Abstract
Complex traits arise from the interplay between genetic and environmental factors. The actions of these factors usually appear to be additive, and few compelling examples of gene-environment synergy have been documented. Here we show that adiposity significantly amplifies the effect of three sequence variants (PNPLA3-I148M, TM6SF2-E167K and GCKR-P446L) associated with nonalcoholic fatty liver disease (NAFLD). Synergy between adiposity and genotype promoted the full spectrum of NAFLD, from steatosis to hepatic inflammation to cirrhosis. We found no evidence of strong interactions between adiposity and sequence variants influencing other adiposity-associated traits. These results indicate that adiposity may augment genetic risk of NAFLD at multiple loci through at least three different metabolic mechanisms.
Introduction
For most complex traits, sequence variations identified by genome-wide association studies (GWAS) account for only a minor fraction of the heritable variation estimated from family studies 1. The missing heritability has been attributed to rare variants that are not represented on commercial SNP arrays 2,3, common variants that do not reach genome-wide significance 4, and gene-gene and gene-environment interactions that amplify the phenotypic effects of individual sequence variations5–7. The contribution of gene-environment interactions remains controversial. Genetic variants are usually assumed to act in an additive manner 5,8, such that the combined effect of two or more sequence variations equals the sum of their individual effects. Compelling examples of synergistic or context-dependent relationships between genetic variants and environmental exposures have been described, including susceptibility to adverse drug reactions 9, infectious diseases 10, and sun exposure 11, but most reports of gene-environment interactions have proved poorly reproducible8,12,13.
Obesity has emerged as a major cause of morbidity due to its role in metabolic disorders such as type 2 diabetes mellitus, hypertension and dyslipidemia. More recently, obesity has been associated with nonalcoholic fatty liver disease (NAFLD), a spectrum of disorders that includes excess liver fat (steatosis), inflammation (steatohepatitis), fibrosis (cirrhosis), and ultimately malignant transformation (hepatocellular carcinoma) 14. Susceptibility to NAFLD is highly variable; not all obese individuals develop steatosis and most cases of steatosis do not progress to chronic liver disease. Expression of the disorder is strongly influenced by heritable factors. One of the most powerful genetic risk factors for NAFLD is a single nucleotide polymorphism (SNP) that changes residue 148 of the patatin-like phospholipase 3 gene (PNPLA3) from isoleucine to methionine (referred to here as the M variant) 15. Here we show that adiposity influences the effect of the M variant on liver fat content, as well as on serum ALT (a marker of hepatocellular injury) and cirrhosis. Interactions with obesity were also observed for sequence variants in two other genes (TM6SF2 and GCKR) that contribute to NAFLD by different mechanisms16–18. We did not observe interactions of similar magnitude for a range of other traits that associate strongly with adiposity. Thus, gene × adiposity interaction on NAFLD appears to be a rather specific and robust phenomenon.
Results
The effect of the M variant on HTGC is highly dependent on adiposity
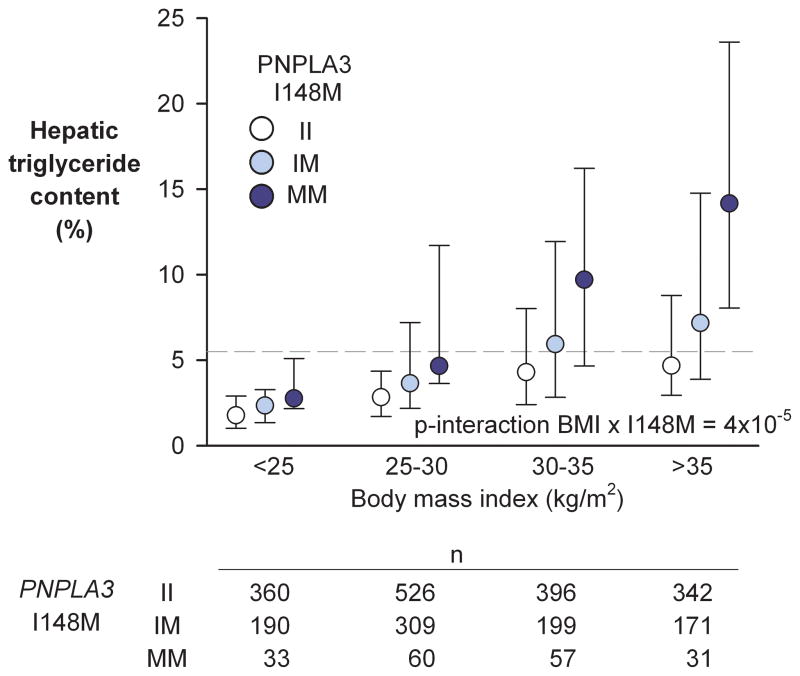
Previously, we showed that steatosis (HTGC>5.5%) was present in 33% of the participants in the Dallas Heart Study and that BMI was strongly associated with increased HTGC (Spearman’s rho=0.4) 19. To determine if the effect of the M variant on HTGC is modified by adiposity, we analyzed the relationship between PNPLA3 genotype and HTGC after stratifying the DHS participants into four bins based on BMI (Fig. 1). Among lean persons (BMI<25 kg/m2), median HTGC increased modestly, though significantly in a stepwise fashion in the II, IM and MM individuals (1.8%, 2.3%, and 2.8%, respectively; P=0.0003). Steatosis was less common in all three genotype groups (II, 9%; IM, 8%; MM, 18%) than in the general population (33%).
Fig. 1.
Hepatic triglyceride content by body mass index and PNPLA3 I148M genotype in the Dallas Heart Study. Hepatic triglyceride content was measured by magnetic resonance spectroscopy. Circles and error bars depict medians and interquartile ranges of HTGC. The HTGC-increasing effect of the 148M-allele was amplified by increasing adiposity (p-interaction I148M × BMI on HTGC=4×10−5). The dashed line marks the 95th percentile of HTGC in the general population. Abbreviations: HTGC, hepatic triglyceride content.
