Abstract
Gliomas are the most common primary central nervous system tumors and account for approximately 80% of malignant brain tumors. MicroRNAs (miRNAs) are a class of small non-coding, regulatory RNA molecules that mediate the expression levels of specific proteins. As a member of the miRNA family, miR-543 plays a tumor suppressive or an oncogenic role in different types of tumors. However, the expression and role of miR-543 in glioma remain unknown. In the present study, the expression level of miR-543 in glioma cell lines and tissues was investigated. A series of in vitro and in vivo experiments was then performed to elucidate the function of miR-543 in glioma. Moreover, proteomic profiling was applied in this study to determine the landscape of differentially expressed proteins associated with miR-543-mediated carcinogenesis in glioma. We found that the expression level of miR-543 was greatly downregulated in glioma cell lines and tissues. Furthermore, the expression level of miR-543 was negatively associated with high-grade glioma. Functional studies demonstrated that miR-543 in glioma cells induced apoptosis and inhibited growth, the cell cycle, migration and invasion. In addition, the in vivo study showed that miR-543 suppressed tumorigenicity of glioma cells. In the present study, a label-free quantitative proteomic approach was performed and 339 proteins were identified as dysregulated after miR-543 was overexpressed. Among these dysregulated proteins, 165 were upregulated and 174 were downregulated. Moreover, multiple pathways were significantly enriched and were probably involved in miR-543-mediated tumorigenesis, including RNA degradation and the inositol phosphate metabolism pathway. In conclusion, miR-543 may function as a tumor suppressor in glioma and may serve as a future therapeutic target in therapy for patients with glioma.
Keywords: miR-543, glioma, tumor suppressor, proteomic profiling
Introduction
Gliomas are the most common primary central nervous system tumors and account for approximately 80% of malignant brain tumors (1). Despite recent improvements in both diagnostic methods and therapeutic interventions, including surgical resection combined with postoperative radiation and chemotherapy, overall survival has not markedly improved for glioma patients (2), especially for high-grade gliomas (grades III and IV). Therefore, it is necessary to obtain a better understanding of the molecular mechanisms involved in carcinogenesis and progression to develop novel and effective therapeutic strategies for this malignancy.
MicroRNAs (miRNAs) are a class of small non-coding, regulatory RNA molecules that mediate the expression levels of specific proteins (3). Through modulating the protein levels of their target genes, miRNAs have been implicated in various biological processes, including cell proliferation, cell cycle progression, apoptosis, migration and differentiation (4). Moreover, mounting evidence suggests that the deregulation of miRNA expression plays a pivotal role in the initiation and progression of numerous cancers (5–7), including gliomas (8–10). For example, miR-217 inhibited cell proliferation and invasion by targeting Runx2 in human glioma (8). In addition, miR-184 inhibited cell proliferation and invasion and specifically targeted TNFAIP2 in glioma (9). miR-195 inhibited the proliferation of human glioma cells by directly targeting cyclin D1 and cyclin E1 (10). Consequently, the use of miRNAs in clinical anticancer therapy is both promising and expected (5–10).
miR-543 is a member of a miRNA cluster located in the imprinted DLK1-DIO3 region on human chromosome 14 (11). As a member of the miRNA family, miR-543 is aberrantly expressed in several human cancers, such as hepatocellular carcinoma and colorectal cancer (12,13), demonstrating the role of miR-543 in tumorigenesis. However, the role of miR-543 in glioma remains unknown. In the present study, the expression level of miR-543 in glioma cell lines and tissues was investigated. A series of in vitro and in vivo experiments were performed to elucidate the function of miR-543 in glioma. In addition, for further exploration, a label-free quantitative proteomic approach was applied in the present study to determine the landscape of differentially expressed proteins associated with miR-543-mediated carcinogenesis in glioma.
