1 (SOS1), neuroblastoma RAS viral (v-ras) oncogene homolog (NRAS), and vitamin D (1,25-dihydroxyvitamin D3) receptor (VDR) were targets of miR-326.
Figure 1.
Regulatory network of miRNA-targets. Red triangles represent miRNA and blue rectangles represent targets of miRNA. miRNA, microRNA.
Functional enrichment analysis of miRNA targets
In order to determine the regulatory functions of has-miR-326, GO function and pathway enrichment analysis were performed for the target genes. The top 5 GO terms and pathways are presented in Table II. The findings revealed that targets of miR-326 were significantly enriched in various functions, including neuron development, neuron differentiation and regulation of cell proliferation. Additionally, targets of miR-326 were significantly enriched in cancer pathways, and the epithelial growth factor receptor (EGFR) and nerve growth factor (NGF) signaling pathways.
Table II.
Functional annotation of hsa-miR-326 targets.
| A, Biological processes | |||
|---|---|---|---|
| Term | Function | Count | P-value |
| GO:0048666 | Neuron development | 20 | 1.10×10-4 |
| GO:0030182 | Neuron differentiation | 22 | 4.20×10−4 |
| GO:0042127 | Regulation of cell proliferation | 32 | 6.13×10-4 |
| GO:0045197 | Establishment or maintenance of epithelial cell apical/basal polarity | 4 | 7.15×10−4 |
| GO:0044057 | Regulation of system process | 17 | 9.04×10-4 |
| B, Cellular components | |||
| Term | Function | Count | P-value |
| GO:0044459 | Plasma membrane part | 72 | 3.26×10−5 |
| GO:0005886 | Plasma membrane | 107 | 7.61×10-5 |
| GO:0005911 | Cell-cell junction | 12 | 1.82×10−3 |
| GO:0031965 | Nuclear membrane | 7 | 3.72×10-3 |
| GO:0030054 | Cell junction | 21 | 4.78×10−3 |
| C, Molecular function | |||
| Term | Function | Count | P-value |
| GO:0019904 | Protein domain specific binding | 18 | 1.00×10-3 |
| GO:0003707 | Steroid hormone receptor activity | 6 | 4.20×10−3 |
| GO:0004879 | Ligand-dependent nuclear receptor activity | 6 | 8.61×10-3 |
| GO:0016247 | Channel regulator activity | 6 | 9.24×10−3 |
| GO:0005249 | Voltage-gated potassium channel activity | 7 | 2.24×10-2 |
| D, KEGG pathways | |||
| Term | Function | Count | P-value |
| hsa05200 | Pathways in cancer | 19 | 5.37×10−4 |
| hsa04360 | Axon guidance | 10 | 3.02×10-3 |
| hsa04720 | Long-term potentiation | 7 | 4.86×10−3 |
| hsa05211 | Renal cell carcinoma | 7 | 5.60×10-3 |
| hsa05223 | Non-small cell lung cancer | 6 | 8.18×10−3 |
| E, REACTOME pathways | |||
| Term | Function | Count | P-value |
| REACT_9417 | Signaling by epidermal growth factor receptor | 6 | 4.27×10-3 |
| REACT_11061 | Signaling by nerve growth factor | 11 | 6.89×10−3 |
GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
PPI interaction network
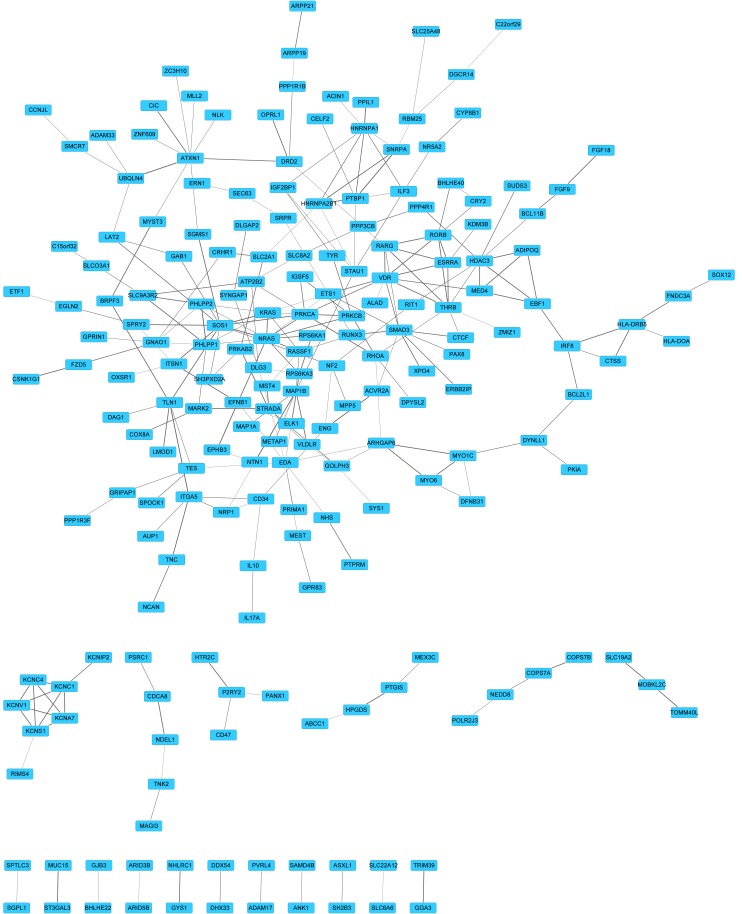
