Abstract
Angiogenesis is a key event in the progression of gliomas. Exosomes, as signaling extracellular organelles, modulate the tumor microenvironment and promote angiogenesis and tumor progression. We previously demonstrated that long intergenic non-coding RNA CCAT2 (linc-CCAT2) was overexpressed in glioma tissues and functioned to promote glioma progression. Therefore, this study aimed to explore an underlying mechanism of glioma cell-affected angiogenesis. First, qRT-PCR was used to determine the expression level of linc-CCAT2 in 4 glioma cell lines and 293T cells, and the results revealed that the U87-MG cells exhibited the highest expression level. Subsequently, the pro-angiogenesis function of exosomes that were derived from negative control shRNA-treated U87-MG cells (ncU87-Exo) and linc-CCAT2 shRNA-treated U87-MG cells (shU87-Exo) was evaluated in vitro and in vivo. We found that ncU87-Exo, which was enriched in linc-CCAT2, could be taken up by HUVECs. ncU87-Exo improved the linc-CCAT2 expression level in HUVECs and more strongly promoted HUVEC migration, proliferation, tubular-like structure formation in vitro and arteriole formation in vivo as well as inhibited HUVEC apoptosis induced by hypoxia. Further mechanistic studies revealed that ncU87-Exo could upregulate VEGFA and TGFβ expression in HUVECs as well as promote Bcl-2 expression and inhibit Bax and caspase-3 expression. Finally, gain-/loss-of-function studies revealed that the overexpression of linc-CCAT2 in HUVECs activated VEGFA and TGFβ, promoted angiogenesis, promoted Bcl-2 expression and inhibited Bax and caspase-3 expression, thus decreasing apoptosis. Downregulation of linc-CCAT2 revealed the opposite effect. Thus, our results revealed a new exosome-mediated mechanism by which glioma cells could promote angiogenesis through the transfer of linc-CCAT2 by exosomes to endothelial cells. Moreover, we suggest that exosomes and linc-CCAT2 are putative therapeutic targets in glioma.
Keywords: exosomes, linc-CCAT2, glioma cells, angiogenesis, apoptosis
Introduction
Angiogenesis is a complex process by which new vessels sprout from existing vasculature to form a vascular network that supplies nutrients and/or metabolites to tissues, thus playing a fundamental role in many physiological and pathological conditions (1–3). Several lines of direct evidence show that angiogenesis is essential for the growth and persistence of solid tumors and their metastases (4–6). Algire (7) demonstrated that the growth rate of tumors implanted in subcutaneous transparent chambers in mice was slow and linear before vascularization and was rapid and nearly exponential after vascularization. Studies also revealed that angiogenesis is a key event in the progression of gliomas, the most common and deadly type of primary brain tumor (8,9), and vascular density in high grade gliomas is markedly higher than in low-grade tumors (10). Therefore, it is essential to elucidate the potential mechanisms of angiogenesis in gliomas for the purpose of developing new anti-angiogenesis therapeutic strategies for the treatment of gliomas.
Exosomes are a subcategory of microvesicles defined as cup-shaped vesicles that are 40–100 nm in size. They are formed by the inward budding of the multivesicular body (MVB) membrane (11) and contain bioactive cargo from the cellular cytoplasm, including proteins, mRNA and non-coding RNA (12,13). Studies have demonstrated that tumor cell-derived exosomes mirror the characteristics of the cell and are suggested to play an important role in cancer growth, angiogenesis, cancer invasion and metastasis, and tumor immunity (14). Recent studies have also suggested that cancer exosomes can communicate with tumor cells and different cell types in the tumor stroma and can affect the formation of the pre-metastatic niche (15,16). As for gliomas, there is accumulating evidence that has revealed that glioma cell-derived exosomes may control the interplay between glioma cells and the microenvironment (17,18). For example, Skog et al (18) reported that glioma cell-derived exosomes contained mRNA, miRNA and angiogenic proteins, which can be taken up by brain microvascular endothelial cells and stimulate tubule formation and angiogenesis. However, the precise mechanism of how glioma cell-derived exosomes affect angiogenesis remains largely unknown.
Long non-coding RNAs (lncRNAs) are non-protein coding transcripts that are longer than 200 nucleotides and regulate gene expression at epigenetic transcriptional and post-transcriptional levels (19). As a subtype of lncRNAs, the long intergenic non-coding RNAs (lincRNAs) have been demonstrated to be transcript units located within genomic intervals between two protein coding genes (20). Increasing evidence has indicated that the aberrant expression of lincRNAs plays a critical role in tumor biology, including tumor initiation, progression, and metastasis (21,22). Our previous research (23) demonstrated that lincRNA-CCAT2 (linc-CCAT2) was overexpressed in glioma and was significantly associated with the tumor WHO grade. Furthermore, knockdown of linc-CCAT2 was demonstrated to inhibit proliferation, cell cycle progression and migration of glioma cells. As Conigliaro et al (24) demonstrated, exosomes released by CD90+ cancer cells that were enriched in lincRNA H19, could be taken up by endothelial cells and could promote an angiogenic phenotype and cell-to-cell adhesion. Thus, we hypothesized that glioma cells could transfer linc-CCAT2 to endothelial cells by exosomes and impact endothelial cell angiogenesis.
In the present study, we demonstrated that exosomes that were released by glioma cell lines U87-MG (U87-Exo) were enriched in linc-CCAT2 and could be internalized by human umbilical vein endothelial cells (HUVECs). The exosomes were able to promote HUVEC angiogenesis by stimulating angiogenesis-related gene and protein expression. In addition, we found that U87-Exo could alleviate HUVEC apoptosis that was induced by hypoxia. Furthermore, we employed gain-/loss-of-function experiments to reveal that the overexpression of linc-CCAT2 in HUVECs activated VEGFA and TGFβ and promoted angiogenesis as well as Bcl-2 expression and inhibited Bax and caspase-3 expression to decrease apoptosis. Downregulation of linc-CCAT2 revealed the opposite effect. These findings demonstrated that glioma cells could transfer linc-CCAT2 via exosomes to endothelial cells to promote angiogenesis, which sheds new light on the progression of gliomas. Therefore, exosomes and linc-CCAT2 may be used as putative therapeutic targets in the treatment of glioma.
Materials and methods
Ethics statement
The protocols employed in this study and the use of human tissues were approved by the Ethics Committee of the Second Affiliated Hospital of Nanchang University. This study was conducted in full accordance with ethical principles, including the World Medical Association Declaration of Helsinki, and the local legislation. All experimental protocols were carried out in accordance with the relevant guidelines and regulations.
Cell lines and culture conditions
Human glioma cell lines (A172, U87-MG, U251, and T98G) were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). All glioma cell lines and 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (vol/vol) fetal bovine serum (FBS) (both from Gibco, Grand Island, NY, USA). HUVECs were isolated from human umbilical cords and cultured in medium 200 (M200) supplemented with 2% low serum growth supplement (M200+LSGS; Cascade Biologics, Portland, OR, USA), as previously described (25). HUVECs at passage 2–10 were used in the experiments as described below. Routine culture was performed in a humidified incubator maintained at 37°C with 5% CO2 and 95% air.
Lentivirus transfection
To obtain the shCCAT2-expressing U87-MG cells, pGV248-CCAT2 shRNA and scramble shRNA from GenePharma (Shanghai, China) were transfected into 293T cells along with the packaging plasmids. The lentivirus partials were harvested and the knockdown efficiency was determined by qRT-PCR 48 h after co-transfection. The lentiviruses with pGV248-CCAT2 shRNA or scramble shRNA were used to infect U87-MG cells to construct stable expression in the cell lines for the following experiments. On the other hand, the full-length complementary DNA of human linc-CCAT2 was cloned into the lentiviral expression vector pLVX-IRES-neo; the shRNA of the human linc-CCAT2 was provided by GenePharma. The lentiviruses with pLVXIRES-neo-linc-CCAT2 and pLVX-IRES-neo-shRNA were used to infect HUVECs to construct stable expression in the cell lines for the following experiments (26).
Exosome isolation and purification
U87-MG glioma cells were grown with 10% depleted FBS (FBS was pre-depleted of bovine exosomes by ultracentrifugation at 4°C, 100,000 × g, 16 h). When cells were 70% confluent, they were rinsed three times with phosphate-buffered saline (PBS) and cultured for 48 h with 10% depleted FBS. Exosomes were isolated and purified from the U87-MG cell supernatant as previously described (25). Exosomes derived from the negative control shRNA-treated U87-MG cells were named ‘ncU87-Exo’, and exosomes derived from the linc-CCAT2 shRNA-treated U87-MG cells were named ‘shU87-Exo’. An equal volume of medium without cultured cells was obtained via the same method that was used to collect the exosomes and was referred to as the ‘control medium’. Exosome protein content was determined with the bicinchoninic acid assay (Thermo Fisher, Waltham, MA, USA) as previously described (27). BSA ranged from 2 mg/ml to 25 µg/ml to generate a calibration curve. The absorbance was read at 562 nm with a microplate reader (Bio-Rad, Berkeley, CA, USA).
Exosome morphology and size analysis
Transmission electron microscopy (TEM) was used to identify the morphology of exosomes as previously described (25). U87-MG cell-derived exosomes were visualized on a Hitachi H-7650 TEM (Hitachi, Tokyo, Japan), and images were captured with a digital camera (Olympus, Tokyo, Japan). Nanoparticle Tracking Analysis (NTA) measurements were performed with the Tunable Resistive Pulse Sensing (TRPS, IZON qNano; Christchurch, New Zealand) to investigate the size and number of the exosomes, as previously reported (28).
RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR) assay
RNAzol RT (Molecular Research Center, USA) was used to extract total RNA from glioma cell lines, HUVECs and exosomal sources (29). qRT-PCR for human-specific repeat sequences was performed as previously described (25). To identify the gene expression in HUVECs treated with ncU87-Exo, shU87-Exo, and control medium, 8×105 HUVECs were seeded into 6-well plates and cultured with fresh medium containing 100 µg/ml of exosomes for 48 h. HUVEC-negative control, HUVEC-linc-CCAT2 overexpression, and downregulated HUVEC-linc-CCAT2 were cultured with M200+LSGS for 48 h. The primers were obtained from GenePharma, and the following human primers were used: linc-CCAT2, basic fibroblast growth factor (bFGF), bFGF receptor (bFGFR), vascular endothelial growth factor A (VEGFA), VEGF receptor 2 (KDR), transforming growth factor β 1 (TGFβ1), intercellular adhesion molecule-1 (ICAM-1), angiogenin (Angio), B-cell leukemia 2 (Bcl-2), Bcl-2 associated X protein (Bax), caspase-3, and GAPDH. The primer sequences used in this study are summarized in Table I. Each sample was analyzed in triplicate for yield validation. The 2−ΔΔCt method was used to determine the relative gene expression levels.
Table I.
Primers used for qRT-PCR.
| Genes | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| h-linc-CCAT2 | CCCTGGTCAAATTGCTTAACCT | TTATTCGTCCCTCTGTTTTATGGAT |
| h-FGF | CAATTCCCATGTGCTGTGAC | ACCTTGACCTCTCAGCCTCA |
| h-FGFR | GACGGCTCCTACCTCAA | GCTGTAGCCCATGGTGTTG |
| h-VEGF | CGCTCGGTGCTGGAATTTGA | AGTGGGGAATGGCAAGCAAA |
| h-KDR | GTGATCGGAAATGACACTGGAG | CATGTTGGTCACTAACAGAAGCA |
| h-TGFβ | TTGAGGGCTTTCGCCTTAGC | TGAACCCTGCGTTGATGTCC |
| h-ICAM | AACCCATTGCCCGAGC | GGTGAGGATTGCATTAGGTC |
| h-Angio | CTCGCTTCGGCAGCACA | GGTGGTCGGAGATTCGTAGC |
| h-Bcl-2 | TTTGAGTTCGGTGGGGTCAT | TGACTTCACTTGTGGCCCAG |
| h-Bax | TGGCAGCTGACATGTTTTCTGAC | TCACCCAACCACCCTGGTCTT |
| h-caspase-3 | GCACTGGAATGTCAGCTCGCA | GCCACCTTCCGGTTAACACGA |
| h-GAPDH | ATCCCATCACCATCTTCC | GAGTCCTTCCACGATACCA |
qRT-PCR, quantitative reverse-transcriptase polymerase chain reaction; VEGF, vascular endothelial growth factor; TGFβ, transforming growth factor β; ICAM-1, intercellular adhesion molecule-1; Angio, angiogenin; Bcl-2, B-cell leukemia 2; Bax, Bcl-2 associated X protein.
Protein extraction and western blotting
Western blotting was used to identify the U87-MG exosome markers Alix, Tsg101, CD9 (30), and Bcl-2 as well as Bax, and caspase-3 protein expression in HUVECs. Cells and exosome pellets were lysed in lysis buffer (Roche Diagnostics, Mannheim, Germany). Subsequently, 5X protein loading buffer was added directly to the protein sample, and the sample was heated at 95°C for 5 min. Next, equal amounts of protein were loaded and separated on SDS-PAGE polyacrylamide gels, and proteins were blotted onto a nitrocellulose membrane (Whatman, Maidstone, Kent, UK). Primary antibodies included Alix (mouse monoclonal anti-Alix, 1:1,000), Tsg101 (mouse monoclonal anti-Tsg101, 1:200), CD9 (rabbit monoclonal anti-CD9, 1:1,000), Bax (rabbit monoclonal anti-Bax, 1:2,000), Bcl-2 (rabbit monoclonal anti-Bcl-2, 1:1,000), and caspase-3 (mouse monoclonal anti-caspase-3, 1:500) (all from Abcam, Cambridge, UK). The primary antibodies were incubated overnight at 4°C, followed by washing and the application of HRP-conjugated goat anti-rabbit secondary antibodies (1:2,000) and goat anti-mouse secondary antibodies (1:2,000) (both from Thermo Fisher). Proteins were detected via enhanced chemiluminescence (Thermo Fisher) and imaged with a Molecular Imager VersaDoc 4000 system (Bio-Rad).
Uptake of exosomes by HUVECs
Immunofluorescence staining was performed to confirm that HUVECs could take up exosomes. U87-MG cells were labeled with 3,3′-dihexadecyloxacarbocyanine perchlorate (CM-Dio; Invitrogen, Grand Island, NY, USA), according to the supplier's instructions. Exosomes derived from Dio-labeled U87-MG cells were routinely collected, and then HUVECs were incubated with 100 µg/ml of exosomes for 6 h. Next, HUVECs were rinsed with PBS and fixed with 4% paraformaldehyde at room temperature for 30 min and then pre-incubated with sodium borohydride (1 mg/ml in PBS) to decrease autofluorescence. The cells were then incubated overnight at 4°C with a CD31 primary antibody (mouse monoclonal anti-CD31, 1:100; Abcam) and then incubated for 1 h with a secondary antibody conjugated to Alexa Fluor 594 (1:200; Abcam). Nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI, 0.5 µg/ml; Invitrogen). Images were obtained with a fluorescence microscope (Leica, Solms, Germany).
Endothelial cell migration assay
The scratch-wound assay was used to analyze the migration ability of HUVECs. Briefly, 2×105 cells were seeded into 12-well plates and were maintained at 37°C to permit cell adhesion and the formation of a confluent monolayer. Next, these confluent monolayers were ‘scratch’-wounded with a p200 pipet tip. The medium was removed and rinsed once with PBS to remove the debris and to smooth the edge of the scratch. The medium was then was replaced with fresh M200+LSGS medium that contained 100 µg/ml of ncU87-Exo, shU87-Exo, or control medium. HUVEC-negative control, HUVEC-linc-CCAT2 overexpression, and dowregulated HUVEC-linc-CCAT2 cells were cultured with M200+LSGS. Wound closure was monitored via the collection of digital images at 0 and 24 h. The wound closure was analyzed with MetaMorph software (Molecular Devices, Sunnyvale, CA, USA), and the wound area at each time-point was normalized to its corresponding area at 0 h.
Endothelial cell proliferation assay
A Cell Counting Kit-8 assay (CCK-8; Dojindo) was used to assess cell proliferation. Briefly, HUVECs were seeded at 5×104 cells/ml (100 µl/well) in a 96-well plate. After quiescence for 12 h, the cells were treated with fresh M200+LSGS medium containing 100 µg/ml ncU87-Exo, shU87-Exo, or control medium. HUVEC-negative control, HUVEC-linc-CCAT2 overexpression, and downregulated HUVEC-linc-CCAT2 cells were cultured with M200+LSGS. At days 0, 1, 2, 3, 4, and 5, 10 µl of CCK-8 solution was added to the HUVECs and incubated for 3 h at 37°C. The absorbance was assessed at 450 nm with a microplate reader. All experiments were performed in triplicate and repeated at least three times.
Endothelial cell capillary-like tube formation assay
In vitro capillary-like tube formation was evaluated on growth factor-reduced Matrigel (BD Biosciences, Sparks, MD, USA). In total, 2×104 HUVECs were seeded onto the plated Matrigel with fresh M200+LSGS medium containing 100 µg/ml ncU87-Exo, shU87-Exo, or control medium. HUVEC-negative control, HUVEC-linc-CCAT2 overexpression, and downregulated HUVECs-linc-CCAT2 cells were cultured with M200+LSGS. Tube formation was quantified at 6 h. The total branching points, total tube length, and total loops per image were assessed by a blinded independent observer.
Chick chorioallantoic membrane assay (CAM)
The CAM assay was performed to confirm the effect of exosomes on blood vessel formation in developing fertilized chicken eggs (31,32). A gelatin sponge with a 5-mm diameter that carried 200 µg of ncU87-Exo, 200 µg of shU87-Exo, and control medium was implanted in CAMs. After incubation for 72 h, CAMs were photographed with a camera (Nikon, Japan). Tertiary arterioles (number of tertiary vessels divided by the number of branches) were quantified, and the data collection was repeated three times.
Enzyme-linked immunosorbent assay (ELISA)
To determine the level of VEGF, TGF-β, and FGF secreted by HUVECs, 8×105 cells were seeded into 6-well plates and cultured with fresh M200+LSGS medium containing 100 µg/ml ncU87-Exo, shU87-Exo, or control medium for 48 h. HUVECs-negative control, HUVEC-linc-CCAT2 overexpression, and downregulated HUVEC-linc-CCAT2 cells were cultured with M200+LSGS. Then, the cell supernatants were collected, centrifuged to remove the cells and stored at −80°C. Specific VEGF, TGF-β, and FGF ELISA kits (all from Westang Bio-Tech, Shanghai, China) were used to identify protein levels.
Hypoxia treatment
For cell hypoxia experiments, 8×105 HUVECs were seeded into 6-well plates and quiescence followed for 12 h. The cells were then cultured with M200+LSGS medium and exposed to normoxia with 21% O2 and 5% CO2 or cultured with fresh M200+LSGS medium containing 100 µg/ml ncU87-Exo, shU87-Exo, or control medium in a hypoxic incubator with 1% O2, 94% N2, and 5% CO2 (Forma Series II Water Jacketed CO2 incubator; Thermo Fisher) for 48 h. Concomitantly, HUVEC-negative control, HUVECs-linc-CCAT2 overexpression, and downregulated HUVEC-linc-CCAT2 cells were also cultured with M200+LSGS under hypoxic conditions for 48 h as described aforementioned.
Flow cytometric analysis
Flow cytometry was performed according to the operation instructions of the Annexin V-FITC/PI apoptosis detection kit (Dojindo, Kumamoto, Japan). Briefly, 5×105 cells were harvested and suspended in 200 µl of binding buffer, and then 10 µl of Annexin V-FITC and 5 µl of PI were added for a 15-min light cycle, avoiding incubation at room temperature. Finally, the cells were resuspended in 250 µl of binding buffer and analyzed with the Guava® easyCyte™ system (Millipore, Billerica, MA, USA). All of these experiments were performed in triplicate and repeated at least three times.