The effect of the M variant increased with increasing BMI. Among the most obese participants (BMI>35 kg/m2), the median HTGC was 3-fold higher in MM than in II individuals (14.2% versus 4.7%) and a significantly greater proportion of the MM homozygotes had hepatic steatosis (84% versus 42%; P=0.001). In all 4 BMI groups, the median HTGC of IM heterozygotes was intermediate between the levels in the two groups of homozygotes.
Regression analysis using BMI as an ordered categorical variable revealed highly significant differences in the effect of PNPLA3 genotype on HTGC among the 4 BMI groups (t-test; P=0.0004). Significant interaction was also observed using BMI as a continuous variable in the regression model (t-test; P = 4×10−5).
Interactive effects on continuous traits are influenced by the scale on which they are analyzed 20,21. Therefore, we repeated the interaction tests using the untransformed data, other transformations of HTGC (inverse normalized, or logarithmically transformed), and after dichotomizing the data (normal vs steatosis). The BMI×PNPLA3 interaction remained significant in all of these analyses (Supplementary Fig. 1). To assess whether observed interaction effects were a result of heteroscedasticity, we repeated the analysis using model-robust estimates of standard errors 22. The BMI×PNPLA3 interactions remained significant (Supplementary Fig. 1). These data indicate that the effect of PNPLA3 genotype on HTGC is strongly influenced by adiposity.
Adiposity amplifies effect of other NAFLD-associated genetic variants
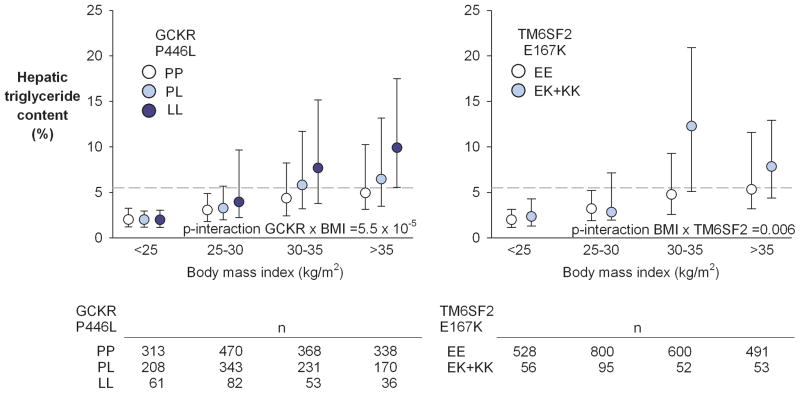
To determine whether the interaction between PNPLA3 genotype and BMI on HTGC is peculiar to the M variant, we examined the relationship between HTGC, BMI, and other NAFLD-related variants. We first tested whether variants in loci that associated with HTGC in the Genetics of Liver Disease (GOLD) study were associated with HTGC in the DHS (Supplementary Table 1) 18. Two of these SNPs, TM6SF2-E167K and GCKR-P446L, were associated with HTGC in the DHS 16.
We examined whether adiposity influenced the effects of the risk alleles at the TM6SF2 and GCKR loci on HTGC in the DHS. As was observed with the M variant, the effect of the GCKR risk allele (P446L) was significantly amplified by increasing BMI (Fig. 2) (P-interaction =5.5×10−5). A similar amplifying effect of BMI on HTGC was seen for the TM6SF2 risk variant (E167K), though the interaction was less significant (P-interaction =0.006), likely due to the lower frequency of the risk allele when compared to the GCKR risk allele (MAF=0.05 versus 0.25). As was observed for PNPLA3, the interactions remained significant after different transformations of HTGC, and when using a heteroscedasticity-robust model (Supplementary Fig. 1). Thus, the interaction between genotype and BMI on HTGC is not unique to PNPLA3. This finding suggests that obesity promotes development of NAFLD through interactions with at least three genes that act in different metabolic pathways.
Fig. 2.
Hepatic triglyceride content by body mass index and GCKR P446L and TM6SF2 E167K genotypes in the Dallas Heart Study. Circles and error bars depict medians and interquartile ranges of HTGC. The dashed line marks the 95th percentile of HTGC in the general population. Abbreviations: HTGC, hepatic triglyceride content.
PNPLA3-148M × adiposity interaction on serum levels of liver enzymes
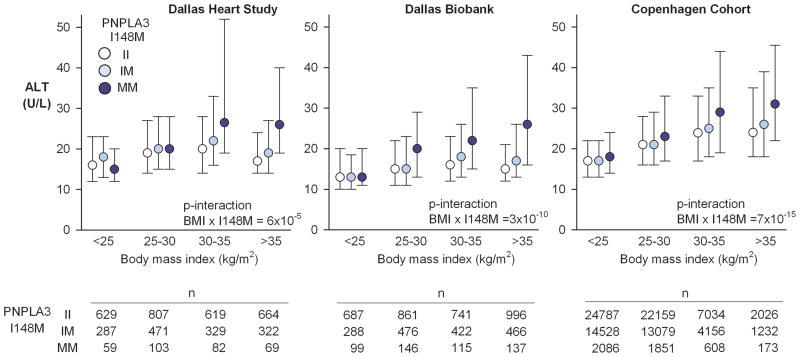
Hepatic steatosis per se is considered to be benign 23. A subset of individuals with steatosis develops hepatic inflammation, which can result in elevated serum levels of liver enzymes, especially alanine aminotransferase (ALT) 24. To determine if adiposity exacerbated the effect of the M variant on liver inflammation, we tested for interaction between the PNPLA3 genotype and BMI on serum ALT levels (Fig. 3). In the DHS, serum ALT levels were increased by the M variant as previously reported 15. Median ALT was 18 U/L in II-homozygotes, 20 U/L in IM-heterozygotes, and 22 U/L in MM-homozygotes (P-trend=9×10−5). As with HTGC, the effect of the M variant on ALT increased with increasing BMI. The M variant was not associated with increased ALT in the lean (BMI<25 kg/m2) or the overweight (BMI 25–30 kg/m2) groups, but increased ALT in the obese (BMI 30–35 kg/m2) and massively obese (BMI >35 kg/m2) groups. In the Dallas Biobank and in the Copenhagen Cohort, which are larger cohorts than the DHS, the effect of the M variant on ALT was also apparent in the overweight group. Nonetheless, in all three cohorts the interaction between BMI and PNPLA3 genotype on ALT was highly statistically significant.