A PPI network was constructed to predict novel interactions for targets of has-miR-326. It was determined that 203 proteins shared 262 links (Fig. 2). According to the degree of targets, the top 10 proteins were with highest degree (Table III) were SOS1, NRAS, VDR and mothers against decapentaplegic family member 3 (SMAD3). Along with the combined score, the top five protein pairs were polypyrimidine tract binding protein 1-small nuclear ribonucleoprotein polypeptide A (PTBP1-SNRPA; 0.999), PH domain and leucine rich repeat protein phosphatase 1–2 (PHLPP1-PHLPP2; 0.992), microtubule-associated protein 1B-netrin 1 (MAP1B-NTN1; 0.985), ubiquilin 4-ataxin 1 (UBQLN4-ATXN1; 0.983) and talin 1-integrin, α 5 (TLN1-ITGA5; 0.982).
Figure 2.
Protein-protein interaction network of hsa-miR-326 targets. Blue rectangles represent targets of miR-326 and lines represent the interaction between miR targets. The extent of thickness of edges is in proportion to the combined score of proteins. miR, microRNA.
Table III.
Degree of the top 10 targets of miR-326 in the protein-protein interaction network.
| Gene | Degree |
|---|---|
| SOS1 | 11 |
| NRAS | 10 |
| VDR | 9 |
| SMAD3 | 9 |
| ATXN1 | 9 |
| THRB | 8 |
| PRKCA | 8 |
| KRAS | 8 |
| HDAC3 | 8 |
| EDA | 8 |
Identification of risk sub-pathway
In order to determine the association between the enriched pathways of miR-326 targets and cerebral LGGs, risk sub-pathways were identified for targets of miR-326 (Table IV). A total of 4 sub-pathways were obtained, involving 8 targets of miR-326. Sphingomyelin synthase 1 (SGMS1), serine palmitoyltransferase, long chain base subunit 3 (SPTLC3) and sphingosine-1-phosphate lyase 1 (SGPL1) were significantly enriched in the sphingolipid metabolism pathway. Hematopoietic prostaglandin D synthase (HPGDS) and prostaglandin I2 (prostacyclin) synthase (PTGIS) were involved in the arachidonic acid metabolism. adenosylhomocysteinase-like 2 (AHCYL2) and gamma-glutamyltransferase 7 (GGT7) were primarily involved in the selenoamino acid metabolism. Additionally, aldehyde dehydrogenase 1 family, member A2 (ALDH1A2) was significantly enriched in the retinol metabolism sub-pathway.
Table IV.
Enriched sub-pathways of miR-326 targets.
| Pathway ID | Pathway name | P | Gene |
|---|---|---|---|
| path:00600_3 | Sphingolipid metabolism | 0.01024568 | SGMS1; SPTLC3; SGPL1 |
| path:00590_9 | Arachidonic acid metabolism | 0.01632627 | HPGDS; PTGIS |
| path:00450_3 | Selenoamino acid metabolism | 0.01935802 | AHCYL2; GGT7 |
| path:00830_2 | Retinol metabolism | 0.03609776 | ALDH1A2 |
Discussion
Abnormal miRNA expression and alterations are frequently associated with progression and prognosis of cancers (2,3). Specific miRNAs may be classified as tumor suppressors or oncogenes. However, a further analysis of their functions in LGGs is necessary. In the present study, three risk miRNAs, including has-miR-326 were identified by means of survival analysis and Cox proportional hazard regression model. Additionally, the PPI network revealed that SOS1, NRAS, VDR and SMAD3 were with a higher degree. Additionally, 8 target genes of miR-326 including SGMS1, SPTLC2, HPGDS and PTGIS were significantly enriched in metabolic sub-pathways.
