Abstract
Metabolic changes induced by oncogenic drivers of cancer contribute to tumor growth and are attractive targets for cancer treatment. Here, we found that increased growth of PTEN mutant cells was dependent on glutamine flux through the de novo pyrimidine synthesis pathway, which created sensitivity to inhibition of dihydroorotate dehydrogenase, a rate limiting enzyme for pyrimidine ring synthesis. S-phase PTEN mutant cells showed increased numbers of replication forks, and inhibitors of dihydroorotate dehydrogenase led to chromosome breaks and cell death due to inadequate ATR activation and DNA damage at replication forks. Our findings indicate that enhanced glutamine flux generates vulnerability to dihydroorotate dehydrogenase inhibition, which then causes synthetic lethality in PTEN deficient cells due to inherent defects in ATR activation. Inhibition of dihydroorotate dehydrogenase could thus be a promising therapy for patients with PTEN mutant cancers.
Keywords: PTEN, metabolism, glutamine, pyrimidine synthesis, DNA damage
Introduction
The Warburg effect, describing heightened aerobic glycolysis in tumors, played a key role in launching the field of cancer metabolism. Subsequent studies have found that glutamine is also vital for growth by fueling tricarboxylic acid cycle intermediates, phospholipid and nucleotide synthesis, and NADPH (1). Oncogenic signaling pathways have been shown to play a major role in reprogramming glucose and glutamine metabolism, thus connecting genetic mutations with metabolic alterations (2–5). PTEN (phosphatase and tensin homolog deleted on chromosome 10) is one of the most commonly mutated tumor suppressors and is a fulcrum of multiple cellular functions (6,7). PTEN’s canonical role is as a lipid phosphatase for phosphatidylinositol-3,4,5-trisphosphate, central to the phosphoinositide-3 kinase (PI3K) pathway, limiting AKT, mTOR, and RAC signaling (8–11). Inactivation of PTEN enhances glucose metabolism and diminishes DNA repair and DNA damage checkpoint pathways (12–14). Furthermore, deficient homologous recombination in PTEN mutant cells leads to sensitivity to gamma-irradiation and PARP inhibitors (13,15). The role of PTEN in metabolism, however, is incompletely understood, and in this study we examine the metabolic consequences of PTEN loss and the resulting vulnerability of PTEN mutant tumors.
Results
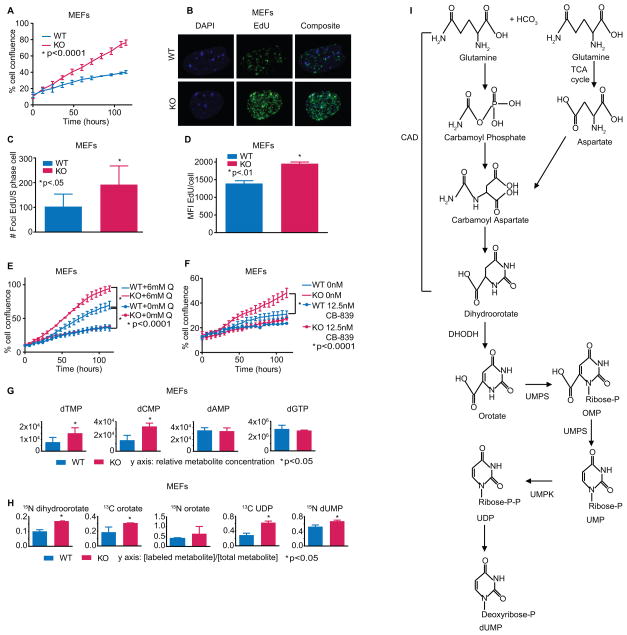
To better understand the relationship between PTEN, cell growth, and cellular metabolism, we generated Pten flox/flox primary mouse embryonic fibroblasts. Pten−/− MEFs proliferated at a higher rate than WT MEFs but showed no difference in cell death (Fig. 1A; Supplementary Fig. S1, A–C). This increased proliferation was associated with an increase in the proportion of cells within S-phase and higher numbers of replication forks per S-phase cell (Fig. 1, B–D; Supplementary Fig. S1D). There was no difference in mitochondrial function between Pten−/− and WT MEFs, suggesting a different source of altered replication (Supplementary Fig. S1, E–F).
Fig. 1.
(A) Growth of Pten WT and KO MEFs (one-way ANOVA, *p<.0001, n=3). (B) MEFs labeled with EdU. Representative confocal microscopy images. (C) Quantification of Fig.1B (Student’s t-test, *p<.05, n=6). (D) MEFs labeled with EdU; flow cytometry determined the mean fluorescence intensity among cells positively stained (Student’s t-test, *p<.01, n=3). (E) Pten WT and KO MEFs in media containing full glutamine (6mM) or no added glutamine (one-way ANOVA, *p<.0001, n=3). (F) MEFs treated with 12.5nM CB-839 or control (one-way ANOVA, *p<.0001, n=3). (G) Relative metabolite concentrations of DNA nucleotide precursors (dGMP was unable to be measured so dGTP was used) (Student’s t-test, *p<.05, n=3). (H) Relative metabolite levels of glutamine-labeled de novo pyrimidine synthesis intermediates (Student’s t-test, *p<.05, n=3). Data were also analyzed with IMPaLA: 13C glutamine-derived pyrimidine metabolism enrichment in PTEN−/− MEFs q-value = 3.92×10−09. (I) Schematic of the de novo pyrimidine synthesis pathway. Not every intermediate was measured in our mass spec panel. Data shown as means + SD.