Fig. 3.
Serum levels of alanine aminotransferase by body mass index and PNPLA3 I148M genotype in the Dallas Heart Study, the Dallas Biobank, and the Copenhagen cohort. Circles and error bars depict medians and interquartile ranges of ALT. The ALT-increasing effect of the 148M-allele was amplified by increasing adiposity (p-interaction I148M × BMI on ALT <0.001 in all three cohorts). Abbreviations: ALT, alanine aminotransferase.
As for HTGC, we retested the BMI×PNPLA3 interaction in the DHS after various transformations of ALT. The interaction remained robust regardless of transformation applied (Supplementary Fig. 2).
We also tested for interaction between BMI and the risk alleles at TM6SF2 and GCKR on ALT levels in the DHS, Biobank, and Copenhagen cohorts. The effect of the TM6SF2-E167K variant on ALT was significantly affected by BMI in the Copenhagen Cohort (P=10−4), but not in the DHS (P=0.14) or Dallas Biobank (P=0.39)(Supplementary Fig. 3). GCKR showed marginal evidence for interaction with BMI on ALT in the DHS (P=0.01), but not in the two larger cohorts (Supplementary Fig. 4). Thus, in contrast to what we observed with the M variant, we did not find reproducible evidence of an interaction between risk alleles at TM6SF2/GCKR and BMI on ALT levels. This may be explained by a reduced effect size on HTGC (e.g., GCKR) or a lower allele frequency (e.g., TM6SF2), resulting in a reduced power to detect interactions.
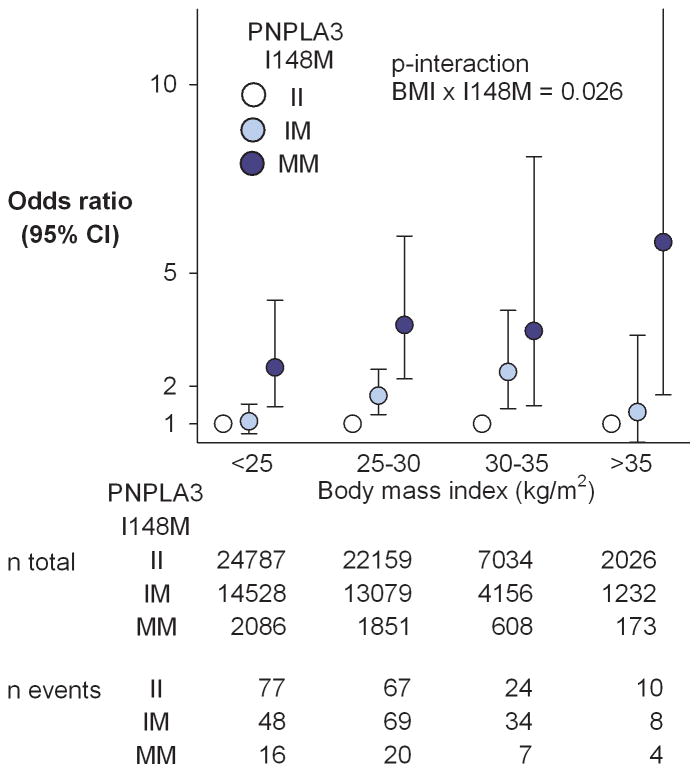
Adiposity augments the effect of M variant on the prevalence of cirrhosis in the Copenhagen Cohort
The DHS and Dallas Biobank both contain too few subjects with cirrhosis to examine the gene-environment interactions for this phenotype. The Copenhagen cohort included 384 participants with cirrhosis due either to alcoholism or NAFLD (see Supplementary Table 2 for baseline characteristics). The effect of the PNPLA3 risk variant on the prevalence of cirrhosis increased with increasing BMI (Fig. 4). The risk of cirrhosis was higher among MM homozygotes in each BMI category, even those with a BMI <25 kg/m2. Among persons with BMI>35 kg/m2, the odds ratio for cirrhosis was 5.8 in homozygotes for the M variant versus those homozygous for the I variant. The corresponding odds ratio in those with BMI<25 kg/m2 was 2.4. Additional adjustments for alcohol×BMI or alcohol×PNPLA3 (individually or simultaneously) did not materially change results. Thus, interaction between adiposity and the 148M isoform appears to promote chronic liver disease as well as steatosis.
Fig. 4.
Risk of cirrhosis by body mass index and PNPLA3 I148M genotype in the Copenhagen cohort. Circles and error bars depict odds ratios and 95% confidence intervals. The II-genotype acted as the reference group within each BMI-group. The risk-increasing effect of the 148M-allele was amplified by increasing adiposity (p-interaction I148M × BMI on risk of cirrhosis=0.026).
Adiposity as a causal risk factor for NAFLD: Mendelian randomization analysis
If adiposity contributes to NAFLD, then SNPs that are associated with BMI would be expected to also associate with HTGC. Since the individual effects on BMI of these SNPs are small, we constructed a genetic risk score using 30 SNPs that were associated with adiposity in a previous GWAS (Supplementary Table 3) 25, and tested for association with BMI and HTGC in the DHS. As expected, an increasing genetic risk score was associated with a modest, though significant increase in BMI (P for trend across SNP-score=0.001) (Supplementary Fig. 5). Subjects in the first quintile had a median BMI of 27.5, whereas those in the fifth quintile of the risk score had a median BMI of 29.3 kg/m2. The BMI risk score was also associated with an increase in HTGC (p=0.02). The modest effect of the obesogenic risk score on HTGC was consistent with the small increase in BMI associated with these variants.