Statistical analysis
Statistical analysis was performed with the SPSS Graduate Pack, version 11.0, statistical software (SPSS). Differences between two groups were analyzed by Student's t-test. Data are expressed as the means ± standard deviations (SDs) of three independent experiments. P-values of <0.05 were considered to be statistically significant.
Results
Characterization of exosomes released by ncU87-MG and shU87-MG cells
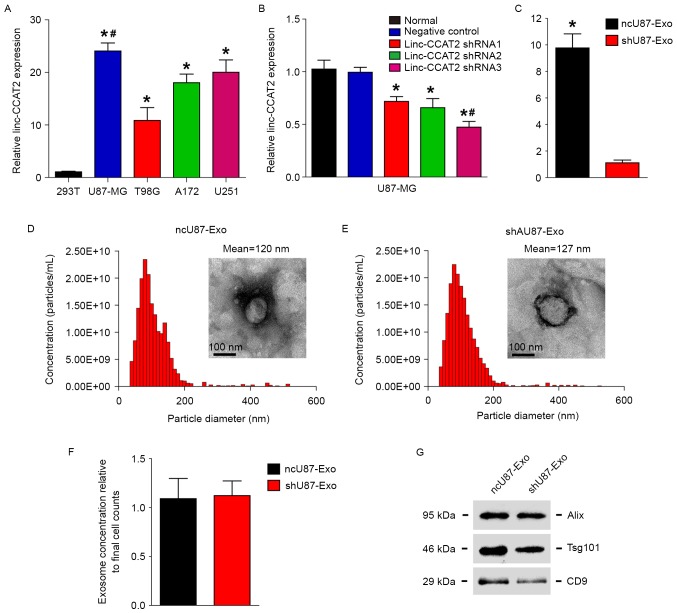
Our previous research (23) demonstrated that linc-CCAT2 was abundantly expressed in glioma tissues and that the expression was positively correlated with advanced tumor stage. In addition, a loss of function assay revealed that knockdown of linc-CCAT2 significantly inhibited glioma cell proliferation and tumorigenesis. We therefore hypothesized that linc-CCAT2 could affect tumor angiogenesis, and we explored how linc-CCAT2 in gliomas affected angiogenesis. First, we detected linc-CCAT2 expression in the following four glioma cell lines: U87-MG, U251, A172, and T98G and found that the linc-CCAT2 expression level in all of the four glioma cell lines was higher than in non-glioma 293T cells (Fig. 1A, P<0.01). The linc-CCAT2 expression level was the highest and lowest in the U87-MG cells (24.077-fold higher compared to the 293T cells) and T98G cells (10.874-fold higher compared to the 293T cells), respectively.
Figure 1.
Characterization of exosomes released by ncU87-MG cells and shU87-MG cells. (A) linc-CCAT2 expression levels were evaluated by qRT-PCR in the following four glioma cell lines: U87-MG, U251, A172, and T98G; non-glioma 293T cells were used as controls. U87-MG cells exhibited the highest expression level of linc-CCAT2 (compared with the 293T cells, *P<0.01; compared with the U251, A172, and T98G cells, #P<0.05). (B) The knockdown effects of linc-CCAT2 were assessed by qRT-PCR in U87-MG cells transfected with the shRNA or negative controls; the linc-CCAT2 shRNA3 was the most efficient in silencing linc-CCAT2 mRNA (compared with the normal and negative controls, *P<0.01; compared with shRNA1 and shRNA2, #P<0.05). (C) ncU87-Exo cells were highly enriched in linc-CCAT2 transcripts compared with shU87-Exo (*P<0.01). (D and E) The nanoparticle size distribution and concentrations for ncU87-Exo and shU87-Exo were obtained via NTA, and the morphology of ncU87-Exo and shU87-Exo was observed by TEM. (F) The ncU87-Exo and shU87-Exo concentrations were normalized to final cell counts; there were no significant differences observed between ncU87-Exo and shU87-Exo cells (P>0.05). (G) Western blot analysis of exosomal markers Alix, Tsg101, and CD9 in ncU87-Exo and shU87-Exo; equal amounts of exosomes (300 ng) were used for the assay. qRT-PCR, quantitative reverse-transcriptase polymerase chain reaction; NTA, Nanoparticle Tracking Analysis; TEM, transmission electron microscopy.
The glioma cell line U87-MG exhibited the highest expression level of linc-CCAT2 among the four cell lines, thus we used U87-MG cells in the following experiment. Fig. 1B shows that linc-CCAT2 was efficiently silenced with shRNAs (shRNA1, shRNA2 and shRNA3) in U87-MG cells compared with negative controls and untreated cells (normal). The linc-CCAT2 shRNA3 cells were the most efficient in silencing linc-CCAT2 mRNA and thus were chosen to be used in the following experiments.
Furthermore, the morphology of ncU87-Exo and shU87-Exo cells was observed by TEM; ncU87-Exo was similar to shU87-Exo (50–100 nm in diameter), and each vesicle revealed a classical cup- or round-shaped appearance (Fig. 1D and E). Next, we performed NTA with the TRPS system to investigate the size distribution profile of ncU87-Exo and shU87-Exo. The nanoparticle size distribution of ncU87-Exo was similar to the shU87-Exo, and the peaks of the particle size ~100 nm were within the expected size of the exosomes (Fig. 1D and E). When nanoparticle concentrations were normalized to cell numbers at the time of harvest, there were no significant differences between ncU87-Exo and shU87-Exo (Fig. 1F). Finally, we performed western blot analyses to identify the presence of exosomal markers Alix, Tsg101 and CD9 in exosomes. As shown in Fig. 1G, both types of exosomes were expressed in exosomal markers Alix, Tsg101 and CD9. These results indicate that the size and concentration of exosomes secreted from U87-MG cells were not affected when the cells were treated with the negative control shRNA and linc-CCAT2 shRNA.
In addition, qRT-PCR was used to identify the expression level of linc-CCAT2 in ncU87-Exo and shU87-Exo, as shown in Fig. 1C; ncU87-Exo was significantly highly enriched in linc-CCAT2 when compared with shU87-Exo (9.773-fold change, P<0.01), which indicated that the inhibition of linc-CCAT2 expression in U87-MG cells decreased the quantity of linc-CCAT2 in exosomes.
ncU87-Exo and shU87-Exo are internalized by HUVECs
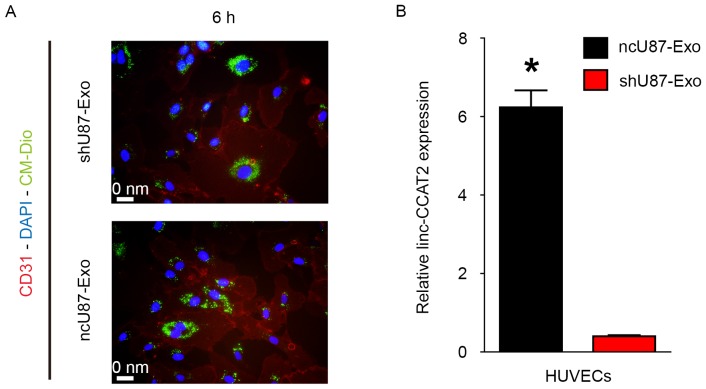
Exosomes must be internalized by recipient cells and release their cargo to achieve their function. Therefore, we explored whether ncU87-Exo and shU87-Exo could be internalized by HUVECs. As shown in Fig. 2A, plasma membrane fluorescent dye Dio-labeled ncU87-Exo and shU87-Exo could be internalized into HUVECs. Furthermore, we determined the linc-CCAT2 expression level in HUVECs when incubated with 100 µg/ml ncU87-Exo and shU87-Exo for 24 h; qRT-PCR results revealed that the linc-CCAT2 expression level in the ncU87-Exo-treated HUVECs was significantly higher than in the shU87-Exo-treated HUVECs (Fig. 2B, P<0.01). These results revealed the transportation of linc-CCAT2 from U87-MG cells to HUVECs by exosomes, even though we cannot exclude the stimulation of endogenous lincRNA.
Figure 2.
ncU87-Exo and shU87-Exo are internalized by HUVECs. (A) Immunofluorescence images of DAPI (blue)-CD31 (red) HBMECs co-cultured with CM-Dio (green) labeled ncU87-Exo and shU87-Exo at 6 h. (B) qRT-PCR was applied to determine linc-CCAT2 expression levels in HUVECs when incubated with 100 µg/ml ncU87-Exo and shU87-Exo for 24 h; the linc-CCAT2 expression level in ncU87-Exo-treated HUVECs was significantly higher than that in the shU87-Exo-treated HUVECs (*P<0.01). HUVECs, human umbilical vein endothelial cells; qRT-PCR, quantitative reverse-transcriptase polymerase chain reaction.