Upon testing the potential role of glutamine for explaining the increased growth of Pten−/− cells, we found that the growth advantage of Pten−/− MEFs was dependent on glutamine: depletion of glutamine or addition of the glutaminase inhibitor CB-839 collapsed the growth difference between Pten−/− and WT MEFs (Fig. 1, E–F). To better understand the relationship between PTEN and glutamine, we performed steady state metabolomic profiling of 292 aqueous metabolites to determine if loss of PTEN triggers abnormal cellular metabolism to increase growth. Unbiased global metabolic assessment of WT and Pten−/− MEFs revealed that seven of the ten most upregulated pathways in Pten−/− MEFs involved nucleotide synthesis and DNA metabolism, including a higher concentration of pyrimidine 2-deoxyribonucleotides in Pten−/− MEFs (Fig. 1G; Supplementary Fig. S2, A–B). Because glutamine contributes both nitrogen and carbon to pyrimidines (16), we performed metabolic flux analysis with heavy-isotope 15N or 13C-labeled glutamine, which showed increased synthesis of dihydroorotate, orotate, and other components of the de novo pyrimidine synthesis pathway in Pten−/− MEFs relative to WT (Fig. 1, H–I; Supplementary Fig. S2C). In addition, the pyrimidine metabolism gene set was upregulated in mRNA from Pten−/− MEFs (Supplementary Fig. S2D). Although Pten−/− fibroblasts had somewhat elevated stead-state glucose metabolism and glycolytic flux relative to WT, depletion of glucose from the medium did not rescue the differences in cell growth, suggesting that glutamine was more critical for the growth advantage of Pten−/− cells (Supplementary Fig. S2, A, E–F). Nucleotide synthesis is a prerequisite for cellular growth, and Pten−/− MEFs appear to channel glutamine for this purpose.
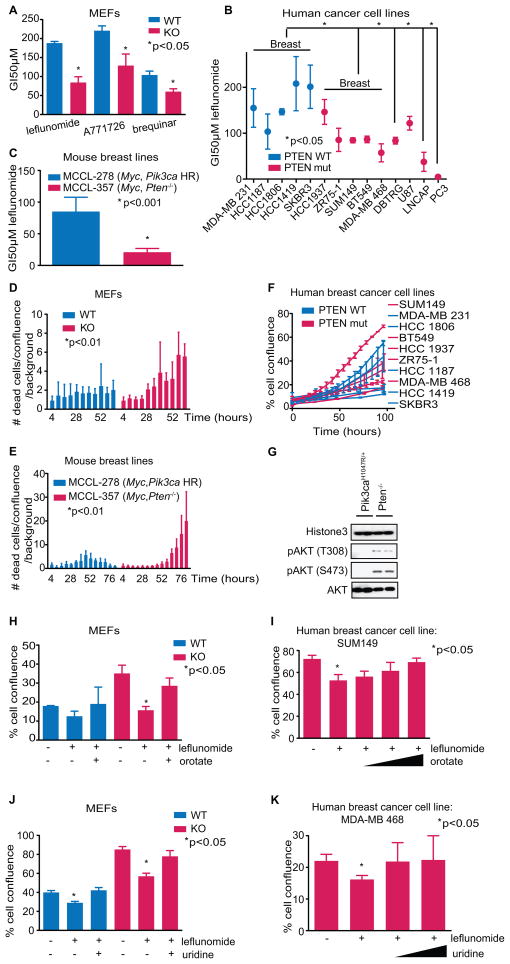
The fourth step of de novo pyrimidine synthesis in mammals is the conversion of dihydroorotate to orotate, catalyzed by dihydroorotate dehydrogenase (DHODH) (17). To see if orotate contributes to the growth effects observed, the effect of DHODH inhibitors on cell proliferation was examined. Pten−/− MEFs were about 3-fold more sensitive to leflunomide, a DHODH inhibitor, (18) than WT MEFs were (Fig. 2A; Supplementary Fig. S3, A–B). Pten−/− MEFs were also more sensitive to A771726, the active metabolite of leflunomide (18,19), as well as another DHODH inhibitor, brequinar (18), indicating that the observed effects were likely through DHODH (Fig. 2A).
Fig. 2.
(A) Pten WT and KO cells treated with dose titrations of leflunomide, A771726, or brequinar to determine GI50s (Student’s t-test, *<.05, n=3). (B, C) Cells treated with dose titrations of leflunomide to determine GI50s (Student’s t-test, *p-values on figures, n=3). (D, E) Cells treated with 100μM leflunomide and DRAQ7 to monitor accumulation of cell death, in intervals of 6 hours (one-way ANOVA, *p-values on the figures). (F) Human breast cancer cell line growth rates. (G) Immunoblots of pAKT in nuclear fractions of Pten−/− and Pik3ca mutant MEFs. (H) Cells treated with 50μM leflunomide in combination with 0 or 640μM orotate. Confluence of cells after 5 days of treatment was measured (Student’s t-test, *p<.05, n=3). (I) Cells treated with 50μM leflunomide in combination with 0, 31.25, 62.5 or 125μM orotate. Confluence of cells after 5 days was measured (Student’s t-test, *p<.05, n=3). (J) Cells treated with 100μM leflunomide in combination with 0 or 3.125mM uridine. Confluence of cells after 5 days of treatment was measured (Student’s t-test, *p<.05, n=3). (K) Cells treated with 100μM leflunomide in combination with 0, 3.125, or 6.25mM uridine. Confluence of cells after 5 days was measured (Student’s t-test, *p<.05, n=3). Data shown as means + SD.