Gene × adiposity interaction on other BMI-associated traits
To assess whether gene-environment interactions were commonly observed with genetic predictors of other metabolic traits that are related to obesity, we screened the DHS database for phenotypes that showed a correlation with BMI with an absolute r-value of more than 0.2 after adjusting for age, gender and ethnicity (Supplementary Table 4). We then determined if these traits were associated with any SNP assayed using the Illumina Exome BeadChip array at an exome-wide significance level (P<3.6×10−7). For comparison we included SNPs found to associate with NAFLD in previous studies 18,26. A total of 13 traits and 21 SNPs meeting these criteria were identified (Supplementary Table 5). As an example, BMI was strongly associated with plasma levels of leptin (Partial r=0.74; P<1×10−300). A SNP located in the leptin gene was strongly associated with leptin levels (per allele change in standardized leptin levels= −0.22 s.d. units; P=2.28×10−11). Despite the strengths of the SNP-leptin and BMI-leptin associations, no SNP×BMI interaction was seen (P=0.60). A similar lack of SNP×BMI interaction was observed for CRP, the second most strongly BMI-correlated trait. Of the 13 traits examined, only HTGC showed robust and highly statistically significant interactions between trait-associated SNPs and BMI.
For the remaining 12 BMI-associated traits, only 3 nominally significant interactions were identified (between BMI and SNPs in APOA5, LPL, and GCKR on plasma TG levels). None of these three interactions were observed in the Dallas Biobank, and only one was replicated (p=0.02) in the large Copenhagen cohort (Supplementary Table 6).
Discussion
The major finding of this paper is that adiposity amplifies the genetic risk of NAFLD. In a cohort from the general population (the DHS), the prevalence of hepatic steatosis ranged from 9% in lean individuals who did not carry the M variant to 84% in very obese individuals who were homozygous for the M variant. Adiposity also amplified the effects of the M variant on serum ALT activity and the risk of cirrhosis. Taken together, these results indicate that gene × adiposity interaction plays a major role in the development and progression of NAFLD in humans. Other traits that are strongly correlated with adiposity (e.g., plasma leptin and CRP levels) were not influenced by gene × BMI interactions in our study, despite having associations with genetic variants that were comparable in magnitude to that of the M variant on HTGC.
The interaction with adiposity was not specific to the M isoform of PNPLA3. We found similar gene × adiposity relationships for two other steatogenic alleles, GCKR-446L and TM6SF2-167K. These three variants promote steatosis by distinct metabolic mechanisms. The M isoform of PNPLA3 accumulates on cytoplasmic lipid droplets and likely compromises TG mobilization 27. GCKR is a negative regulator of glucokinase; the loss-of-function variant (446L) results in increases in phosphorylation of glucose 28, glycolysis, and fatty acid synthesis 17 in the liver. TM6SF2 is a polytopic ER protein that is required for VLDL secretion from the liver 16. The E167K substitution is a loss-of-function mutation that results in impaired hepatic TG secretion and accumulation of hepatic fat 16. Thus, obesity may augment genetic risk of NAFLD through at least three different metabolic mechanisms.
One possibility is that obesity may amplify the effect of the three risk alleles by altering their expression. PNPLA3 is a direct target of the insulin-regulated transcription factor sterol regulatory element binding protein-1c (SREBP-1c) and is highly regulated by fasting and refeeding 29. GCKR expression is also increased by glucose and insulin 30. The insulin resistance associated with obesity may therefore increase expression of these two genes. However, TM6SF2 does not respond to food intake 31. Thus, the gene × adiposity interaction appears not to be due simply to an enhancement in the expression of the risk allele at these three loci.
Do the variants in PNPLA3, TM6SF2, and GCKR interact with other environmental risk factors associated with obesity? Increasing visceral fat content augmented the effect of the M-variant on hepatic fat content in a previous study of 2,257 non-diabetic European-Americans 32. A diet rich in carbohydrates is associated with an increase in risk of NAFLD 33. Among 158 Hispanic children, a high carbohydrate intake increased HTGC in those homozygous for the PNPLA3 M-variant, but not in IM-heterozygotes or II-homozygotes 34. We observed a similar phenomenon in mice. Knockin mice expressing PNPLA3-148M do not develop hepatic steatosis on a low-fat chow diet, whereas on a high-sucrose diet, which dramatically increases the levels of insulin, the mice have 2–3 fold increased HTGC compared to wildtype mice 27. These findings support the hypothesis that gene × diet interactions play a role in NAFLD. We speculate that energy surplus is an absolute requirement for the deposition of fat in the liver. In the absence of an energy surplus, there is no driver for hepatic fat accumulation, irrespective of genotype. This situation is analogous to a pharmacogenetics interaction, in which the effect of a genetic variant is contingent on the action of a drug.
An estimated 30% of those individuals who develop steatosis have associated hepatic inflammation 35. In a subset of these individuals, liver enzymes are released into the blood. PNPLA3-I148M was the SNP most strongly associated with serum ALT levels in the first GWAS on serum liver enzyme levels 36, and this result has been confirmed in several subsequent studies 15,37. We found that adiposity amplified the effect of the M variant on ALT levels in a manner that was similar to the effect on steatosis. This finding contrasts with Larrieta-Carrasco et. al., who reported that the odds ratio of elevated ALT associated with the M variant was greater in normal-weight children than in obese children 38, and with Giudice et. al.39, who reported that the effect of the M variant increased with waist to hip ratio, but not with BMI, in obese children. The reasons for these discrepancies are not known.
We also found a gene×adiposity interaction when we analyzed the effect of PNPLA3-I148M on cirrhosis due to NAFLD or to alcoholic liver disease. Among massively obese individuals, MM-homozygotes had a 5.8-fold increased risk of cirrhosis compared to II-homozygotes. Among lean persons (BMI<25 kg/m2), the MM-homozygotes had a 2.4-fold increased risk of cirrhosis compared to II-homozygotes. Thus, adiposity appears to amplify the effect of the PNPLA3-148M variant on the entire spectrum of NAFLD, from steatosis, to steatohepatitis, to end-stage liver disease. A limitation to the observed interaction on cirrhosis is that the number of cases was relatively modest, and that cirrhosis was defined by registry-based ICD-codes. Pending independent replication in larger patient cohorts, the interaction on cirrhosis should therefore be viewed as preliminary.