Hsa-miR-326 has been downregulated in gliomas by suppression of the Notch signaling pathway, and is in turn inhibited by Notch (30). The Notch signaling pathway is a candidate pathway which may contribute to glioma progression (31). Additionally, miR-326 may be a potential tumor suppressor in glioma cells, and transfection of miR-326 into glioma cells may reduce tumorigenicity (30). As one grade of glioma, LGGs may be affected by miR-326. From the results of survival analysis, patients with high expression of miR-326 had reduced survival compared with those with low expression of it. Accordingly, miR-326 may suppress some oncogenes and lead to tumor development. Targets of miR-326 were regulated, including SOS1, NRAS, VDR, SMAD3 and SGMS1.
SOS1, as a dual guanine nucleotide exchange factor for Ras and Rac1, may convert inactive Ras-guanosine diphosphate into active Ras-guanosine triphosphate in various cells (32). Additionally, Ras was stimulated by EGFR and its close relative, erb-b2 receptor tyrosine kinase 2 (33). EGFR signaling has been determined to be involved in cell survival, tumorigenesis and metastasis (34). Notably, EGFR is one of the targets of several therapeutic agents in colorectal and non-small-cell lung cancers (34). Additionally, NGF may stimulate SOS1 and activate Ras signaling to exert various functions in cell proliferation (35). A previous study indicated that NGF may lead to proliferation and migration of endothelial cells and had a vital role in angiogenesis associated with tumors and cardiovascular diseases (35). NRAS, a member of the Ras family, is widely expressed in several cell types. Therefore, activation of NRAS may be stimulated by SOS1 and be involved in EGFR and NGF signaling. Consistent with a previous study (35), in the present study, SOS1 and NRAS were enriched in the EGFR and NGF signaling pathway. Furthermore, NRAS participated in cell proliferation in GO function analysis. Therefore, SOS1 and NRAS may regulate cell proliferation and angiogenesis via the EGFR and NGF signaling pathway in LGGs.
VDR is a transcription factor expressed in the brain (36) and mediates the effects of 1,25(OH)2D3. The vitamin D metabolite 1,25(OH)2D3 has been demonstrated to protect against cancer by inducing apoptosis and inhibiting cell proliferation and angiogenesis (37). SMAD3, the effector of transforming growth factor β (TGF-β), may directly bind Akt and inhibit TGF-β-induced apoptosis (38). SMAD3 may additionally interact with phosphatase and tensin homologue, which is a tumor suppressor in glioblastomas, to downregulate TGF-β signaling and decrease TGF-β-mediated tumor invasion (39). Consistent with this, the function annotation in the presents study revealed that VDR and SMAD3 were significantly enriched in the cell proliferation GO term. Therefore, it is possible that targets of miR-326, VDR and SMAD3 may regulate tumor cell proliferation in LGGs to increase tumor growth and invasion.
SGMS1, a sphingomyelin synthase, may produce sphingomyelin in the Golgi apparatus. The sphingomyelin levels have been previously reported to reduce the variety of tumor cells and reduce sphingomyelin by negatively regulating SGMS1 induction of cell proliferation of cancer cells (40). A previous study has reported that sphingolipids metabolism may influence cell cycle progression and cell migration (41). Additionally, ceramide and sphingosine-1-phosphate, two major sphingolipid metabolites, have been determined to be involved in process of apoptosis, cell proliferation and differentiation (42). Therefore, the miR-326 target SGMS1 may regulate cerebral LGG cell proliferation and apoptosis via the sphingolipids metabolism signaling pathway.
HPGDS, a prostaglandin D (PGD) synthase, catalyzes the synthesis of PDG2 from endogenous arachidonic acid. Additionally, arachidonic acid may be metabolized by cyclooxygenases (COX), cytochrome P450 and lipoxygenases (LOX) (43). Previous studies have revealed that COX and LOX inhibition induces apoptosis in several tumor cells (44–46). These were consistent with the current finding that HPGDS was primarily enriched in the arachidonic acid metabolism signaling pathway. Accordingly, HPGDS may regulate LGG cell apoptosis via this pathway.