To determine whether PTEN genotype is predictive of sensitivity to DHODH inhibition in cancer cells, we tested human breast, glioblastoma, and prostate cell lines with DHODH inhibitors. Consistently, the GI50 of the PTEN mutant lines was lower than that of WT (Fig. 2B; Supplementary Fig. S3C). Mouse cancer lines MCCL-357 (Myc, Pten−/−) and CaP8 (Pten−/−) were also more sensitive than mouse cancer lines MCCL-278 (Myc, Pik3ca H1047R) and Myc-CaP (Myc) were (Fig. 2C; Supplementary Fig. S3, D–E) (20,21). Moreover, Pten−/− MEFs, PTEN mutant human breast cancer cell lines, and Pten−/− mouse breast lines displayed an increased accumulation of dead cells over time upon treatment with leflunomide (Fig. 2, D–E; Supplementary Fig. S3F). It is important to note that sensitivity to leflunomide was not associated with the proliferation rates of human breast, mouse breast, or mouse prostate tumor cell lines (Fig. 2F; Supplementary Fig. S3, G-H). Additionally, consistent with previous reports (22), we found that Pten homozygous deletion caused greater AKT phosphorylation than Pik3ca missense mutation did. This was particularly prominent in the nuclear fractions, where AKT may phosphorylate nuclear substrates (Fig 2G; Supplementary Fig. S3, I–J).
To independently test if DHODH inhibition is detrimental to PTEN deficient cells, we performed a rescue experiment with orotate, the metabolite directly downstream of DHODH, as well as with uridine. Increasing concentrations of orotate or uridine rescued growth inhibition by leflunomide (Fig. 2, H-K; Supplementary Fig. S4, A–C). In addition, siRNA against DHODH preferentially killed PTEN mutant cells, verifying that DHODH was the target of the small molecule inhibitors (Supplementary Fig. S4D). There was no endogenous difference in DHODH protein level between Pten−/− and WT MEFs, and A771726 did not affect PI3K signaling (Supplementary Fig. S4, E–F). Consistent with prior reports, CAD phosphorylation downstream of mTORC1 was increased in Pten−/− cells, likely contributing to the push of glutamine flux into the pyrimidine synthesis pathway (Supplementary Fig. S4, G–H) (5). Pten−/− cells were more sensitive than WT cells to the mTOR inhibitor RAD001 as expected, but RAD001 did not synergize with leflunomide (Supplementary Fig. S4, I–K) (10,23). Interestingly, treatment with nucleotide analog inhibitors – 5-flurouracil or mercaptopurine – did not show a differential sensitivity, demonstrating that Pten−/− MEFs are selectively vulnerable to inhibition of de novo pyrimidine synthesis (Supplementary Fig. S4, L–M).
Myc activation is known to cause glutamine addiction (4). CaP8 (Pten−/−) cells were nearly as sensitive to glutamine deprivation as Myc-CaP (Myc oncogene transformed) cells were, substantiating that a notable level of glutamine dependency is also elicited by PTEN loss (Supplementary Fig. S4N). Since Myc-CaP cells were resistant to leflunomide, it seems it is not the entry alone of glutamine but its flux into pyrimidines that is important (Supplementary Fig. S3D). While MYC is known to largely direct glutamine to the TCA cycle and phospholipid synthesis (4), our data suggest that Pten loss in MEFs causes glutamine to cascade through the de novo pyrimidine synthesis pathway, creating the point of vulnerability to DHODH inhibition.
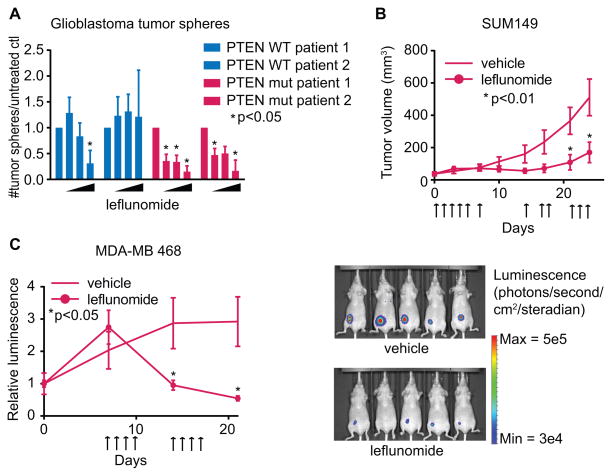
To determine how clinically relevant leflunomide may be as a targeted cancer therapy, we grew patient-derived glioblastomas as 3-dimensional neurospheres. Re-formation of neurospheres was inhibited at lower concentrations of leflunomide in PTEN deficient samples (Fig. 3A; Supplementary Fig. S5A). Additionally, we treated two PTEN mutant triple negative breast cancer xenografts with leflunomide, dosing orally as is done clinically. Tumors slowed or regressed upon treatment; remarkably, even very large tumors (4×107 photons) regressed after only 1 week of treatment, indicating that leflunomide may have use for neoadjuvant therapy (Fig. 3, B–C; Supplementary Fig. S5B). To ensure the effect in vivo is specific to PTEN loss, MCCL-357 and MCCL-278 xenografts were treated with leflunomide; MCCL-357 xenografts had a 4-fold better response than MCCL-278 xenografts did (Supplementary Fig. S5C).
Fig. 3.
(A) Dispersed (single cell suspension) glioblastomas were treated with DMSO or 50, 100, or 200μM leflunomide for 5 days. The number of re-formed 3-dimensional tumor spheres was quantified and normalized to untreated samples. (Student’s t-test, *p<.05, n=3). (B) SUM149 xenografts. Mice were treated with 100mg/kg leflunomide or vehicle on days indicated with arrows (one-way ANOVA with multiple t-tests, corrected for multiple comparisons, *p<.01 for ANOVA and t-tests, n=6). (C) MDA-MB 468 xenografts expressing luciferase, normalized to control. Treatment was started on day 7, with 100mg/kg leflunomide or vehicle for four consecutive days each week (one-way ANOVA with multiple t-tests, corrected for multiple comparisons, *p<.05 for ANOVA and t-tests, n=5). Right panel: luminescence of treated and control mice after 2 weeks of treatment. Data shown as means + SD for (A) and + SEM for (B)-(C).