ncU87-Exo and shU87-Exo regulate HUVEC migration, proliferation, and tube formation in vitro and angiogenesis in vivo
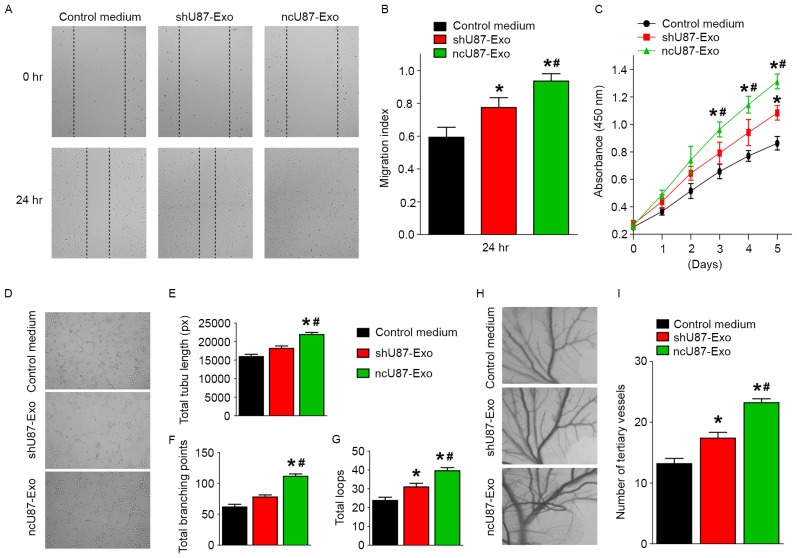
Tumor angiogenesis is important for tumor progression, so we further investigated the function of ncU87-Exo and shU87-Exo in the regulation of HUVEC migration, proliferation, and tube formation in vitro and angiogenesis in vivo to determine the effects of glioma exosomes on angiogenesis. First, HUVEC migration, proliferation, and tube formation capacities were assessed with a series of in vitro angiogenic assays. Markedly, the scratch-wound assay revealed that both ncU87-Exo and shU87-Exo significantly enhanced the motility of HUVECs, while ncU87-Exo exhibited better pro-migration ability (P<0.05, Fig. 3A and B). The CCK-8 cell counting analysis revealed that ncU87-Exo significantly stimulated HUVEC proliferation, while shU87-Exo only slightly promoted HUVEC proliferation when compared to the control medium group (P<0.05, Fig. 3C). To determine the effects on tube formation, the cells were seeded on Matrigel and cultured for 6 h. The total branching points, total tube length, and total loops were assessed to quantify the ability of HUVECs to form tubes. Compared with the control medium group and the shU87-Exo group, all indicators of tube formation capacity were significantly increased in HUVECs incubated with ncU87-Exo, while there was no significant difference in total branching points and total tube length between the shU87-Exo group and the control medium group. These findings indicated that ncU87-Exo exhibited better pro-tube formation function (P<0.05, Fig. 3D-G). Next, we performed the CAM assay to estimate the pro-angiogenesis function of exosomes in vivo. As shown in Fig. 3H and I, the number of newly formed arterioles was well correlated with the effect of exosomes, and samples treated with ncU87-Exo clearly exhibited more new arterioles than samples treated with shU87-Exo (P<0.05), which was also evident in our in vitro data. These data indicated that exosomes released from glioma cells play positive roles in angiogenesis, and linc-CCAT2 may act as a possible mediator of pro-angiogenesis properties in exosomes released by glioma cells.
Figure 3.
ncU87-Exo and shU87-Exo regulates HUVEC migration, proliferation, and tube formation in vitro and angiogenesis in vivo. (A and B) The migration ability was assessed with a scratch-wound assay, and both ncU87-Exo and shU87-Exo significantly enhanced the motility of HUVECs, while ncU87-Exo exhibited a more efficient pro-migration ability. (C) The proliferation was assessed with the CCK-8 assay; ncU87-Exo significantly stimulated HUVEC proliferation, while shU87-Exo only slightly promoted HUVEC proliferation. (D-G) The tube formation assay was performed on growth factor-reduced Matrigel. The number of total branching points, total tube length, and total loops at 6 h in the ncU87-Exo-treated HUVECs was higher than that in the shU87-Exo-treated HUVECs. (H and I) Similar to the in vitro experiment, ncU87-Exo induced CAM arteriole formation to a greater extent, in contrast to shU87-Exo. Representative images of the CAM assays are presented in the left panel, and the number of vessels is presented in the right panel (n=3). (*P<0.05 when compared to the control medium; #P<0.05 when compared to shU87-Exo). HUVECs, human umbilical vein endothelial cells; CCK-8, Cell Counting Kit-8 assay.
ncU87-Exo and shU87-Exo promote the expression of angiogenesis-related molecules in HUVECs
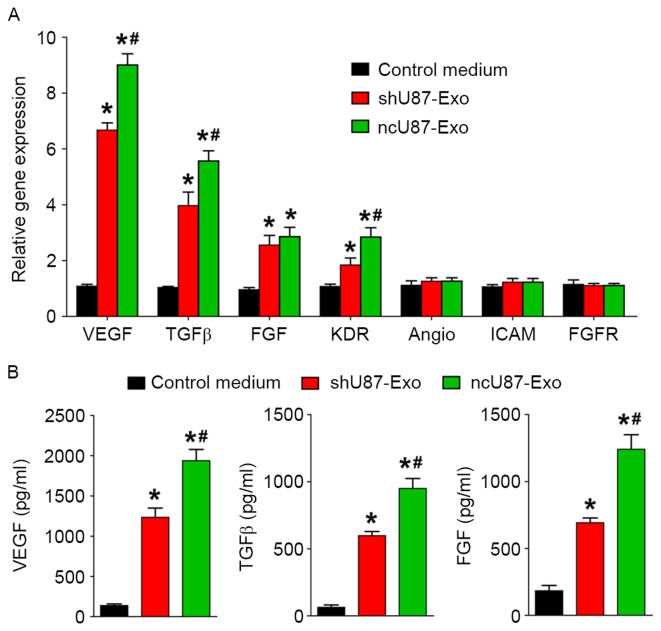
To investigate the possible underlying mechanism in the promotion of angiogenesis from exosomes derived from glioma cells, we incubated HUVECs with 100 µg/ml ncU87-Exo, shU87-Exo, and control medium and applied qRT-PCR and ELISA to detect the expression level of angiogenesis-related genes (VEGF, TGFβ, FGF, KDR, Angio, ICAM, and FGFR) and proteins (VEGF, TGFβ, and FGF) in HUVECs. As shown in Fig. 4A, ncU87-Exo induced a significantly higher expression of VEGF, TGFβ, FGF, and KDR transcripts (P<0.05), while no modulation was observed for the transcription of Angio, ICAM, and FGFR in the three groups (P>0.05). The ELISA assay (Fig. 4B) found a substantial increase in VEGF, TGFβ, and FGF secretion in HUVECs incubated with ncU87-Exo and shU87-Exo when compared with the control medium group (P<0.05), and HUVECs incubated with ncU87-Exo secreted much more VEGF, TGFβ, and FGF when compared with the shU87-Exo group (P<0.05). Collectively, these results demonstrated that ncU87-Exo increased VEGF, TGFβ, and FGF expression in HUVECs to promote angiogenesis, and linc-CCTA2 may play a critical role in the process.
Figure 4.
ncU87-Exo and shU87-Exo regulate angiogenesis related-genes and protein expression in HUVECs. (A) qRT-PCR analysis of the expression level of angiogenesis-related genes in HUVECs treated by ncU87-Exo and shU87-Exo. Compared with the shU87-Exo group, ncU87-Exo significantly upregulated VEGF, TGFβ, FGF and KDR gene expression. (B) ELISA analysis of the secretion level of angiogenesis-related proteins in HUVECs treated by ncU87-Exo and shU87-Exo. Compared with the shU87-Exo group, ncU87-Exo significantly increased VEGF, TGFβ, and FGF protein secretion. (*P<0.05 when compared to the control medium; #P<0.05 when compared to shU87-Exo). HUVECs, human umbilical vein endothelial cells; qRT-PCR, quantitative reverse-transcriptase polymerase chain reaction; VEGF, vascular endothelial growth factor; TGFβ, transforming growth factor β.
ncU87-Exo and shU87-Exo decrease HUVEC apoptosis in response to hypoxia in vitro
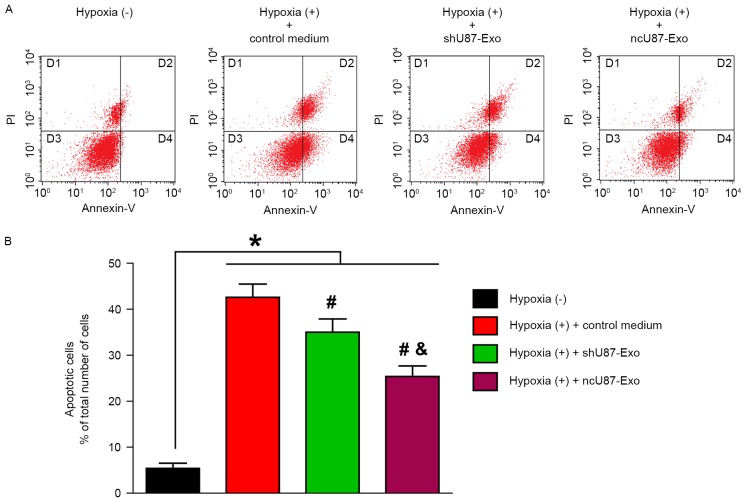
In the next step, we performed flow cytometric analysis to quantify the number of apoptotic or necrotic HUVECs after treatment with hypoxia. As shown in Fig. 5A, the D3 quadrant exhibited viable cells that did not bind Annexin V or PI; the D4 quadrant exhibited cells in the early stages of apoptosis that bound Annexin V but still had intact cell membranes and excluded PI; the D2 quadrant exhibited cells with advanced stages of apoptosis or necrosis that were both Annexin V- and PI-positive; and the D1 quadrant exhibited cells that lost their intact cell membranes, that bound PI and excluded Annexin V. The results are presented in the form of percentages. As shown in Fig. 5B. the apoptosis rate (the D2 and D4 quadrants) of HUVECs in the hypoxia (+) group treated with control medium, ncU87-Exo and shU87-Exo was significantly higher than in the hypoxia (−) group (42.85, 35.30, 25.64 vs. 5.95%, respectively; P<0.05). Compared with the control medium group, both ncU87-Exo and shU87-Exo decreased the HUVEC apoptosis rate induced by hypoxia (P<0.05), however ncU87-Exo was more efficient (P<0.05).