Adiposity has been found to also amplify the effect of alcohol on liver disease 40. Among obese men, those who drank >15 units of alcohol per week had an 18.9 fold increased risk of death from liver disease compared to non-drinkers40. The corresponding relative risk among lean men was 3.2. Taken together, our data and the results of Hart et al.40 indicate that adiposity exacerbates the effects on fatty liver disease of both genetic and nongenetic factors. It is possible that adiposity exacerbates alcoholic liver disease through its actions on PNPLA3, since PNPLA3-148M has been shown to confer risk of cirrhosis among alcoholics41.
Whereas the burgeoning of obesity in the population is a result of secular changes in lifestyle factors (presumably diet and exercise), inter-individual differences in adiposity are also partly heritable. We show here that genetic variants that associate with increased BMI also associate with increased hepatic fat content. This finding indicates that the sequence variants at nearly 100 loci that have been associated with BMI 42 would be predicted to be associated with liver fat content in a PNPLA3/TM6SF2/GCKR genotype-dependent manner. Thus, the heritability of HTGC is determined not only by the primary effect of PNPLA3/TM6SF2/GCKR genotype but also by the secondary effects of variation at nearly 100 loci that influence HTGC indirectly via their effects on adiposity. Thus the interaction between BMI and NAFLD-variants reported here should in fact be viewed as a mixture of gene×gene and gene×environment interactions.
Obesity increases susceptibility to a wide variety of common complex diseases ranging from cancer (e.g., breast and colon cancer) and hypertension to metabolic disorders (e.g., type 2 diabetes mellitus). Few single gene×adiposity interactions have been robustly documented for any of these conditions 12. In an effort to probe the contribution of gene×adiposity interactions to adiposity-associated traits more generally, we screened the DHS database for metabolic phenotypes that are strongly correlated with BMI, and then for SNPs that are strongly associated with those traits. Of the phenotypes tested, 13 met both criteria, but apart from the BMI×SNP interactions on HTGC, only three SNPs showed nominally significant interactions with BMI (variants in LPL, APOA5, and GCKR interacted with BMI in their effect on plasma TG levels). The interactions were not robustly replicated in two larger cohorts. Limitations of this screen include the relatively modest sample size and the comparison of candidate gene SNPs with those identified by an agnostic exome-wide approach. Nevertheless, these findings support the hypothesis that gene × adiposity interactions of comparable magnitude to those observed for PNPLA3, TM6SF2, and GCKR on HTGC are uncommon.
What distinguishes NAFLD from other adiposity-associated phenotypes, such as hypertension and blood glucose levels? First, the common alleles that contribute to hypertension and blood glucose levels all have smaller phenotypic effects than do the fatty liver susceptibility alleles 43,44. For example, homozygotes for the M variant have a 2-fold increase in HTGC compared to II-homozygotes, and the variant explains ~5–10% of the variance in HTGC in different ethnicities. In contrast, the alleles most robustly associated with blood pressure or with blood glucose only increase these traits by ~1%, and each explains less than 1% of the total trait variance 43,44. These modest effect sizes limit the power to detect interactions with adiposity.
A second major difference between blood pressure and blood glucose, and liver fat content is that blood pressure and blood glucose are both under homeostatic control. Consequently, the effect of any genetic variant on the levels of blood glucose, or on blood pressure, will be opposed by counter-regulatory effects. In contrast, there is no evidence that the concentration of TG in the liver is subject to feedback regulation. Therefore, sequence variants or environmental factors (such as increased food consumption) can promote the accumulation of large amounts of TG within lipid droplets in the liver without eliciting a counter-regulatory response. The frequency of the M variant increases from sub-Saharan Africa to South America in a pattern that reflects human migration. This pattern suggests that the variant may have been under positive selective pressure. Could the M variant be part of the “thrifty genome”, as has been suggested previously 45,46?
The findings reported here raise the possibility that consideration of adiposity and genotype jointly may provide improved prediction of individuals at highest risk of progressing from simple steatosis to chronic liver disease. The risk alleles of the three strongest NAFLD risk variants confer only moderate risk in lean individuals, but are major risk factors in the obese, suggesting that genetic screening would be especially valuable in this subgroup. Similarly, while all obese individuals would benefit from a weight-loss intervention, our data suggest that individuals at high genetic risk of NAFLD are likely to benefit the most.
Online Methods
Studies were approved by institutional review boards and ethics committees of the University of Texas Southwestern Medical Center and by Danish institutional review boards and ethic committees, and were conducted according to the Declaration of Helsinki. Written informed consent was obtained from participants. There was no overlap of individuals between the studies.
Participants
We included participants from four studies: the Dallas Heart Study (DHS), the Dallas Biobank, the Copenhagen City Heart Study (CCHS), and the Copenhagen General Population Study (CGPS). The DHS is a multiethnic, probability-based sample of Dallas County that was collected between 2000 and 2002, and between 2007 and 2009 15,19. Ethnicity was self-reported in accordance with U.S. census categories. From the DHS, we included 2,675 participants in whom hepatic triglyceride content (HTGC) was measured 47 and up to 1,786 additional individuals were added to the sample size for the analysis of other traits. We included 5,434 persons from the Dallas Biobank, a general population cohort of African Americans and Hispanic Americans from Dallas, TX 16. The CCHS and CGPS are prospective studies of the Danish general population initiated in 1976 and 2003, respectively 16,48. All participants from the CCHS and CGPS were white and of Danish descent, as determined by the National Danish Civil Registration System. We combined the CCHS and CGPS into one cohort, totaling 93,719 persons, here referred to as the Copenhagen cohort.