Materials and methods
Human specimens
The human glioma and normal brain specimens from patients were collected at the Department of Neurosurgery, the Second Affiliated Hospital of Soochow University. All glioma specimens were confirmed by pathological diagnosis and classified according to the World Health Organization (WHO) criteria. The study comprised of 6 WHO I, 22 WHO II, 16 WHO III and 18 WHO IV glioma samples. Ten normal brain tissues were obtained from patients with cerebral injury. Specimens were immediately snap-frozen in liquid nitrogen after surgical removal and stored at −80°C before use. Informed consent was obtained from all patients, and the study was approved by the Ethics Committee of the Second Affiliated Hospital of Soochow University.
Cell culture
Human glioma cell lines (U87, U251, U343, A172, T98G and LN229) were purchased from the Cell Bank of Chinese Academy of Science. Primary normal human astrocytes (NHA) were kindly provided by Professor Ming Li (Department of Neurosurgery, The Second Affiliated Hospital of Soochow University, Suzhou, China). The glioma cells were maintained in Dulbeccos modified Eagles medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and antibiotics (100 U/ml penicillin and 100 U/ml streptomycin; Gibco, Grand Island, NY, USA). Cells were grown in a 37°C incubator with 5% CO2. Primary human astrocytes were maintained in astrocyte media (Sciencell Research Laboratories, Carlsbad, CA, USA) containing 10% FBS, 1% astrocyte growth supplement and 1% penicillin/streptomycin.
RNA extraction and real-time quantitative PCR (qRT- PCR)
Total RNA was extracted from cell lines and frozen tissues using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Complementary DNA (cDNA) was synthesized using an miRNA reverse transcription kit (Shanghai GenePharma Co., Ltd., Shanghai, China) according to the manufacturers instructions. Then, the expression levels of miR-543 were quantified using a QuantiNova SYBR-Green PCR kit (Qiagen, Hilden, Germany) with the ABI 7900HT Fast system (Applied Biosystems, Foster City, CA, USA). Primer sequences of miR-543 were CAGTGCTAAAACATTCGCGG (forward) and TATGGTTGTTCACGACTCCTTCAC (reverse). The U6 gene was used as an internal reference. The primer sequences were synthesized by Shanghai GenePharma. The reaction was performed for 40 cycles under following conditions: denaturation at 95°C for 3 min, 95°C for 10 sec and 55°C for 30 sec. The relative expression ratio of miR-543 was calculated using the 2−ΔΔCT method.
Cell transfection
U87 and U251 cells were transfected with the miR-543 mimic and the corresponding negative control (miR-NC), which were obtained from Shanghai GenePharma. The sequences were as follows: miR-543 mimic, 5-UGGCG GAGAACUGAUAAGGGUCCUUAUCAGUUCUCCGUCC AUU-3; negative control (miR-NC), 5-UUCUCCGAACGUG UCACGUTTACGUGACACGUUCGGAGAATT-3.
miR-543 mimic or miR-NC was transfected into the glioma cells when the cells reached 60–70% confluence using Lipofectamine 3000 (Invitrogen) according to the manufacturers instructions. Transfection efficiencies were determined in every experiment at 48 h after transfection by qRT-PCR.
Cell proliferation assay
The number of viable cells was assayed using a Cell Counting kit-8 (CCK-8; Dojindo Laboratories, Kumamoto, Japan) per the manufacturers protocol. The cells were seeded into 96-well plates at a density of 2×103 cells/well. Then, 10 µl of the CCK-8 solution was added to each well every day for 6 days, and the plates were incubated for 1 h at 37°C. The optical density (OD) at 450 nm was measured using a microplate reader (Bio-Rad Laboratories, Hercules, CA, USA).
Colony formation assay
The colony formation rate was measured using the plate colony formation assay. Then, 1,000 cells of each group were added to each well of the 6-well plates and cultured for 10 days in DMEM medium containing 10% FBS. Then, the cells were washed with phosphate-buffered saline (PBS) and fixed using a crystal violet cell colony staining kit (Genemed, Shanghai, China). Images of the colonies were taken and the colonies were counted under a microscope (Olympus, Tokyo, Japan).