Figure 5.
ncU87-Exo and shU87-Exo decrease HUVEC apoptosis induced by hypoxia in vitro. Both ncU87-Exo and shU87-Exo decreased apoptosis of HUVECs induced by hypoxia, and ncU87-Exo exhibited more efficient anti-apoptosis. (A) Representative FCM images of PI and Annexin V-FITC double-stained HUVEC apoptosis. (B) The bar chart of PI and Annexin V-FITC double-stained HUVEC apoptosis. (*P<0.01, hypoxia (−) group vs. hypoxia (+) control medium, ncU87-Exo and shU87-Exo group; #P<0.05, hypoxia (+) ncU87-Exo and shU87-Exo group vs. hypoxia (+) control medium; &P<0.05, hypoxia (+) ncU87-Exo group vs. hypoxia (+) shU87-Exo group). HUVECs, human umbilical vein endothelial cells.
ncU87-Exo and shU87-Exo regulate apoptosis-related factor expression in HUVECs induced by hypoxia
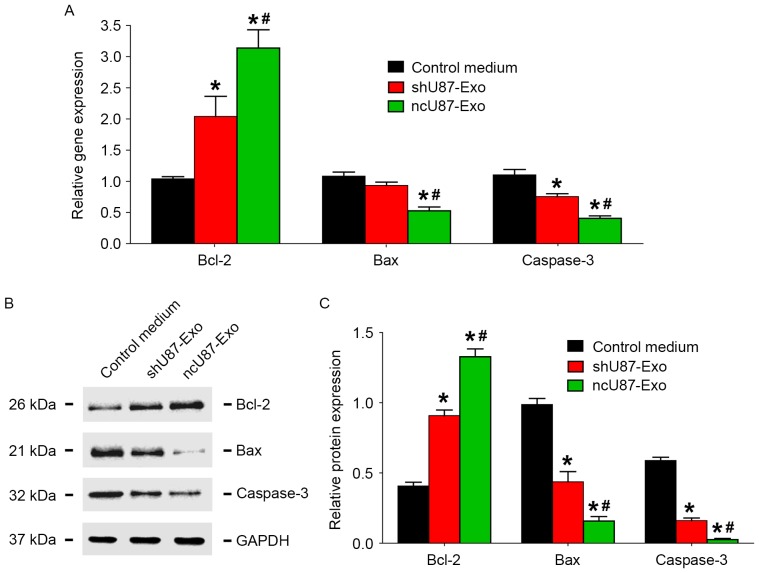
To investigate the possible mechanism of exosome release by glioma cells in the decrease of HUVEC apoptosis induced by hypoxia, we incubated HUVECs with 100 µg/ml ncU87-Exo, shU87-Exo, and control medium under hypoxic conditions for 48 h, and applied qRT-PCR and western blot assays to detect the expression level of apoptosis-related factors (Bcl-2, Bax, and caspase-3) in HUVECs. As shown in Fig. 6A, compared to the control medium group, both ncU87-Exo and shU87-Exo improved Bcl-2 transcript expression and decreased Bax and caspase-3 transcript expression (P<0.05). Compared to the shU87-Exo group, the ncU87-Exo group more strongly promoted Bcl-2 transcript expression and inhibited Bax and caspase-3 transcript expression (P<0.05). The western blot assay (Fig. 6B and C) revealed a similar tendency as the qRT-PCR results in that both ncU87-Exo and shU87-Exo improved Bcl-2 protein expression and decreased Bax and caspase-3 protein expression (P<0.05), while the ncU87-Exo group more strongly promoted Bcl-2 protein expression and inhibited Bax and caspase-3 protein expression (P<0.05). Collectively, these results demonstrated that ncU87-Exo increased Bcl-2 expression and decreased Bax and caspase-3 expression in HUVECs to anti-apoptosis induced by hypoxia, and linc-CCTA2 may play a critical role in that process.
Figure 6.
ncU87-Exo and shU87-Exo regulate the expression of apoptosis-related factors in HUVECs induced by hypoxia. (A) qRT-PCR analysis of the expression level of apoptosis-related factors Bcl-2, Bax, and caspase-3 in HUVECs treated by ncU87-Exo and shU87-Exo after hypoxia. Compared with the control medium group and the shU87-Exo group, ncU87-Exo significantly upregulated Bcl-2 gene expression and inhibited Bax and caspase-3 gene expression. (B and C) Western blot analysis revealed that both ncU87-Exo and shU87-Exo increased Bcl-2 expression and decreased Bax and caspase-3 expression, while ncU87-Exo was more efficient. (*P<0.05 when compared to the control medium; #P<0.05 when compared to shU87-Exo). HUVECs, human umbilical vein endothelial cells; qRT-PCR, quantitative reverse-transcriptase polymerase chain reaction; Bcl-2, B-cell leukemia 2; Bax, Bcl-2 associated X protein.
Linc-CCAT2 enhances HUVEC migration, proliferation, and tube formation in vitro
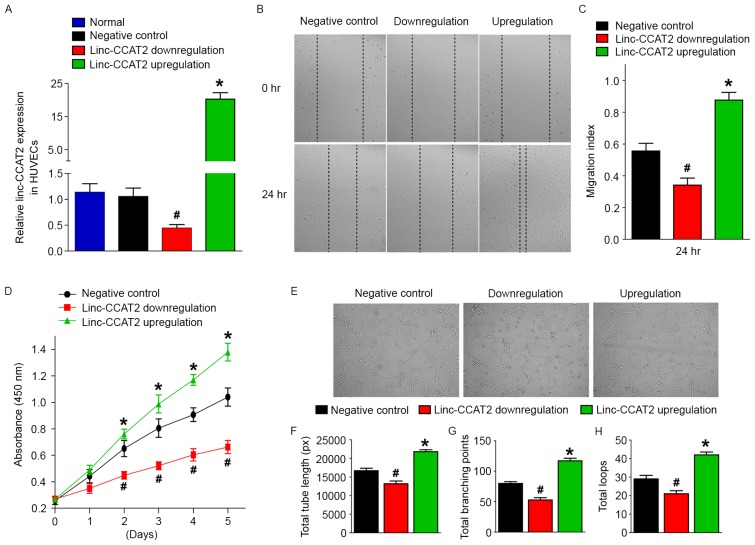
Our research indicated that ncU87-Exo cells were enriched in linc-CCAT2 and could upregulate their expression level in HUVECs as well as stimulate VEGF, TGFβ, FGF, and KDR gene expression and VEGF, TGFβ, and FGF protein secretion, which promoted HUVEC angiogenesis in vitro and in vivo. These results indicated that ncU87-Exo promoted angiogenesis by transferring linc-CCAT2 into HUVECs to stimulate angiogenesis-related molecule expression. To confirm these hypotheses, we constructed HUVECs with a stable overexpression of linc-CCAT2 and a stable downregulation of linc-CCAT2 with lentiviruses. The qRT-PCR analysis revealed that lentiviral-mediated silencing of linc-CCAT2 resulted in a downregulation of 0.44-fold in HUVECs, and lentiviral-mediated overexpression of linc-CCAT2 resulted in an upregulation of 20.28-fold in HUVECs when compared to the negative control (P<0.05; Fig. 7A). Furthermore, we investigated the angiogenic function of linc-CCAT2 in HUVECs with the scratch wound, CCK-8, and tube formation assays. Compared with the negative control, linc-CCAT2 overexpression increased HUVEC migration (P<0.05, Fig. 7B and C) and proliferation (P<0.05, Fig. 7D). In contrast, downregulation of linc-CCAT2 significantly decreased HUVEC migration (P<0.05, Fig. 7B and C) and proliferation (P<0.05, Fig. 7D). Furthermore, compared with the negative control, the total branching points, total tube length, and total loops were significantly increased in the overexpressed linc-CCAT2 HUVECs and were markedly decreased in the downregulated linc-CCAT2 HUVECs (P<0.05, Fig. 7E-H). These results indicated that linc-CCAT2 participated in and exhibited a positive function in the angiogenic process.
Figure 7.
linc-CCAT2 regulates HUVEC migration, proliferation, and tube formation in vitro. (A) The linc-CCAT2 expression levels were evaluated by qRT-PCR in linc-CCAT2 overexpression, downregulation, negative control, and normal HUVECs. linc-CCAT2 overexpression in HUVECs exhibited the highest expression level of linc-CCAT2, while downregulation of linc-CCAT2 in HUVECs exhibited the lowest expression level of linc-CCAT2. (B and C) The migration ability was assessed with the scratch-wound assay; linc-CCAT2 overexpression increased HUVEC migration, while downregulation of linc-CCAT2 significantly decreased HUVEC migration. (D) The proliferation was assessed with the CCK-8 assay; linc-CCAT2 overexpression promoted HUVEC proliferation, while linc-CCAT2 downexpression inhibited HUVEC proliferation. (E-H) The tube formation assay was performed on growth factor-reduced Matrigel. The total number of branching points, total tube length, and total loops at 6 h in the linc-CCAT2 overexpression in HUVECs was higher than in the negative control HUVECs, while downregulation of linc-CCAT2 in HUVECs was lower than the negative control HUVECs. (*P<0.05 when compared to the negative control; #P<0.05 when compared to the negative control). HUVECs, human umbilical vein endothelial cells; qRT-PCR, quantitative reverse-transcriptase polymerase chain reaction; CCK-8, Cell Counting Kit-8 assay.
Linc-CCAT2 regulates angiogenesis-related molecule expression in HUVECs
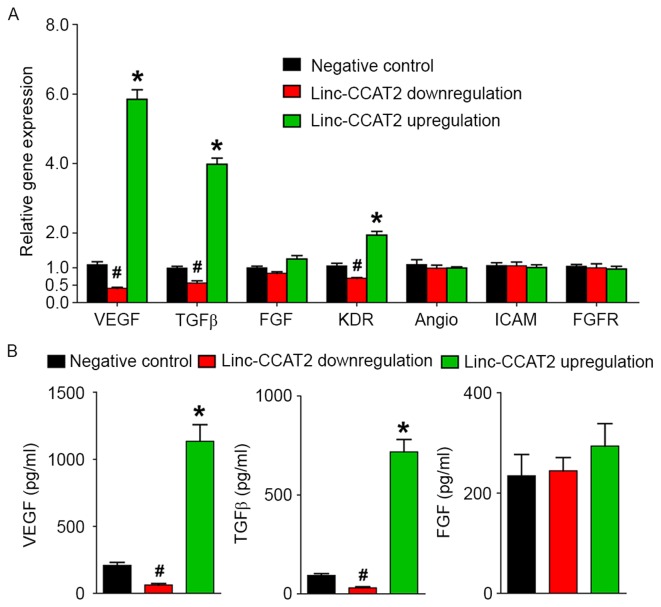
Furthermore, we investigated the expression level of angiogenesis-related genes (VEGF, TGFβ, FGF, KDR, Angio, ICAM and FGFR) and proteins (VEGF, TGFβ and FGF) for linc-CCAT2 overexpression and downregulation in HUVECs. As shown in Fig. 8A, compared to the negative control, linc-CCAT2 overexpression in HUVECs resulted in a significant increase in VEGF, TGFβ, and KDR transcript expression (P<0.05), while downregulation of linc-CCAT2 inhibited VEGF, TGFβ, and KDR transcript expression. There were no significant differences in the transcription levels of FGF, Angio, ICAM, and FGFR in the three groups (P>0.05). The ELISA assay (Fig. 8B) revealed a higher level of VEGF and TGFβ secretion in the overexpressed linc-CCAT2 HUVECs, and a lower secretion level in the downregulated linc-CCAT2 HUVECs (P<0.05) when compared with the negative control. The difference in the FGF secretion level was not statistically significant in the three groups (P>0.05). Collectively, these results demonstrated that linc-CCAT2 promoted angiogenesis partly by activating VEGF and TGFβ.