Measurements
Body mass index was measured as weight in kilograms divided by measured height in meters squared. Hepatic triglyceride content (HTGC) was measured in the DHS using proton magnetic resonance spectrometry47. Hepatic steatosis was defined as an HTGC of 5.5% or greater; 5.5% represents the 95th percentile of the distribution of HTGC in a population with no risk factors for steatosis 47. Serum levels of ALT were measured as described 49. PNPLA3 I148M (rs738409; NC_000022.11:g.43928847C>G, p.Ile148Met), TM6SF2 E167K (rs58542926; NC_000019.10:g.19268740C>T, p.Glu167Lys), and GCKR P446L (rs1260326; NC_000002.11:g.27730940C>T, p.Pro446Leu), the 30 BMI-associated variants, and the exome-wide variants used to screen for associations and interactions with other phenotypes were genotyped in the Dallas Heart Study by an exome chip as previously described 16. PNPLA3 I148M, TM6SF2 E167K, and GCKR P446L were genotyped by Taqman in the Dallas Biobank, and by Taqman and PCR-based KASP genotyping in the Copenhagen cohort. Alcohol intake in the Copenhagen cohort was self-reported.
Cirrhosis
In the Copenhagen cohort, diagnoses of cirrhosis (ICD8: 57109 [alcoholic cirrhosis], 57192 [unspecified cirrhosis], 57199 [non-alcoholic cirrhosis] and ICD10: K70.3 [alcoholic cirrhosis], K74.0 [hepatic fibrosis], K74.6 [unspecified cirrhosis]) were collected from the National Danish Patient Registry, and the National Danish Causes of Death Registry from January 1, 1977 to November 10th, 2014. The National Danish Patient Registry has information on all patient contacts with all clinical hospital departments in Denmark, including emergency wards and outpatient clinics (from 1994). The National Danish Causes of Death Registry contains data on the causes of all deaths in Denmark, as reported by hospitals and general practitioners. A validation study in the Danish registry found that 85.4% of patients with an ICD-code for cirrhosis fulfilled the diagnostic criteria for cirrhosis 50.
Statistical analysis
All analyses were performed using Stata SE 12 (Stata Corp., College Station, Texas) and/or R statistical analysis software v 3.2.3 (https://www.R-project.org). A two-sided p-value <0.05 was considered statistically significant in all main analyses, whereas p<3.6×107 was considered significant in the exome-wide screen. For statistical tests, genotypes were coded 0, 1, 2. Body mass index was entered as a continuous variable in all analyses (apart from a sensitivity test for interaction, in which BMI groups were entered as an ordered categorical variable, encoded 0–3). To depict the interaction between genotype and BMI visually, participants were divided into four groups of BMI: lean (≤25 kg/m2), overweight (25–30 kg/m2), obese (30–35 kg/m2) and very obese (>35 kg/m2). The distributions of HTGC and ALT were highly skewed to the right 19. Therefore, prior to entering these variables into regression analyses, we transformed them to HTGC^0.3 and 1/(ALT^0.25), in order to approximate normality and constant variance of the residuals. These transformations were selected by using Tukey’s ladder of power transformations, and by visual inspection of Q-Q plots of residuals after the transformation. To assess the robustness of the interactions on different scales, we also used untransformed, inverse normally transformed, logarithmically transformed and dichotomized HTGC or ALT. For each transformation, we plotted distributions of the variable, the normal Q-Q plot of the residuals, and distribution of the residuals by BMI-category, and tested for BMI×SNP interactions (Supplementary Figs. 1 and 2). To account for a higher variance in HTGC in the most obese compared to lean subjects (heteroscedasticity), we repeated all interaction tests using a heteroscedasticity-robust model 22. We considered whether adjusting for BMI and PNPLA3/GCKR/TM6SF2 in the models could introduce collider bias 51. This was deemed unlikely, given that none of the 3 genetic variants associate with BMI, and that NAFLD is not known to causally influence adiposity. Prevalence of cirrhosis and steatosis were evaluated by logistic regression models adjusted for sex and age (and ethnicity in the DHS).
We evaluated the interactions between BMI and SNPs by the inclusion of interaction terms between BMI and SNPs in the linear or logistic regression models, adjusted for sex, age, and ethnicity (encoded African American=1, European American=2, Hispanic American=3, and entered as a factorial variable in the regression). Body mass index and SNPs were entered as continuous variables in the interaction term (ie. all interaction tests are 1 degree of freedom). In a sensitivity analysis, the interaction on cirrhosis was retested after further adjustment for alcohol×BMI and alcohol×PNPLA3 interaction, entered individually or simultaneously into the regression.
To test whether adiposity is a likely causal risk factor for increased HTGC, we genotyped 30 SNPs known to be associated with BMI in Whites (Supplementary Table 1) 25. For each SNP, the BMI-increasing alleles were weighted by the per-allele effect size reported in the GWAS 25. A gene score was calculated for each European American participant of the DHS by summation of weighted alleles across all 30 BMI-associated SNPs. The gene score was tested for association with BMI and HTGC using linear regression, with the gene score included as a continuous variable. To depict the association between the genotype score and BMI and HTGC visually, the genotype score was divided into quintiles (Supplementary Fig. 6). Instrumental variable analysis was conducted in order to compare the observational association between BMI and HTGC with the effect of genetically increased BMI on HTGC52. The observational association between BMI and HTGC^0.3 was determined using linear regression, adjusted for age and sex. For the genetic, causal analysis, two-stage least squares regression was used to assess the effect of a 1 kg/m2 increase in genetically modeled BMI on HTGC^0.352. Strength of the genetic instrument was evaluated by F-statistics and R2, where F>10 was considered sufficient to avoid weak-instrument bias, and R2 indicates the fraction of variation in BMI explained by the instrument.