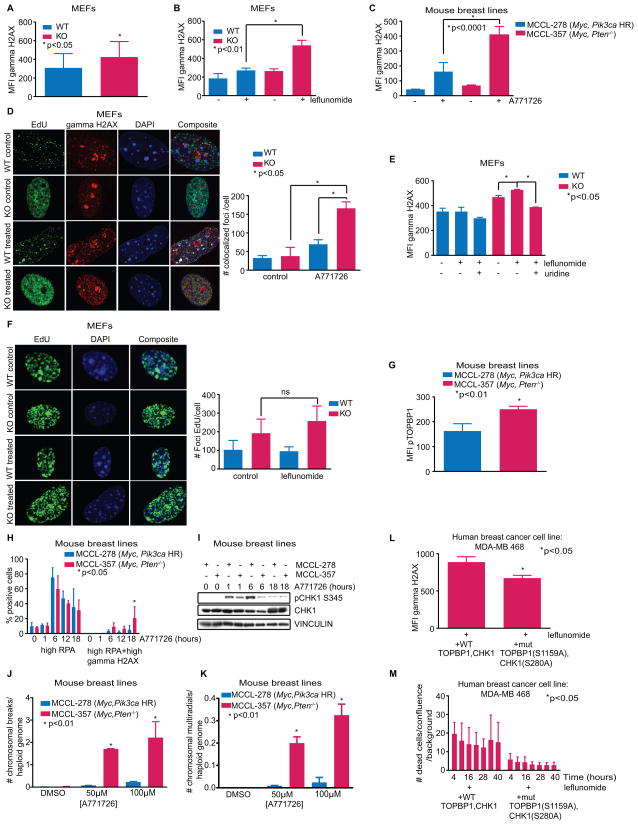
It is logical that a blockade of pyrimidine synthesis would stop cells from dividing, and leflunomide has been previously established as a cytostatic drug (18). What is more enigmatic, however, is why it would cause PTEN−/− cells to die. Consistent with prior reports (24), Pten−/− MEFs had a higher level of gamma-H2AX, an indicator of DNA damage (Fig. 4A). We hypothesized that the dearth of pyrimidine deoxynucleotides caused by DHODH inhibition would exacerbate this defect, and discovered that leflunomide (or A771726) augmented DNA damage to a significantly greater degree in PTEN deficient cells and that this damage co-localized with replication forks labeled with EdU (Fig. 4, B–D; Supplementary Fig. S6, A–B). Leflunomide-induced DNA damage was rescued by uridine, demonstrating that damage is likely instigated by pyrimidine depletion (Fig. 4E). The greater number of replication forks we described in Pten−/− MEFs remained intact after 24h of treatment with leflunomide, showing that the cells continue to replicate despite the presence of DNA damage (Fig. 1B, 4F; Supplementary Fig. S6, C–D).
Fig. 4.
(A) Cells were labeled with a gamma-H2AX antibody. Flow cytometry determined the mean fluorescence intensity (MFI) (Student’s t-test, *p<.05, n=3). (B-C) Cells treated with 100μM leflunomide or A771726 were labeled with a gamma-H2AX antibody. Flow cytometry determined the mean fluorescence intensity (MFI) (Student’s t-test, *p-values on figures, n=3). (D) MEFs treated with 150μM A771726 for 24h, labeled with EdU and gamma-H2AX. Left: representative confocal microscopy images. Right: quantified EdU and gamma-H2AX colocalized foci (Student’s t-test, *p<.05, n=3). (E) Cells treated with 100μM leflunomide with or without uridine and labeled with a gamma-H2AX antibody. Flow cytometry determined the mean fluorescence intensity (MFI) (Student’s t-test, *p-values on figures, n=3). (F) MEFs treated with 100μM leflunomide or control for 48h and labeled with EdU. Left: representative confocal microscopy images. Right: quantification of the number of foci per cell (Student’s t-test, p>.05, n=6). (G) Cells were labeled with a pTOPBP1 S1159 antibody. Flow cytometry determined the mean fluorescence intensity (MFI) (Student’s t-test, *p<.05, n=3). (H) Cells treated with 150μM A771726 for times indicated and labeled with antibodies to RPA and gamma-H2AX. Flow cytometry determined the percentage of the cell population positively-stained for RPA alone or both RPA and gamma-H2AX (Student’s t-test, *p<.05, n=4). (I) pCHK1 immunoblot after 150μM A771726 treatment for times indicated. (J-K) Quantified chromosomal breaks and multiradial formations per haploid genome (Student’s t-test, *p-values on figure, cells scored/replicate>100). (L) PTEN mutant cells were transfected with either WT TOPBP1 and CHK1, or mutants incapable of being phosphorylated by AKT, and labeled with a gamma-H2AX antibody after 100μM leflunomide treatment. Flow cytometry determined the mean fluorescence intensity (MFI) (Student’s t-test, *p<.05, n=3). (M) PTEN mutant cells were transfected with either WT TOPBP1 and CHK1, or mutants incapable of being phosphorylated by AKT, and DRAQ7 was used to monitor accumulation of cell death in intervals of 6 hours (one-way ANOVA, *p<.05, n=3). Data shown as means + SD.