Figure 8.
linc-CCAT2 regulates angiogenesis-related genes and protein expression in HUVECs. (A) qRT-PCR analysis of the expression level of angiogenesis-related genes in linc-CCAT2 overexpression, downregulation and negative control HUVECs. linc-CCAT2 overexpression in HUVECs increased the gene expression of VEGF, TGFβ, and KDR, while downregulation of linc-CCAT2 inhibited VEGF, TGFβ, and KDR expression. (B) ELISA analysis of the level of secreted angiogenesis-related proteins in linc-CCAT2 overexpression, downregulation and negative control HUVECs. linc-CCAT2 overexpression in HUVECs increased VEGF and TGFβ protein secretion, while downregulation of linc-CCAT2 inhibited VEGF and TGFβ protein secretion. (*P<0.05 when compared to the negative control; #P<0.05 when compared to the negative control). HUVECs, human umbilical vein endothelial cells; qRT-PCR, quantitative reverse-transcriptase polymerase chain reaction; VEGF, vascular endothelial growth factor; TGFβ, transforming growth factor β.
linc-CCAT2 decreases apoptosis of HUVECs induced by hypoxia in vitro
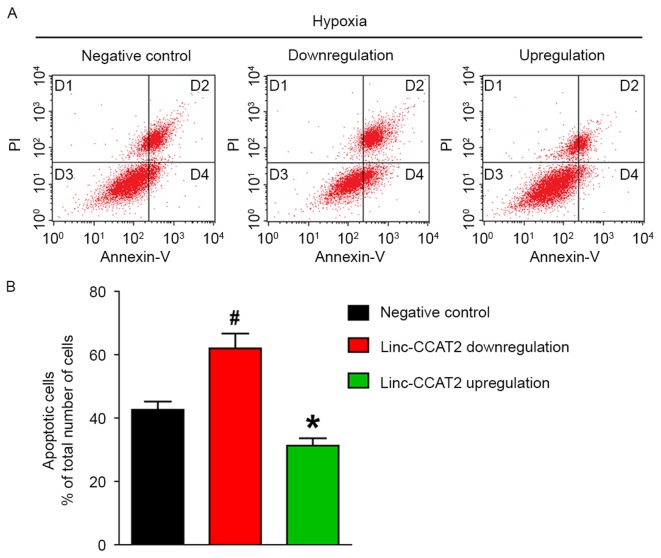
Flow cytometric analysis was also used to quantify the number of apoptotic or necrotic HUVECs in the negative control, downregulated and overexpressed groups when exposed to hypoxia. As shown in Fig. 9A and B, the apoptosis rate (the D2 and D4 quadrants) for HUVECs with downregulation was significantly higher than in the negative control HUVECs (62.06 vs. 42.61%, respectively; P<0.05). The apoptosis rate in HUVECs with overexpression was markedly lower than in the negative control HUVECs (31.28 vs. 42.61%, respectively; P<0.05). These results demonstrated that linc-CCAT2 overexpression decreased HUVEC apoptosis.
Figure 9.
linc-CCAT2 decreases HUVEC apoptosis induced by hypoxia in vitro. (A) Representative FCM images of PI and Annexin V-FITC double-stained HUVEC apoptosis. (B) The bar chart of PI and Annexin V-FITC double-stained HUVEC apoptosis. The apoptosis percentage in the downregulated lin-CCAT2 HUVECs was significantly higher than that in the negative control HUVECs, while the apoptosis percentage in the overexpressed linc-CCAT2 HUVECs was markedly lower than that in the negative control HUVECs. (*P<0.05 when compared to negative control; #P<0.05 when compared to negative control). HUVECs, human umbilical vein endothelial cells.
linc-CCAT2 regulates apoptosis-related factor expression in HUVECs induced by hypoxia
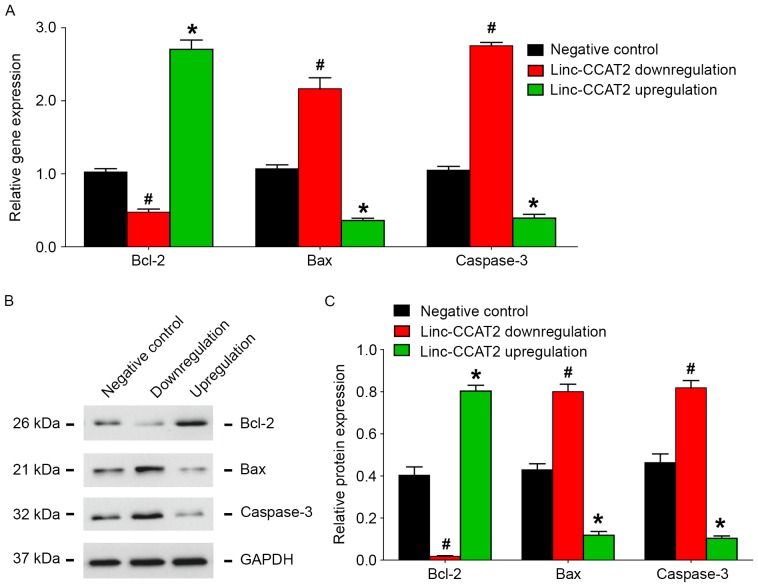
Similar to our ncU87-Exo results, as shown in Fig. 10A, when compared to the negative control, HUVECs with linc-CCAT2 overexpression exhibited higher Bcl-2 transcript expression and lower Bax and caspase-3 transcript expression (P<0.05), while HUVECs with downregulation of CCAT2 exhibited lower Bcl-2 transcript expression and higher Bax and caspase-3 transcript expression (P<0.05). The western blot assay (Fig. 10B and C) revealed a similar tendency as the qRT-PCR results, linc-CCAT2 overexpression improved Bcl-2 protein expression and decreased Bax and caspase-3 protein expression, while downregulation of linc-CCAT2 decreased Bcl-2 protein expression and increased Bax and caspase-3 protein expression. Collectively, these results demonstrated that linc-CCAT2 decreased HUVEC hypoxia-related apoptosis by promoting Bcl-2 expression and inhibiting Bax and caspase-3 expression.
Figure 10.
linc-CCAT2 regulates apoptosis-related factor expression in HUVECs induced by hypoxia. (A) qRT-PCR analysis of the expression level of apoptosis-related factors Bcl-2, Bax and caspase-3 in linc-CCAT2 overexpression, downregulation and negative control HUVECs. Compared with the negative control HUVECs, overexpression promoted Bcl-2 gene expression and inhibited Bax and caspase-3 gene expression, while downregulation of linc-CCAT2 in HUVECs inhibited Bcl-2 gene expression and promoted Bax and caspase-3 gene expression. (B and C) Western blot analysis of the expression level of Bcl-2, Bax, and caspase-3 in HUVECs. Compared with the negative control HUVECs, linc-CCAT2 overexpression improved Bcl-2 protein expression and decreased Bax and caspase-3 protein expression, while downregulation of linc-CCAT2 decreased Bcl-2 protein expression and increased Bax and caspase-3 protein expression. (*P<0.05 when compared to the negative control; #P<0.05 when compared to the negative control). HUVECs, human umbilical vein endothelial cells; qRT-PCR, quantitative reverse-transcriptase polymerase chain reaction; Bcl-2, B-cell leukemia 2; Bax, Bcl-2 associated X protein.
Discussion
Despite advances in surgical and medical therapy for glioma in recent years, the prognosis of patients with malignant glioma is extremely poor (33). The formation of abnormal tumor vasculature is believed to be one of the major reasons for glioma development, but the underlying mechanism of tumor vascularization is quite complex and still unclear (34). Tumor-derived exosomes can act as mediators in intercellular communication of the tumor microenvironment as well as construct a fertile environment to support tumor proliferation, angiogenesis, invasion and premetastatic niche preparation through the intercellular transfer of proteins, mRNA, miRNA, and lncRNA (35,36). Therefore, we conducted the present study to clarify the possible relationships between glioma and angiogenesis and to explore the potential application of glioma-derived exosomes in the diagnosis and treatment of glioma.
It has been recently reported that lncRNAs demonstrate incredible functions in the control of gene expression and chromatin structure, including the recruitment of chromatin-modifying proteins, the modulation of protein-DNA binding, the organization of nuclear architecture, the regulation of mRNA stability and translation, and the modulation of mRNA levels, among other functions (19,37–40). Moreover, accumulating studies have demonstrated the aberrant expression of lncRNAs in various human tumors, including colorectal cancer (41), renal cell carcinoma (42), and breast cancer (43). As for gliomas, genome-wide profiling studies have revealed that lncRNA expression profiles between normal brain tissue and gliomas are significantly different. Certain lncRNAs are involved in cancer progression, and gliomas of different malignancy grades have also been shown to have differential lncRNA expression (24,44–46). For instance, our previous research (26) demonstrated that linc-POU3F3 expression was increased in glioma tissues and was significantly associated with WHO grade. We also demonstrated that linc-CCAT2 was overexpressed in glioma tissues and was positively correlated with tumor WHO grade, and that the decrease in linc-CCAT2 expression led to the inhibition of cell proliferation, colony formation, and tumor formation (23). These studies indicated that lncRNAs are involved in glioma progression.