In conclusion, the present study determined that hsa-miR-326 may be a potential risk miRNA for diagnosis and prognosis of LGG. Hsa-miR-326 may regulate cell proliferation and apoptosis of cancer cells by targeting certain genes including SOS1, NRAS, VDR, SMAD3, SGMS1 and HPGDS. However, further empirical investigations are required to confirm these findings.
References
- 1.Cavaliere R, Lopes MB, Schiff D. Low-grade gliomas: An update on pathology and therapy. Lancet Neurol. 2005;4:760–770. doi: 10.1016/S1474-4422(05)70222-2. [DOI] [PubMed] [Google Scholar]
- 2.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 3.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 6.Giovannetti E, van der Velde A, Funel N, Vasile E, Perrone V, Leon LG, de Lio N, Avan A, Caponi S, Pollina LE, et al. High-throughput microRNA (miRNAs) arrays unravel the prognostic role of MiR-211 in pancreatic cancer. PLoS One. 2012;7:e49145. doi: 10.1371/journal.pone.0049145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, Zeng YX, Shao JY. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348–2360. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Markou A, Tsaroucha EG, Kaklamanis L, Fotinou M, Georgoulias V, Lianidou ES. Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non-small cell lung cancer by quantitative real-time RT-PCR. Clin Chem. 2008;54:1696–1704. doi: 10.1373/clinchem.2007.101741. [DOI] [PubMed] [Google Scholar]
- 9.Hu X, Macdonald DM, Huettner PC, Feng Z, El Naqa IM, Schwarz JK, Mutch DG, Grigsby PW, Powell SN, Wang X. A miR-200 microRNA cluster as prognostic marker in advanced ovarian cancer. Gynecol Oncol. 2009;114:457–464. doi: 10.1016/j.ygyno.2009.05.022. [DOI] [PubMed] [Google Scholar]
- 10.Huse JT, Brennan C, Hambardzumyan D, Wee B, Pena J, Rouhanifard SH, Sohn-Lee C, le Sage C, Agami R, Tuschl T, Holland EC. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev. 2009;23:1327–1337. doi: 10.1101/gad.1777409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.le Sage C, Nagel R, Egan DA, Schrier M, Mesman E, Mangiola A, Anile C, Maira G, Mercatelli N, Ciafrè SA, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26:3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kefas B, Godlewski J, Comeau L, Li Y, Abounader R, Hawkinson M, Lee J, Fine H, Chiocca EA, Lawler S, Purow B. microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res. 2008;68:3566–3572. doi: 10.1158/0008-5472.CAN-07-6639. [DOI] [PubMed] [Google Scholar]
- 13.Hou SX, Ding BJ, Li HZ, Wang L, Xia F, Du F, Liu LJ, Liu YH, Liu XD, Jia JF, et al. Identification of microRNA-205 as a potential prognostic indicator for human glioma. J Clin Neurosci. 2013;20:933–937. doi: 10.1016/j.jocn.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 14.Zhang C, Zhang J, Hao J, Shi Z, Wang Y, Han L, Yu S, You Y, Jiang T, Wang J, et al. High level of miR-221/222 confers increased cell invasion and poor prognosis in glioma. J Transl Med. 2012;10:119. doi: 10.1186/1479-5876-10-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Therneau TM, Grambsch PM. Modeling survival data: Extending the Cox model. Springer; 2000. [DOI] [Google Scholar]
- 16.Jun Yan. KMsurv: Data sets from Klein and Moeschberger (1997), Survival Analysis. R package version 0.1–5 2012 Original by Klein, Moeschberger and modifications by. [Google Scholar]
- 17.Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: An integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37:D105–D110. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk-database: Prediction of possible miRNA binding sites by ‘walking’ the genes of three genomes. J Biomed Inform. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 19.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, El Naqa IM. Prediction of both conserved and nonconserved microRNA targets in animals. Bioinformatics. 2008;24:325–332. doi: 10.1093/bioinformatics/btm595. [DOI] [PubMed] [Google Scholar]
- 21.Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 22.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 23.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 24.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 28.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li C. iSubpathwayMiner: The package can implement the graph-based reconstruction and analyses of the KEGG pathways. R package version 3.0. 2012 [Google Scholar]