Cell cycle assay
For cell cycle analysis, cells in each group were fixed with cold 70% ethanol at 4°C for 12 h. The cells were washed twice with PBS and then stained with propidium iodide (PI) with RNase in PBS. The treated cells were then evaluated by flow cytometry (Beckman Coulter, Inc., Brea, CA, USA).
Cell apoptosis assay
Cellular apoptosis levels were detected using an Annexin V-PE apoptosis detection kit PE (BD Biosciences, San Diego, CA, USA) by flow cytometer (Beckman Coulter). Cells in each group were collected and resuspended in 400 µl of 1X binding buffer containing 50 µl Annexin V-PE and 7-AAD (7-amino-actinomycin D) at 4°C in the dark for 15 min.
Wound healing assay
The wound healing assay was conducted to detect cell migration. Cells were seeded in 6-well plates and cultured to 80% confluence. A 10-µl pipette tip was applied to generate a linear wound. Then, cells were cultured for 24 h and observed under a microscope (Olympus). The widths of wounds were determined at 0 and 24 h.
Cell invasion assay
For the cell invasion assay, Transwell chambers (Corning, Inc., Corning, NY, USA) were covered with Matrigel (BD Bioscience), and 3×104 cells suspended in 150 µl of serum-free medium were added to the upper chambers. The lower chambers were filled with 600 µl of DMEM with 10% FBS. After 24 h of incubation, the cells on the upper surface were removed, and the cells on the lower surface were stained with 0.1% crystal violet for 15 min. The stained cells were counted in 10 randomly selected fields under a microscope (magnification, ×400; Olympus).
Tumor xenograft growth assay
All procedures and experiments involving animals in this study were approved by the Institutional Animal Care and Use Committee and the Local Ethics Board. Female immunodeficient nude mice of 4–5 weeks of age were purchased from Shanghai SLAC Laboratory Animal Co., Ltd. (Shanghai, China) and maintained under specific pathogen-free (SPF) conditions. U87 and U251 cells and glioma cells (2×106) harboring miR-543 or miR-NC were separately injected subcutaneously (s.c.) into the flanks of nude mice (n=5). Growth curves and tumor volumes were measured every other day. The tumor volume was determined according to the following formula: TV (mm3) = 1/2 × width2 × length. On the 35th day after the injection, the mice were sacrificed and the tumors were removed, weighed and photographed.
Protein extraction
U87 cells were transfected with miR-543 mimic or miR-NC. Prepared samples were first frozen to a dry powder with a vacuum freeze drier. The freeze-dried powder was dissolved in 200 µl of TEAB (tetraethylammonium bromide) dissolution buffer and 800 µl of cold acetone containing 10 mM DTT (dithiothreitol). Then, the resuspended powder was incubated for ~2 h. After centrifugation at 13,000 r/min for 20 min at 15°C, the precipitate was collected and mixed with 800 µl of cold acetone containing a solution of 10 mM DTT for 1 h at 56°C to break the protein disulfide bonds. The sample was centrifuged at 13,000 r/min for 20 min at 15°C. The precipitate was collected, dried and stored at −80°C for later use.
Protein quantification and digestion
Total protein concentration was measured using the Bradford method (14). For each sample, 100 µg of protein was dissolved in 500 µl of dissolution buffer and then diluted with 500 µl of 50 mM NH4HCO3. After reduction and alkylation, 2 µg of trypsin was added and then incubated overnight at 37°C for protein digestion. After protein digestion, an equal volume of 0.1% FA (formic acid) was added for acidification. Peptides were purified thrice using a Strata-X C18 pillar, washed with 0.1% FA + 5% ACN (acetonitrile) twice, and eluted with 1 ml of 0.1% FA + 80% ACN. Eluted peptides were dried with a vacuum concentration meter.