Depletion of nucleotide pools normally activates the ATR checkpoint at replication forks in S-phase cells (25). ATR checkpoint activation at stalled forks requires two signals, one through single-strand DNA binding protein (RPA) interaction with single-strand DNA to recruit the ATRIP-ATR complex, and a second signal through TOPBP1 interaction with the ATR activation domain (26,27). Deletion of PTEN in cells is known cause poor ATR checkpoint activation, and AKT phosphorylation of TOPBP1 on serine 1159 and CHK1 on serine 280 inhibits their function (14,24,28,29). Prior work showed increased CHK1 serine 280 phosphorylation in PTEN−/− cells, which reduced CHK1 activity (24). We found greater TOPBP1 S1159 phosphorylation and concomitantly less TOPBP1 localization to replication forks in Pten−/− cells (Fig. 4G; Supplementary Fig. S6, E–F). Diminished AKT activity through PI3K inhibition also reduced leflunomide-induced DNA damage (Supplementary Fig. S6G). To further investigate the response to DNA damage occurring at Pten−/− forks, we examined the interaction of RPA and gamma-H2AX by flow cytometry. An increase in RPA signal was first achieved regardless of PTEN genotype in the presence of A771726, followed by a shift toward both RPA and gamma-H2AX-positive cells in Pten−/− MCCL-357 but not in Pten WT MCCL-278 cells (Fig. 4H). Moreover, gamma-H2AX appeared almost exclusively in RPA-positive MCCL-357 cells treated with A771726 (Supplementary Fig. S6H). A771726 also triggered ATR phosphorylation of CHK1 at serine 345 in Pten WT but to a much lesser extent in Pten−/− cells (Fig. 4I; Supplementary Fig. S6I). Thus, Pten−/− cells appear to be incapable of generating an appropriate activation of the ATR-CHK1 checkpoint at replication forks. Activation of CHK1 in MCCL-278 cells declined as RPA declined, suggesting that Pten WT cells eventually recovered from DHODH inhibition, while Pten−/− cells instead accumulated damage at 18h (Fig. 4I). By 48h this genomic stress manifested in a greater number of chromosome gaps, breaks, and multiradial formations in MCCL-357 cells treated with A771726 compared to MCCL-278 cells (Fig. 4, J–K; Supplementary Fig. S6, J–K). These findings are consistent with the sensitivity to hydroxyurea that occurs in the setting of an ATR inhibitor (30). Furthermore, we were able to rescue DNA damage and cell death in leflunomide-treated PTEN mutant cells by transfecting cells with TOPBP1 and CHK1 mutants incapable of being phosphorylated by AKT (S1159A and S280A, respectively), demonstrating that the synthetic lethality between pyrimidine depletion and mutation of PTEN is due to the AKT-mediated defects in the ATR pathway (Fig. 4, L–M).
Based on our data, we propose that inhibition of DHODH in PTEN deficient cells first causes stalled forks due to inadequate nucleotide pools required to support replication, and that sustained treatment leads to insufficient ATR activation due to AKT phosphorylation of TOPBP1 and CHK1, leading to a buildup of DNA damage and cell death. PTEN WT cells do not exhibit this dependency on pyrimidine synthesis and have fewer forks per cell, perhaps because ATR-CHK1 coordinates origin firing during S-phase (31). In PTEN WT cells, treatment initially increased the RPA signal and triggered transient phosphorylation of CHK1, while longer treatment led to abated RPA with little concurrent increase in gamma-H2AX, explaining the largely unaffected WT population upon DHODH inhibition (Supplementary Fig. S7). While Pik3ca mutant cells also exhibit AKT signaling, their relative resistance to DHODH inhibitors suggests that a PI3K signaling dosage-effect due to their lower level of AKT activation may be important.
Discussion
PTEN loss leads to chemoresistance in prostate cancer, a poorer response to trastuzumab in triple negative breast cancer, and a shorter survival time in patients with gliomas (32). Targeting consequences of PTEN loss could be beneficial, particularly since the standard of care for the aforementioned cancers is primarily chemotherapy and radiation. Inhibiting DHODH has the advantage of affecting a specific pathway of glutamine flux downstream of glutaminase, thus preserving glutamine’s other important functions in the cell. This increases the specificity of DHODH inhibitors to cells which are dependent on glutamine’s role in pyrimidine synthesis per se, and is perhaps why their toxicity is low enough to be taken as a daily medication by rheumatoid arthritis and multiple sclerosis patients (19). It has been shown that leflunomide inhibits B-cell and T-cell proliferation, contributing to its immunomodulating effects (33,34). It is possible that preexisting B and T cells can still function in the presence of DHODH inhibitors, thus potentially arguing for the benefit of immunotherapy in combination with a DHODH inhibitor, perhaps in a metronomic therapy pattern. Here, we show that high activation of AKT toward TOPBP1 and CHK1 that down regulates ATR activation at replication forks compounded with enhanced pyrimidine flux that both occur as a consequence of PTEN inactivation contributes to the observed synthetic lethality between PTEN mutation and DHODH inhibition. We hope that DHODH inhibitors will be a promising therapy for patients with PTEN deficient cancers.
Methods
Immunoblotting
Samples were lysed in 2x Laemelli sample buffer before separation by SDS-PAGE and transferring to PVDF membranes, blocked with 10% milk in TBST for 1 hour, and incubated with primary antibodies overnight. Antibodies: PTEN 6H2.1 (Millipore 04-035), DHODH (Protein Tech 14877-1-AP), vinculin (Sigma), pCHK1 (Cell Signaling 2341), CHK1 G-4 (Santa Cruz sc-8408), pCAD (Cell Signaling 12662), CAD (Cell Signaling 11933). HRP conjugated secondary antibodies were used to detect protein signals.
Cell culture
MEFs and mouse breast tumor lines: DMEM (Corning mt10013cv) supplemented with 10% FBS (Atlanta Biologicals), 1% pen/strep (Fisher 30002ci) and 2mM L-glutamine (total 6mM) (Fisher MT25005CI). MDA-MB468, MDA-MB 231, Myc-CaP (2015), and U87: DMEM supplemented with 10% FBS and 1% pen/strep. HCC1419, HCC1187, HCC 1937, HCC 1806, BT549, ZR75-1, PC3, LNCAP, DBTRG: RPMI (Fisher 10040cv) supplemented with 10% FBS and 1% pen/strep. CaP8 cells (2015): DMEM with 10% FBS, 1% pen/strep, and 5ug/mL insulin (Sigma I9278). Neurospheres: stem cell media with 10ug/mL FGF (R&D Systems 233-FB-025), 20ug/mL EGF (Peprotech AF-100-15) and heparin (from Dr. Raymund Yong, May 2015). All cells were cultured in a 37°C incubator with humidity and 5% CO2. Cell lines were obtained from ATCC (which authenticate cell lines using several methods including DNA fingerprinting) in 2006, with the exception of MEFs, MCCL-278, and MCCL-357 which were produced in our lab from mice (2012-2016). Cell lines were clear of mycoplasma as determined by the Lonza kit (LT07-418) within 6 months of their use. Cell lines were further authenticated in 2015 by LabCorp using a short tandem repeat method.