It has been demonstrated that most solid tumors rely on angiogenesis for continuous growth, and tumor blood vessels provide nutrition and oxygen to the tumor, resulting in tumor progression (47). Similarly, angiogenesis is also a key event in the progression of gliomas. Vascular density in high grade gliomas is markedly higher than in low grade tumors, and glioblastomas are generally acknowledged to be the most vascularized tumors with the worst prognosis (9,10). However, the mechanism of glioma angiogenesis is still unclear. Currently, exosomes have been demonstrated to act as signaling extracellular organelles that modulate the tumor microenvironment, promote angiogenesis and tumor progression (48,49). Conigliaro et al (24) revealed that exosomes released by CD90+ cancer cells were enriched in lncRNA H19, which can be taken up by endothelial cells and promote angiogenesis in vitro. Based on these studies, we hypothesized that angiogenesis in gliomas may be caused by glioma-derived exosomes that contain linc-CCAT2, which is transferred to endothelial cells to regulate angiogenesis.
In the present study, we first assessed the expression level of linc-CCAT2 in 4 glioma cell lines. We found that U87-MG glioma cells expressed the highest level of linc-CCAT2 and therefore used these cells in our experiments. Subsequently, we applied shRNA transfection technology to inhibit linc-CCAT2 expression in U87-MG cells, and we found that silencing linc-CCAT2 expression in U87-MG cells significantly decreased its expression in exosomes but did not influence exosome size or concentration. Thirdly, we investigated the function of ncU87-Exo and shU87-Exo in the regulation of HUVEC angiogenesis, and we found that both ncU87-Exo and shU87-Exo could be effectively internalized by HUVECs. However, we also found that the linc-CCAT2 expression level in the shU87-Exo-treated HUVECs was significantly lower than in the ncU87-Exo-treated HUVECs, which was possibly due to linc-CCAT2 overexpression in HUVECs. Angiogenesis functional assays revealed that ncU87-Exo cells exhibited an improved ability to promote HUVEC migration, proliferation, and tubular-like structure formation in vitro and arteriole formation in vivo. Moreover, with the use of linc-CCAT2 overexpression and downregulation in HUVECs in experiments, we demonstrated that linc-CCAT2 had a positive function in promoting angiogenesis. These results revealed that ncU87-Exo promoted angiogenesis by transferring linc-CCAT2 to HUVECs, and glioma cells promoted angiogenesis partly by transferring exosomes enriched in linc-CCAT2 to endothelial cells.
Our previous study (23) demonstrated that linc-CCAT2 activated the Wnt/β-catenin pathway by mediating the expression of downstream genes (c-Myc, MMP-7 and cyclin D1), and Wnt signaling was involved in the regulation of vascular development and endothelial cell specification (50,51). We therefore estimated the expression level of angiogenesis-related genes and proteins in HUVECs after treatment with ncU87-Exo and shU87-Exo. We found that both ncU87-Exo and shU87-Exo stimulated angiogenesis-related gene (VEGF, TGFβ, FGF, and KDR) expression and protein (VEGF, TGFβ, and FGF) secretion, while ncU87-Exo cells were more efficient. Meanwhile, we found that the overexpression of linc-CCAT2 in HUVECs promoted VEGF, TGFβ, and KDR transcript expression and VEGF and TGFβ protein secretion; downregulation of linc-CCAT2 inhibited VEGF, TGFβ, and KDR transcript expression and VEGF and TGFβ protein secretion. These results indicated that ncU87-Exo promoted angiogenesis by transferring linc-CCAT2 to HUVECs to stimulate angiogenesis-related gene and protein expression, and glioma cells promoted angiogenesis partly by transferring exosomes enriched in linc-CCAT2 to endothelial cells to activate these angiogenesis-related molecules. However, why ncU87-Exo promotes FGF gene expression and protein secretion in HUVECs while overexpression or downregulation of linc-CCAT2 in HUVECs has no effect on FGF expression, may be due to other cargo in exosomes activating FGF.
The growth of blood vessel networks occurs by angiogenesis; the inhibition of endothelial cell apoptosis that provides endothelial cell survival is thought to be an essential mechanism during angiogenesis (52). Dimmeler and Zeiher (53) demonstrated that the dysregulation of endothelial cell apoptosis markedly disturbs the establishment of the primordial vascular network in the embryo and finally leads to embryonic death. Therefore, we further investigated whether glioma-derived exosomes regulated endothelial cell apoptosis when exposed to hypoxia. Our in vitro study demonstrated that ncU87-Exo was more efficient in decreasing HUVEC apoptosis after hypoxia by upregulating the expression of Bcl-2 and downregulating the expression of Bax and caspase-3. Concomitantly, we also demonstrated that when exposed to hypoxia, HUVECs with linc-CCAT2 overexpression also exhibited higher Bcl-2 expression and lower Bax and caspase-3 expression, while HUVECs with downregulation of linc-CCAT2 exhibited lower Bcl-2 expression and higher Bax and caspase-3 expression. Wnt signaling participated in the regulation of early and late stages of apoptosis in both development and cellular injury in the cell populations of neurons, endothelial cells, vascular smooth muscle cells and cardiomyocytes by promoting anti-apoptotic protein Bcl-2 expression and inhibiting pro-apoptotic protein Bax and caspase-3 expression (54,55). Therefore, these results indicated that ncU87-Exo alleviated apoptosis by transferring linc-CCAT2 to HUVECs to activate Bcl-2 and suppress Bax and caspase-3, and glioma cell anti-apoptosis partly by transferring exosomes enriched in linc-CCAT2 to endothelial cells to activate Bcl-2 and suppress Bax and caspase-3. Furthermore, the Wnt signaling pathway may participate in this process.
In conclusion, the present study demonstrated that glioma cell-derived exosomes transfer linc-CCAT2 as a key mediator in noncontact cell-to-cell communication and in the regulation of glioma angiogenesis. In light of several studies that have highlighted the importance of exosomes in cancer biology and the results described here, we suggest that targeting exosomes and linc-CCAT2 may represent two new therapeutic applications in glioma treatment and recommend that these factors be further explored for future clinical use.
Acknowledgements
This study was supported by the National Scientific Foundation of China grant nos. 81560411 and 81560193.
References
- 1.Carmeliet P. Angiogenesis in life, disease and medicine. Nature. 2005;438:932–936. doi: 10.1038/nature04478. [DOI] [PubMed] [Google Scholar]
- 2.Carmeliet P, Jain RK. Molecular mechanisms and clinical applications of angiogenesis. Nature. 2011;473:298–307. doi: 10.1038/nature10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kerbel RS. Tumor angiogenesis. N Engl J Med. 2008;358:2039–2049. doi: 10.1056/NEJMra0706596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Folkman J. What is the evidence that tumors are angiogenesis dependent? J Natl Cancer Inst. 1990;82:4–6. doi: 10.1093/jnci/82.1.4. [DOI] [PubMed] [Google Scholar]
- 5.Millauer B, Shawver LK, Plate KH, Risau W, Ullrich A. Glioblastoma growth inhibited in vivo by a dominant-negative Flk-1 mutant. Nature. 1994;367:576–579. doi: 10.1038/367576a0. [DOI] [PubMed] [Google Scholar]
- 6.Tsai AC, Pan SL, Liao CH, Guh JH, Wang SW, Sun HL, Liu YN, Chen CC, Shen CC, Chang YL, et al. Moscatilin, a bibenzyl derivative from the India orchid Dendrobium loddigesii, suppresses tumor angiogenesis and growth in vitro and in vivo. Cancer Lett. 2010;292:163–170. doi: 10.1016/j.canlet.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 7.Algire GH. Vascular reactions of normal and malignant tissues in vivo. VII. Observations on vascular reactions in destruction of tumor homografts. J Natl Cancer Inst. 1954;15:483–491. [PubMed] [Google Scholar]
- 8.Ostrom QT, Gittleman H, Fulop J, Liu M, Blanda R, Kromer C, Wolinsky Y, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2008–2012. Neuro Oncol. 2015;17:iv1–iv62. doi: 10.1093/neuonc/nou327. (Suppl 4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rahman R, Smith S, Rahman C, Grundy R. Antiangiogenic therapy and mechanisms of tumor resistance in malignant glioma. J Oncol. 2010;2010:251231. doi: 10.1155/2010/251231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Onishi M, Ichikawa T, Kurozumi K, Date I. Angiogenesis and invasion in glioma. Brain Tumor Pathol. 2011;28:13–24. doi: 10.1007/s10014-010-0007-z. [DOI] [PubMed] [Google Scholar]
- 11.Andaloussi ELS, Mäger I, Breakefield XO, Wood MJ. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013;12:347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 12.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 13.Zheng X, Ye C, Zhao J, Bian P, Zhang Y, Jia Z. Alterations and clinical signifecance of exosome-containing innate immunity related lncRNAs in patients of hemorrhagic fever with renal syndrome. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2016;32:1522–1526. (In Chinese) [PubMed] [Google Scholar]