To determine whether gene-environment interactions were commonly observed with other obesity-associated traits, we screened phenotypes relevant to metabolism (plasma lipids, glucose and insulin homeostasis, blood pressure, liver enzymes, sterols, biomarkers) for correlation with BMI in the DHS. Phenotypes showing a partial correlation with BMI (after adjustment for age, gender and ethnicity) exceeding 0.2 in absolute value were further screened for association with genetic variants present on the Illumina HumanExome BeadChip (12v1_A) 16. Variants exceeding our exome-wide significance threshold (p<3.6×107), or variants in established genetic loci from a previously published NAFLD GWAS 18, were then tested for SNP×BMI interaction, using linear regression adjusted for age, gender and ethnicity. All nominally significant SNP×BMI-interactions (p<0.05) were retested in the Dallas Biobank and in the Copenhagen Cohort (where both phenotype and genotype data were available).
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgments
Supported by grants from the NIH: PO1 HL20948 and RO1 DK090066 (H.H.H and J.C.C), UL1TR001105 (H.H.H), and The Danish Council for Independent Research, Medical Sciences (Sapere Aude 4004-00398) (S.S.). The Copenhagen Cohort is supported by the Danish Council for Independent Research, the Research Fund at Rigshospitalet, Copenhagen University Hospital, Chief Physician Johan Boserup and Lise Boserup’s Fund, Ingeborg and Leo Dannin’s Grant, Henry Hansen and Wife’s Grant, and a grant from the Odd Fellow Order (A.T-H.).
Footnotes
Author contributions
S.S.: Study concept and design, analysis and interpretation of data, drafting of the manuscript, statistical analysis, critical revision of the manuscript. J.K.: Analysis and interpretation of data, statistical analysis, critical revision of the manuscript. A.T-H.: Acquisition of data, critical revision of the manuscript. B.G.N.: Acquisition of data, critical revision of the manuscript. H.H.H.: Study concept and design, analysis and interpretation of data, drafting of the manuscript, critical revision of the manuscript, acquisition of data, study supervision. J.C.C.: Study concept and design, analysis and interpretation of data, drafting of the manuscript, critical revision of the manuscript, acquisition of data, study supervision.
Conflicts of interest
None of the authors had potential conflicts of interest.
References
- 1.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pritchard JK. Are rare variants responsible for susceptibility to complex diseases? Am J Hum Genet. 2001;69:124–37. doi: 10.1086/321272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen JC, et al. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–72. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 4.Yang J, et al. Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat Genet. 2015;47:1114–20. doi: 10.1038/ng.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zuk O, Hechter E, Sunyaev SR, Lander ES. The mystery of missing heritability: Genetic interactions create phantom heritability. Proc Natl Acad Sci U S A. 2012;109:1193–8. doi: 10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Purcell S. Variance components models for gene-environment interaction in twin analysis. Twin Res. 2002;5:554–71. doi: 10.1375/136905202762342026. [DOI] [PubMed] [Google Scholar]
- 7.Kaprio J. Twins and the mystery of missing heritability: the contribution of gene-environment interactions. J Intern Med. 2012;272:440–8. doi: 10.1111/j.1365-2796.2012.02587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sadee W, et al. Missing heritability of common diseases and treatments outside the protein-coding exome. Hum Genet. 2014;133:1199–215. doi: 10.1007/s00439-014-1476-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang L, McLeod HL, Weinshilboum RM. Genomics and drug response. N Engl J Med. 2011;364:1144–53. doi: 10.1056/NEJMra1010600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Samson M, et al. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–5. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 11.Baron AE, et al. Interactions between ultraviolet light and MC1R and OCA2 variants are determinants of childhood nevus and freckle phenotypes. Cancer Epidemiol Biomarkers Prev. 2014;23:2829–39. doi: 10.1158/1055-9965.EPI-14-0633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reddon H, Gueant JL, Meyre D. The importance of gene-environment interactions in human obesity. Clin Sci (Lond) 2016;130:1571–97. doi: 10.1042/CS20160221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manuck SB, McCaffery JM. Gene-environment interaction. Annu Rev Psychol. 2014;65:41–70. doi: 10.1146/annurev-psych-010213-115100. [DOI] [PubMed] [Google Scholar]
- 14.Ludwig J, Viggiano TR, McGill DB, Oh BJ. Nonalcoholic steatohepatitis: Mayo Clinic experiences with a hitherto unnamed disease. Mayo Clin Proc. 1980;55:434–8. [PubMed] [Google Scholar]
- 15.Romeo S, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–5. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozlitina J, et al. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2014;46:352–6. doi: 10.1038/ng.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santoro N, et al. Hepatic De Novo Lipogenesis in Obese Youth Is Modulated by a Common Variant in the GCKR Gene. J Clin Endocrinol Metab. 2015;100:E1125–32. doi: 10.1210/jc.2015-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Speliotes EK, et al. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011;7:e1001324. doi: 10.1371/journal.pgen.1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Browning JD, et al. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology. 2004;40:1387–95. doi: 10.1002/hep.20466. [DOI] [PubMed] [Google Scholar]
- 20.Falconer DS. Introduction to Quantitative Genetics. 1. The Ronald Press Company; New York: 1960. pp. 292–301. [Google Scholar]
- 21.Thompson WD. Effect modification and the limits of biological inference from epidemiologic data. J Clin Epidemiol. 1991;44:221–32. doi: 10.1016/0895-4356(91)90033-6. [DOI] [PubMed] [Google Scholar]