Mouse Embryonic Fibroblasts
Embryos were harvested 14 days after conception from Pten flox/flox mice from Jackson Labs. MEFs were treated with adenovirus diluted in growth media supplemented with polybrene with or without cre recombinase and studied passages 2–5 post infection. Please see supplement for additional details.
Proliferation assay
1500 cells per well (mouse cells) or 3000 cells per well (human cells) were plated in 96 well plates (Corning 720089). Growth rates were determined using the phase-confluency readings on an IncuCyte ZOOM (Essen Biosciences) on live cells over time.
Metabolite labeling
For glutamine flux, media without added glutamine (Corning 17-207-CV) was supplemented with 13C glutamine (fully labeled) or 15N glutamine (amide labeled) (Cambridge Isotope Labs). For glucose flux, media without added glucose (Corning 17-207-CV) was supplemented with 13C glucose (fully labeled) (Cambridge Isotope Labs). Cells were plated in 10cm dishes and grown in normal media. 1 hour prior to metabolite extraction, media was aspirated and replaced with heavy isotope-labeled media.
Metabolic extraction
Metabolites were extracted in methanol. Please see supplement for details.
Targeted Mass Spectrometry
Mass Spec was performed by the core facility at Beth Israel Deaconess Medical Center. Please see supplement for details.
Cell cycle Analysis
Instructions for the FlowCellect™ Bivariate Cell Cycle Kit (Millipore FCCH025102) were followed. Fluorescence was measured on a Guava® flow cytometer. BrdU was pulsed for 18hrs.
Cell death
Instructions for the FlowCellect™ Annexin Red Kit (Millipore FCCH100108) were followed. Fluorescence was measured on a Guava® flow cytometer.
Seahorse Analysis
A Seahorse XF (Agilent) was used to determine the oxygen consumption and extracellular acidification rates. Cells were plated in the Seahorse cartridge and pH-adjusted media was added. Oligomycin, FCCP, and rotenone (Seahorse XF kit, Agilent) were injected into cartridge wells. Controls for calibration and establishing baseline were used.
Drug response assays
Cells were plated in 96-well plates at a density of 1500 or 3000 cells per well. Leflunomide (Sigma PHR1378-1G), A771726 (Sigma SML0936), mercaptopurine (Sigma 852678), brequinar (Sigma SML0113), 5-fluorouracil (Millipore 343922), RAD001 and GDC0491(obtained from Stand Up to Cancer PI3K Dream Team pharmacy), and CB-839 (MedChemexpress HY-12248) were dissolved in DMSO. Sensitivity was determined by a dose-response titration for each cell line, with an equivalent amount of DMSO in each well: 300μL media with drug was added to one column of wells, and 150μL media with equivalent DMSO was added to remaining wells. Serial dilutions of 150μL resulted in a gradient with half the drug concentration as the previous column while maintaining the same amount of DMSO. GI50 values were calculated by linear interpolation: the maximum growth confluence for a cell line prior to growth plateau was divided by 2 to obtain the 50% confluence value. A linear regression curve was calculated using drug concentrations as x-values and confluence as y-values for points surrounding the 50% value. Linear interpolation using the regression line yielded the GI50 concentration. For cell death assays, DRAQ7TM (Cell Signaling 7406S) was added to the media at a 1:200 dilution and red fluorescence was measured in addition to phase in live-cell imaging to measure accumulation of dead cells. An IncuCyte ZOOM was used.
Gamma-H2AX measurement
Instructions for the FlowCellect™ Cell Cycle Checkpoint H2A.X DNA Damage Kit (Millipore FCCH12542) were followed. Briefly, cells were fixed and permeabilized, followed by staining with an anti-phospho-H2A.X antibody and propidium iodide. For co-staining with RPA, an additional step was performed during which cells were incubated with an RPA antibody (Abcam ab79398) for 1 hour and secondary antibody for 1 hour. (Propidium iodide was not used in this setting.) Fluorescence was measured on a Guava® flow cytometer.
EdU detection
Instructions for the EdU cell proliferation Kit (Millipore 17-10525) were followed. Cells were fixed and permeabilized following a 45min EdU pulse and a click chemistry reaction was used to add a fluorescent tag. Fluorescence was measured on a Guava® flow cytometer or by immunofluorescence.
Immunofluorescence
Cells were plated on cover slips in media. For detecting replication forks: following a 45min EdU pulse, cover slip-attached cells were fixed and permeabilized, and detected after azide conjugation to EdU. For detecting gamma-H2AX or pTOPBP1: cells were incubated with primary antibody (Upstate Cell Signaling and Bethyl A300-111A-M, respectively) overnight at 4°C and with secondary antibody for 2 hours at room temperature. Images were taken using a Zeiss LSM880 Airyscan confocal microscope at 63X, and foci number and colocalization was quantified with Image J.
pTOPBP1 measurement
Cells were fixed and permeabilized, followed by incubation with primary antibody (Abgent AP3774a) for 2 hours at room temperature and secondary antibody for 2 hours at room temperature. Fluorescence was measured on a Guava® flow cytometer.
Karyotyping
Chromosomal analysis was performed on cultured cells treated with colcemid by Dr. Murty Vundavalli at Columbia University; please see supplement for details.