- 14.Wang Z, Chen J-Q, Liu JL, Tian L. Exosomes in tumor microenvironment: Novel transporters and biomarkers. J Transl Med. 2016;14:297. doi: 10.1186/s12967-016-1056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peinado H, Alečković M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, Hergueta-Redondo M, Williams C, García-Santos G, Ghajar C, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santos JC, Ribeiro ML, Sarian LO, Ortega MM, Derchain SF. Exosomes-mediate microRNAs transfer in breast cancer chemoresistance regulation. Am J Cancer Res. 2016;6:2129–2139. [PMC free article] [PubMed] [Google Scholar]
- 17.Arscott WT, Tandle AT, Zhao S, Shabason JE, Gordon IK, Schlaff CD, Zhang G, Tofilon PJ, Camphausen KA. Ionizing radiation and glioblastoma exosomes: implications in tumor biology and cell migration. Transl Oncol. 2013;6:638–648. doi: 10.1593/tlo.13640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vance KW, Ponting CP. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014;30:348–355. doi: 10.1016/j.tig.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo H, Bu D, Sun L, Fang S, Liu Z, Zhao Y. Identification and function annotation of long intervening noncoding RNAs. Brief Bioinform: bbw046. 2016 doi: 10.1093/bib/bbw046. doi: 10.1093/bib/bbw046. [DOI] [PubMed] [Google Scholar]
- 21.Meseure D, Alsibai Drak K, Nicolas A, Bieche I, Morillon A. Long noncoding RNAs as new architects in cancer epigenetics, prognostic biomarkers, and potential therapeutic targets. Biomed Res Int. 2015;2015:320214. doi: 10.1155/2015/320214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pandey GK, Kanduri C. Long noncoding RNAs and neuroblastoma. Oncotarget. 2015;6:18265–18275. doi: 10.18632/oncotarget.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo H, Hu G, Yang Q, Zhang P, Kuang W, Zhu X, Wu L. Knockdown of long non-coding RNA CCAT2 suppressed proliferation and migration of glioma cells. Oncotarget. 2016;7:81806–81814. doi: 10.18632/oncotarget.13242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conigliaro A, Costa V, Dico Lo A, Saieva L, Buccheri S, Dieli F, Manno M, Raccosta S, Mancone C, Tripodi M, et al. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol Cancer. 2015;14:155. doi: 10.1186/s12943-015-0426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu GW, Li Q, Niu X, Hu B, Liu J, Zhou SM, Guo SC, Lang HL, Zhang CQ, Wang Y, et al. Exosomes secreted by human-induced pluripotent stem cell-derived mesenchymal stem cells attenuate limb ischemia by promoting angiogenesis in mice. Stem Cell Res Ther. 2015;6:10. doi: 10.1186/scrt546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo H, Wu L, Yang Q, Ye M, Zhu X. Functional linc-POU3F3 is overexpressed and contributes to tumorigenesis in glioma. Gene. 2015;554:114–119. doi: 10.1016/j.gene.2014.10.038. [DOI] [PubMed] [Google Scholar]
- 27.Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Madrid F, Mittelbrunn M. Analysis of microRNA and protein transfer by exosomes during an immune synapse. In: Circulating MicroRNAs Methods Mol Biol. 2013;1024:41–51. doi: 10.1007/978-1-62703-453-1_4. [DOI] [PubMed] [Google Scholar]
- 28.de Vrij J, Maas SL, van Nispen M, Sena-Esteves M, Limpens RW, Koster AJ, Leenstra S, Lamfers ML, Broekman ML. Quantification of nanosized extracellular membrane vesicles with scanning ion occlusion sensing. Nanomedicine (Lond) 2013;8:1443–1458. doi: 10.2217/nnm.12.173. [DOI] [PubMed] [Google Scholar]
- 29.Khoury S, Ajuyah P, Tran N. Isolation of small noncoding RNAs from human serum. J Vis Exp. 2014;88:e51443. doi: 10.3791/51443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guescini M, Genedani S, Stocchi V, Agnati LF. Astrocytes and glioblastoma cells release exosomes carrying mtDNA. J Neural Transm Vienna. 2010;117:1–4. doi: 10.1007/s00702-009-0288-8. [DOI] [PubMed] [Google Scholar]
- 31.Nur H, Rao L, Frassanito MA, De Raeve H, Ribatti D, Mfopou JK, Van Valckenborgh E, De Bruyne E, Vacca A, Vanderkerken K, et al. Stimulation of invariant natural killer T cells by α-galactosylceramide activates the JAK-STAT pathway in endothelial cells and reduces angiogenesis in the 5T33 multiple myeloma model. Br J Haematol. 2014;167:651–663. doi: 10.1111/bjh.13092. [DOI] [PubMed] [Google Scholar]
- 32.Roma-Rodrigues C, Heuer-Jungemann A, Fernandes AR, Kanaras AG, Baptista PV. Peptide-coated gold nanoparticles for modulation of angiogenesis in vivo. Int J Nanomedicine. 2016;11:2633–2639. doi: 10.2147/IJN.S108661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prados MD, Byron SA, Tran NL, Phillips JJ, Molinaro AM, Ligon KL, Wen PY, Kuhn JG, Mellinghoff IK, de Groot JF, et al. Toward precision medicine in glioblastoma: The promise and the challenges. Neuro Oncol. 2015;17:1051–1063. doi: 10.1093/neuonc/nov031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hendrix MJ, Seftor EA, Hess AR, Seftor RE. Vasculogenic mimicry and tumour-cell plasticity: Lessons from melanoma. Nat Rev Cancer. 2003;3:411–421. doi: 10.1038/nrc1092. [DOI] [PubMed] [Google Scholar]
- 35.Balaj L, Lessard R, Dai L, Cho YJ, Pomeroy SL, Breakefield XO, Skog J. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun. 2011;2:180. doi: 10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kogure T, Yan IK, Lin W-L, Patel T. Extracellular vesicle-mediated transfer of a novel long noncoding RNA TUC339: A mechanism of intercellular signaling in human hepatocellular cancer. Genes Cancer. 2013;4:261–272. doi: 10.1177/1947601913499020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paralkar VR, Weiss MJ. Long noncoding RNAs in biology and hematopoiesis. Blood. 2013;121:4842–4846. doi: 10.1182/blood-2013-03-456111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rutenberg-Schoenberg M, Sexton AN, Simon MD. The properties of long noncoding RNAs that regulate chromatin. Annu Rev Genomics Hum Genet. 2016;17:69–94. doi: 10.1146/annurev-genom-090314-024939. [DOI] [PubMed] [Google Scholar]
- 39.Tan JY, Marques AC. The miRNA-mediated cross-talk between transcripts provides a novel layer of posttranscriptional regulation. Adv Genet. 2014;85:149–199. doi: 10.1016/B978-0-12-800271-1.00003-2. [DOI] [PubMed] [Google Scholar]
- 40.Ulitsky I, Bartel DP. lincRNAs: Genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang L, Wei H, Xiao HJ. Long non-coding RNA Loc554202 expression as a prognostic factor in patients with colorectal cancer. Eur Rev Med Pharmacol Sci. 2016;20:4243–4247. [PubMed] [Google Scholar]
- 42.He HT, Xu M, Kuang Y, Han XY, Wang MQ, Yang Q. Biomarker and competing endogenous RNA potential of tumor-specific long noncoding RNA in chromophobe renal cell carcinoma. Onco Targets Ther. 2016;9:6399–6406. doi: 10.2147/OTT.S116392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee JJ, Kim M, Kim HP. Epigenetic regulation of long noncoding RNA UCA1 by SATB1 in breast cancer. BMB Rep. 2016;49:578–583. doi: 10.5483/BMBRep.2016.49.10.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barsyte-Lovejoy D, Lau SK, Boutros PC, Khosravi F, Jurisica I, Andrulis IL, Tsao MS, Penn LZ. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res. 2006;66:5330–5337. doi: 10.1158/0008-5472.CAN-06-0037. [DOI] [PubMed] [Google Scholar]
- 45.Vital AL, Tabernero MD, Castrillo A, Rebelo O, Tão H, Gomes F, Nieto AB, Oliveira Resende C, Lopes MC, Orfao A. Gene expression profiles of human glioblastomas are associated with both tumor cytogenetics and histopathology. Neuro Oncol. 2010;12:991–1003. doi: 10.1093/neuonc/noq050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang X, Sun S, Pu JKS, Tsang AC, Lee D, Man VO, Lui WM, Wong ST, Leung GK. Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol Dis. 2012;48:1–8. doi: 10.1016/j.nbd.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 47.Hida K, Maishi N, Torii C, Hida Y. Tumor angiogenesis - characteristics of tumor endothelial cells. Int J Clin Oncol. 2016;21:206–212. doi: 10.1007/s10147-016-0957-1. [DOI] [PubMed] [Google Scholar]
- 48.Corrado C, Raimondo S, Saieva L, Flugy AM, De Leo G, Alessandro R. Exosome-mediated crosstalk between chronic myelogenous leukemia cells and human bone marrow stromal cells triggers an interleukin 8-dependent survival of leukemia cells. Cancer Lett. 2014;348:71–76. doi: 10.1016/j.canlet.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 49.Kahlert C, Kalluri R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J Mol Med (Berl) 2013;91:431–437. doi: 10.1007/s00109-013-1020-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dejana E. The role of wnt signaling in physiological and pathological angiogenesis. Circ Res. 2010;107:943–952. doi: 10.1161/CIRCRESAHA.110.223750. [DOI] [PubMed] [Google Scholar]
- 51.Parmalee NL, Kitajewski J. Wnt signaling in angiogenesis. Curr Drug Targets. 2008;9:558–564. doi: 10.2174/138945008784911822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chavakis E, Dimmeler S. Regulation of endothelial cell survival and apoptosis during angiogenesis. Arterioscler Thromb Vasc Biol. 2002;22:887–893. doi: 10.1161/01.ATV.0000017728.55907.A9. [DOI] [PubMed] [Google Scholar]
- 53.Dimmeler S, Zeiher AM. Endothelial cell apoptosis in angiogenesis and vessel regression. Circ Res. 2000;87:434–439. doi: 10.1161/01.RES.87.6.434. [DOI] [PubMed] [Google Scholar]
- 54.Gao K, Wang YS, Yuan YJ, Wan ZH, Yao TC, Li HH, Tang PF, Mei XF. Neuroprotective effect of rapamycin on spinal cord injury via activation of the Wnt/β-catenin signaling pathway. Neural Regen Res. 2015;10:951–957. doi: 10.4103/1673-5374.158360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pećina-Slaus N. Wnt signal transduction pathway and apoptosis: A review. Cancer Cell Int. 2010;10:22. doi: 10.1186/1475-2867-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]