- 22.Voorman A, Lumley T, McKnight B, Rice K. Behavior of QQ-plots and genomic control in studies of gene-environment interaction. PLoS One. 2011;6:e19416. doi: 10.1371/journal.pone.0019416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen TA, Sanyal AJ. Pathophysiology guided treatment of nonalcoholic steatohepatitis. J Gastroenterol Hepatol. 2012;27(Suppl 2):58–64. doi: 10.1111/j.1440-1746.2011.07018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohen JC, Horton JD, Hobbs HH. Human fatty liver disease: old questions and new insights. Science. 2011;332:1519–23. doi: 10.1126/science.1204265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Speliotes EK, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–48. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mancina RM, et al. The MBOAT7-TMC4 Variant rs641738 Increases Risk of Nonalcoholic Fatty Liver Disease in Individuals of European Descent. Gastroenterology. 2016;150:1219–1230. e6. doi: 10.1053/j.gastro.2016.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smagris E, et al. Pnpla3I148M knockin mice accumulate PNPLA3 on lipid droplets and develop hepatic steatosis. Hepatology. 2015;61:108–18. doi: 10.1002/hep.27242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rees MG, et al. Cellular characterisation of the GCKR P446L variant associated with type 2 diabetes risk. Diabetologia. 2012;55:114–22. doi: 10.1007/s00125-011-2348-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang Y, et al. A feed-forward loop amplifies nutritional regulation of PNPLA3. Proc Natl Acad Sci U S A. 2010;107:7892–7. doi: 10.1073/pnas.1003585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arden C, et al. Elevated glucose represses liver glucokinase and induces its regulatory protein to safeguard hepatic phosphate homeostasis. Diabetes. 2011;60:3110–20. doi: 10.2337/db11-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smagris E, Gilyard S, BasuRay S, Cohen JC, Hobbs HH. Inactivation of Tm6sf2, a Gene Defective in Fatty Liver Disease, Impairs Lipidation but Not Secretion of Very Low Density Lipoproteins. J Biol Chem. 2016;291:10659–76. doi: 10.1074/jbc.M116.719955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graff M, et al. PNPLA3 gene-by-visceral adipose tissue volume interaction and the pathogenesis of fatty liver disease: the NHLBI family heart study. Int J Obes (Lond) 2013;37:432–8. doi: 10.1038/ijo.2012.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.York LW, Puthalapattu S, Wu GY. Nonalcoholic fatty liver disease and low-carbohydrate diets. Annu Rev Nutr. 2009;29:365–79. doi: 10.1146/annurev-nutr-070208-114232. [DOI] [PubMed] [Google Scholar]
- 34.Davis JN, et al. Increased hepatic fat in overweight Hispanic youth influenced by interaction between genetic variation in PNPLA3 and high dietary carbohydrate and sugar consumption. Am J Clin Nutr. 2010;92:1522–7. doi: 10.3945/ajcn.2010.30185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams CD, et al. Prevalence of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis among a largely middle-aged population utilizing ultrasound and liver biopsy: a prospective study. Gastroenterology. 2011;140:124–31. doi: 10.1053/j.gastro.2010.09.038. [DOI] [PubMed] [Google Scholar]
- 36.Yuan X, et al. Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. Am J Hum Genet. 2008;83:520–8. doi: 10.1016/j.ajhg.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chambers JC, et al. Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat Genet. 2011;43:1131–8. doi: 10.1038/ng.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larrieta-Carrasco E, et al. Association of the I148M/PNPLA3 variant with elevated alanine transaminase levels in normal-weight and overweight/obese Mexican children. Gene. 2013;520:185–8. doi: 10.1016/j.gene.2013.03.038. [DOI] [PubMed] [Google Scholar]
- 39.Giudice EM, et al. The association of PNPLA3 variants with liver enzymes in childhood obesity is driven by the interaction with abdominal fat. PLoS One. 2011;6:e27933. doi: 10.1371/journal.pone.0027933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hart CL, Morrison DS, Batty GD, Mitchell RJ, Davey Smith G. Effect of body mass index and alcohol consumption on liver disease: analysis of data from two prospective cohort studies. BMJ. 2010;340:c1240. doi: 10.1136/bmj.c1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tian C, Stokowski RP, Kershenobich D, Ballinger DG, Hinds DA. Variant in PNPLA3 is associated with alcoholic liver disease. Nat Genet. 2010;42:21–3. doi: 10.1038/ng.488. [DOI] [PubMed] [Google Scholar]
- 42.Locke AE, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.International Consortium for Blood Pressure Genome-Wide Association. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–9. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dupuis J, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–16. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Browning JD, Cohen JC, Hobbs HH. Patatin-like phospholipase domain-containing 3 and the pathogenesis and progression of pediatric nonalcoholic fatty liver disease. Hepatology. 2010;52:1189–92. doi: 10.1002/hep.23946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neel JV. Diabetes mellitus: a “thrifty” genotype rendered detrimental by “progress”? Am J Hum Genet. 1962;14:353–62. [PMC free article] [PubMed] [Google Scholar]
- 47.Szczepaniak LS, et al. Magnetic resonance spectroscopy to measure hepatic triglyceride content: prevalence of hepatic steatosis in the general population. Am J Physiol Endocrinol Metab. 2005;288:E462–8. doi: 10.1152/ajpendo.00064.2004. [DOI] [PubMed] [Google Scholar]
- 48.Stender S, Frikke-Schmidt R, Nordestgaard BG, Tybjaerg-Hansen A. The ABCG5/8 cholesterol transporter and myocardial infarction versus gallstone disease. J Am Coll Cardiol. 2014;63:2121–8. doi: 10.1016/j.jacc.2013.12.055. [DOI] [PubMed] [Google Scholar]
- 49.Victor RG, et al. The Dallas Heart Study: a population-based probability sample for the multidisciplinary study of ethnic differences in cardiovascular health. Am J Cardiol. 2004;93:1473–80. doi: 10.1016/j.amjcard.2004.02.058. [DOI] [PubMed] [Google Scholar]
- 50.Vestberg K, et al. Data quality of administratively collected hospital discharge data for liver cirrhosis epidemiology. J Med Syst. 1997;21:11–20. doi: 10.1023/a:1022835207287. [DOI] [PubMed] [Google Scholar]
- 51.Day FR, Loh PR, Scott RA, Ong KK, Perry JR. A Robust Example of Collider Bias in a Genetic Association Study. Am J Hum Genet. 2016;98:392–3. doi: 10.1016/j.ajhg.2015.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Burgess S, Small DS, Thompson SG. A review of instrumental variable estimators for Mendelian randomization. Stat Methods Med Res. 2015;0:1–26. doi: 10.1177/0962280215597579. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.