Orotate Rescue
Orotate (Sigma O2750) was dissolved in DMSO. Cells were plated at fixed concentration leflunomide with increasing concentrations of orotate, keeping DMSO constant in all wells.
Uridine Rescue
Uridine (Sigma U 3750) was dissolved in media. Cells were plated at fixed concentration leflunomide with increasing concentrations of orotate, keeping DMSO constant in all wells.
RNA interference
siRNA for DHODH was purchased from Qiagen. Cells were transfected using lipofectamine (Invitrogen 11668-019) and knockdown was confirmed at 48 hours. Scrambled siRNA was used as a control.
Transfection
Plasmids were electroporated into cells (1 million) using an Amaxa Nucleofector™ 2b (Lonza) and Cell Line Nucleofector® Kit V (Lonza VCA-1003). An mcherry plasmid was co-transfected to determine transfection efficiency and to gate transfected cells for flow cytometry experiments.
Xenografts
6-week old female nu/nu mice were engrafted orthotopically with either 5 million SUM149, 5 million MDAMB 468-luciferase, 1 million MCCL-357, or .75million MCCL-278 cells. Mice were treated by oral gavage with 100mg/kg leflunomide or vehicle (1% carboxymethylcelluose in water). Animal experiments were approved by an Institutional Animal Care and Use Committee.
Neurosphere sensitivity assay
Neurospheres were disrupted by manual pipetting until single cell suspension was achieved, and 10,000cells/well were plated in low-attachment 6-well plates (Fisher 3471). After 5 days, neurospheres were counted; sphere-forming ability is an indicator of tumorigenicity. Dense clusters >.05mm in diameter were counted as true tumor spheres. Spheres were measured on ImageJ and quantified.
Statistical Analysis
ANOVA or student’s t-tests were used to test means between groups. Correction for multiple comparisons was added where needed. Analysis was done using GraphPad Prism 6 or Microsoft Excel.
Supplementary Material
Statement of significance.
We have found a prospective targeted therapy for PTEN deficient tumors, with efficacy in vitro and in vivo in tumors derived from different tissues. This is based upon changes in glutamine metabolism, DNA replication, and DNA damage response which are consequences of inactivation of PTEN.
Acknowledgments
Financial Support: This work was supported by NCI R01CA082783, R01CA155117, and P01CA97403 (R.P.), and partially supported by NIH grants 5P01CA120964 and 5P30CA006516 (J.MA.), and R01 GM041890 and the Breast Cancer Research Foundation (L.C.).
We’d like to thank the members of the Parsons lab for providing feedback on this manuscript, and Deepti Mathur’s thesis committee for helping shape the project. Confocal experiments were performed at the Microscopy core facility at Icahn School of Medicine at Mount Sinai. We thank Min Yuan and Susanne Breitkopf for help with mass spectrometry work. We also thank Dr. Gerard Karsenty for use of his Seahorse XF and Dr. Grzegorz Sumara for training us to use it. TOPBP1 WT and mutant plasmids were graciously supplied by Dr. Weei-Chin Lin.
Footnotes
Conflict of Interest: The authors disclose no potential conflicts of interest.
References
- 1.Dang CV. Links between metabolism and cancer. Genes & development. 2012;26(9):877–90. doi: 10.1101/gad.189365.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shim H, Chun YS, Lewis BC, Dang CV. A unique glucose-dependent apoptotic pathway induced by c-Myc. Proceedings of the National Academy of Sciences. 1998;95(4):1511–6. doi: 10.1073/pnas.95.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elstrom RL, Bauer DE, Buzzai M, Karnauskas R, Harris MH, Plas DR, et al. Akt stimulates aerobic glycolysis in cancer cells. Cancer research. 2004;64(11):3892–9. doi: 10.1158/0008-5472.CAN-03-2904. [DOI] [PubMed] [Google Scholar]
- 4.Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang X-Y, Pfeiffer HK, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proceedings of the National Academy of Sciences. 2008;105(48):18782–7. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ben-Sahra I, Howell JJ, Asara JM, Manning BD. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science. 2013;339(6125):1323–8. doi: 10.1126/science.1228792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, et al. PTEN, a Putative Protein Tyrosine Phosphatase Gene Mutated in Human Brain, Breast, and Prostate Cancer. Science. 1997;275(5308):1943–7. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 7.Steck PA, Pershouse MA, Jasser SA, Yung WKA, Lin H, Ligon AH, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23. 3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15(4):356–62. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 8.Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3, 4, 5-trisphosphate. Journal of Biological Chemistry. 1998;273(22):13375–8. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 9.Stambolic V, Suzuki A, De La Pompa JL, Brothers GM, Mirtsos C, Sasaki T, et al. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95(1):29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 10.Podsypanina K, Lee RT, Politis C, Hennessy I, Crane A, Puc J, et al. An inhibitor of mTOR reduces neoplasia and normalizes p70/S6 kinase activity in Pten+/− mice. Proceedings of the National Academy of Sciences. 2001;98(18):10320–5. doi: 10.1073/pnas.171060098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mense SM, Barrows D, Hodakoski C, Steinbach N, Schoenfeld D, Su W, et al. PTEN inhibits PREX2-catalyzed activation of RAC1 to restrain tumor cell invasion. Sci Signal. 2015;8(370):ra32-ra. doi: 10.1126/scisignal.2005840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong J, Kim P, Peacock J, Yau T, Mui A-F, Chung S, et al. Pten (phosphatase and tensin homologue gene) haploinsufficiency promotes insulin hypersensitivity. Diabetologia. 2007;50(2):395–403. doi: 10.1007/s00125-006-0531-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mendes-Pereira AM, Martin SA, Brough R, McCarthy A, Taylor JR, Kim JS, et al. Synthetic lethal targeting of PTEN mutant cells with PARP inhibitors. EMBO molecular medicine. 2009;1(6–7):315–22. doi: 10.1002/emmm.200900041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shtivelman E, Sussman J, Stokoe D. A role for PI 3-kinase and PKB activity in the G2/M phase of the cell cycle. Current Biology. 2002;12(11):919–24. doi: 10.1016/s0960-9822(02)00843-6. [DOI] [PubMed] [Google Scholar]