- 30.Kefas B, Comeau L, Floyd DH, Seleverstov O, Godlewski J, Schmittgen T, Jiang J, diPierro CG, Li Y, Chiocca EA, et al. The neuronal microRNA miR-326 acts in a feedback loop with notch and has therapeutic potential against brain tumors. J Neurosci. 2009;29:15161–15168. doi: 10.1523/JNEUROSCI.4966-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong JW. MicroRNA-induced silencing of glioma progression. J Neurosci. 2010;30:3868–3869. doi: 10.1523/JNEUROSCI.0442-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gureasko J, Galush WJ, Boykevisch S, Sondermann H, Bar-Sagi D, Groves JT, Kuriyan J. Membrane-dependent signal integration by the Ras activator Son of sevenless. Nat Struct Mol Biol. 2008;15:452–461. doi: 10.1038/nsmb0608-651a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 34.Goffin JR, Zbuk K. Epidermal growth factor receptor: Pathway, therapies, and pipeline. Clin Ther. 2013;35:1282–1303. doi: 10.1016/j.clinthera.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 35.Nico B, Mangieri D, Benagiano V, Crivellato E, Ribatti D. Nerve growth factor as an angiogenic factor. Microvasc Res. 2008;75:135–141. doi: 10.1016/j.mvr.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 36.Eyles DW, Smith S, Kinobe R, Hewison M, McGrath JJ. Distribution of the vitamin D receptor and 1 alpha-hydroxylase in human brain. J Chem Neuroanat. 2005;29:21–30. doi: 10.1016/j.jchemneu.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 37.Hansen CM, Binderup L, Hamberg KJ, Carlberg C. Vitamin D and cancer: Effects of 1,25(OH)2D3 and its analogs on growth control and tumorigenesis. Front Biosci. 2001;6:D820–D848. doi: 10.2741/A645. [DOI] [PubMed] [Google Scholar]
- 38.Conery AR, Cao Y, Thompson EA, Townsend CM, Jr, Ko TC, Luo K. Akt interacts directly with Smad3 to regulate the sensitivity to TGF-beta induced apoptosis. Nat Cell Biol. 2004;6:366–372. doi: 10.1038/ncb1117. [DOI] [PubMed] [Google Scholar]
- 39.Hjelmeland AB, Hjelmeland MD, Shi Q, Hart JL, Bigner DD, Wang XF, Kontos CD, Rich JN. Loss of phosphatase and tensin homologue increases transforming growth factor beta-mediated invasion with enhanced SMAD3 transcriptional activity. Cancer Res. 2005;65:11276–11281. doi: 10.1158/0008-5472.CAN-05-3016. [DOI] [PubMed] [Google Scholar]
- 40.Tafesse FG, Ternes P, Holthuis JC. The multigenic sphingomyelin synthase family. J Biol Chem. 2006;281:29421–29425. doi: 10.1074/jbc.R600021200. [DOI] [PubMed] [Google Scholar]
- 41.Zeidan YH, Hannun YA. Translational aspects of sphingolipid metabolism. Trends Mol Med. 2007;13:327–336. doi: 10.1016/j.molmed.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 42.Oskouian B, Saba JD. Sphingolipids as Signaling and Regulatory Molecules. Springer; 2010. Cancer treatment strategies targeting sphingolipid metabolism; pp. 185–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pidgeon GP, Lysaght J, Krishnamoorthy S, Reynolds JV, O'Byrne K, Nie D, Honn KV. Lipoxygenase metabolism: Roles in tumor progression and survival. Cancer Metastasis Rev. 2007;26:503–524. doi: 10.1007/s10555-007-9098-3. [DOI] [PubMed] [Google Scholar]
- 44.Jiang WG, Douglas-Jones A, Mansel RE. Levels of expression of lipoxygenases and cyclooxygenase-2 in human breast cancer. Prostaglandins Leukot Essent Fatty Acids. 2003;69:275–281. doi: 10.1016/S0952-3278(03)00110-8. [DOI] [PubMed] [Google Scholar]
- 45.Hoque A, Lippman SM, Wu TT, Xu Y, Liang ZD, Swisher S, Zhang H, Cao L, Ajani JA, Xu XC. Increased 5-lipoxygenase expression and induction of apoptosis by its inhibitors in esophageal cancer: A potential target for prevention. Carcinogenesis. 2005;26:785–791. doi: 10.1093/carcin/bgi026. [DOI] [PubMed] [Google Scholar]
- 46.Leung HW, Yang WH, Lai MY, Lin CJ, Lee HZ. Inhibition of 12-lipoxygenase during baicalein-induced human lung nonsmall carcinoma H460 cell apoptosis. Food Chem Toxicol. 2007;45:403–411. doi: 10.1016/j.fct.2006.08.021. [DOI] [PubMed] [Google Scholar]