- 15.Bassi C, Ho J, Srikumar T, Dowling R, Gorrini C, Miller S, et al. Nuclear PTEN controls DNA repair and sensitivity to genotoxic stress. Science. 2013;341(6144):395–9. doi: 10.1126/science.1236188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cory JG, Cory AH. Critical roles of glutamine as nitrogen donors in purine and pyrimidine nucleotide synthesis: asparaginase treatment in childhood acute lymphoblastic leukemia. In vivo. 2006;20(5):587–9. [PubMed] [Google Scholar]
- 17.Stamato TD, Patterson D. Biochemical genetic analysis of pyrimidine biosynthesis in mammalian cells. II. Isolation and characterization of a mutant of Chinese hamster ovary cells with defective dihydroorotate dehydrogenase (EC 1.3. 3. 1) activity. Journal of cellular physiology. 1979;98(3):459–68. doi: 10.1002/jcp.1040980304. [DOI] [PubMed] [Google Scholar]
- 18.Greene S, Watanabe K, Braatz-Trulson J, Lou L. Inhibition of dihydroorotate dehydrogenase by the immunosuppressive agent leflunomide. Biochemical Pharmacology. 1995;50(6):861–7. doi: 10.1016/0006-2952(95)00255-x. [DOI] [PubMed] [Google Scholar]
- 19.Munier-Lehmann Hln, Vidalain P-O, Tangy Fdr, Janin YL. On dihydroorotate dehydrogenases and their inhibitors and uses. Journal of medicinal chemistry. 2013;56(8):3148–67. doi: 10.1021/jm301848w. [DOI] [PubMed] [Google Scholar]
- 20.Jiao J, Wang S, Qiao R, Vivanco I, Watson PA, Sawyers CL, et al. Murine cell lines derived from Pten null prostate cancer show the critical role of PTEN in hormone refractory prostate cancer development. Cancer research. 2007;67(13):6083–91. doi: 10.1158/0008-5472.CAN-06-4202. [DOI] [PubMed] [Google Scholar]
- 21.Stratikopoulos EE, Dendy M, Szabolcs M, Khaykin AJ, Lefebvre C, Zhou M-M, et al. Kinase and BET inhibitors together clamp inhibition of PI3K signaling and overcome resistance to therapy. Cancer cell. 2015;27(6):837–51. doi: 10.1016/j.ccell.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, Neve RM, Kuo W-L, Davies M, et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer research. 2008;68(15):6084–91. doi: 10.1158/0008-5472.CAN-07-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neshat MS, Mellinghoff IK, Tran C, Stiles B, Thomas G, Petersen R, et al. Enhanced sensitivity of PTEN-deficient tumors to inhibition of FRAP/mTOR. Proceedings of the National Academy of Sciences. 2001;98(18):10314–9. doi: 10.1073/pnas.171076798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Puc J, Parsons R. PTEN loss inhibits CHK1 to cause double stranded-DNA breaks in cells. Cell Cycle. 2005;4(7):927–9. doi: 10.4161/cc.4.7.1795. [DOI] [PubMed] [Google Scholar]
- 25.Tibbetts RS, Cortez D, Brumbaugh KM, Scully R, Livingston D, Elledge SJ, et al. Functional interactions between BRCA1 and the checkpoint kinase ATR during genotoxic stress. Genes & development. 2000;14(23):2989–3002. doi: 10.1101/gad.851000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300(5625):1542–8. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 27.Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124(5):943–55. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 28.Liu K, Graves JD, Scott JD, Li R, Lin W-C. Akt switches TopBP1 function from checkpoint activation to transcriptional regulation through phosphoserine binding-mediated oligomerization. Molecular and cellular biology. 2013;33(23):4685–700. doi: 10.1128/MCB.00373-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu K, Paik JC, Wang B, Lin FT, Lin WC. Regulation of TopBP1 oligomerization by Akt/PKB for cell survival. The EMBO journal. 2006;25(20):4795–807. doi: 10.1038/sj.emboj.7601355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toledo Luis I, Altmeyer M, Rask M-B, Lukas C, Larsen Dorthe H, Povlsen Lou K, et al. ATR Prohibits Replication Catastrophe by Preventing Global Exhaustion of RPA. Cell. 2013;155(5):1088–103. doi: 10.1016/j.cell.2013.10.043. [DOI] [PubMed] [Google Scholar]
- 31.Shechter D, Costanzo V, Gautier J. ATR and ATM regulate the timing of DNA replication origin firing. Nat Cell Biol. 2004;6(7):648–55. doi: 10.1038/ncb1145. [DOI] [PubMed] [Google Scholar]
- 32.Keniry M, Parsons R. The role of PTEN signaling perturbations in cancer and in targeted therapy. Oncogene. 2008;27(41):5477–85. doi: 10.1038/onc.2008.248. [DOI] [PubMed] [Google Scholar]
- 33.Siemasko KF, Chong AS, Williams JW, Bremer EG, Finnegan A. Regulation of B Cell Function By the Immunosuppressive Agent Leflunomide. Transplantation. 1996;61(4):635–42. doi: 10.1097/00007890-199602270-00020. [DOI] [PubMed] [Google Scholar]
- 34.ANITA SC, Finnegan A, Jiang X, Gebel H, Sankary HN, Foster P, et al. Leflunomide, A Novel Immunosuppressive Agent: The Mechanism of Inhibition of T Cell Proliferation. Transplantation. 1993;55(6):1361–6. doi: 10.1097/00007890-199306000-00028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.