LC-MS/MS analysis
LC-MS/MS analysis was performed on an AB SCIEX NanoLC-MS/MS system (Triple TOF 5600 plus; AB Sciex, Framingham, MA, USA). Samples were chromatographed using a 120-min gradient from 2 to 35% (buffer A 0.1% (v/v) FA, 5% (v/v) ACN, buffer B 0.1% (v/v) FA, 95% (v/v) ACN] after direct injection onto a 20-cm PicoFrit emitter (New Objective, Inc., Woburn, MA, USA) packed to 20 cm with a Magic C18 AQ 3-µm 200-Å stationary phase. MS1 spectra were collected in the range from 360 to 1,460 m/z for 250 ms. The 40 most intense precursors with a charge state of 2–5 were selected for fragmentation, and MS2 spectra were collected in the range 50–2,000 m/z for 100 ms. Precursor ions were excluded from reselection for 15 sec.
Data analysis
The original MS/MS file data were submitted to ProteinPilot Software v4.5 for data analysis. For protein identification, the Paragon algorithm (15), which was integrated into ProteinPilot, was compared against the uniprot homo database for database searching. The parameters were set as follows: the instrument was TripleTOF 5600 plus, cysteine modified with iodoacetamide; biological modifications were selected as ID focus and trypsin was selected for protein digestion. For false discovery rate (FDR) calculation, an automatic decoy database search strategy was employed to estimate FDR using the PSPEP (Proteomics System Performance Evaluation Pipeline Software, integrated in the ProteinPilot Software). Only unique peptides with global FDR values of fit <1% were considered for further analysis. Skyline v2.5 software was employed for peptide and protein quantification. For differentially expressed protein (DEP) determination, fold-changes were calculated as the average comparison pairs among biological replicates. Proteins with a fold-change >2 or <0.5 were considered to be significantly differentially expressed.
Bioinformatics and annotations
To determine the biological and functional properties of the identified proteins, the identified protein sequences were mapped with Gene Ontology terms. A homology search was first performed for all the identified sequences with a localized NCBI BLASTP program against the NCBInr Homo database. The e-value was set to <1e-5, and the best hit for each query sequence was considered for Gene Ontology (GO) term matching. The GO term matching was performed with Blast2GO v4.5 pipeline (16). In addition, an enrichment analysis of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways was performed. To identify candidate biomarkers, we employed a hypergeometric test to perform pathway enrichment. A P<0.05 was set as the threshold value.
Statistical analysis
Data from at least three independent experiments are presented as the mean ± standard deviation (SD). Statistical analyses were performed using Statistical Package for the Social Sciences (SPSS) software version 19.0 (SPSS, Inc., Chicago, IL, USA). The differences between the groups were analyzed using the Students t-test or the one-way analysis of variance (ANOVA). A P<0.05 was considered to be statistically significant.
Results
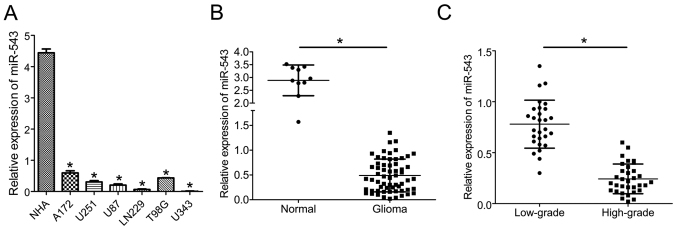
miR-543 expression was downregulated in glioma cell lines and human tissues
To explore the potential role of miR-543 in glioma progression, the expression levels of miR-543 in six glioma cell lines and normal human astrocytes (NHA) were first analyzed by quantitative real-time PCR (qRT-PCR). The expression level of miR-543 was significantly reduced in all of the six analyzed glioma cell lines compared with NHA (P<0.01; Fig. 1A). Furthermore, miR-543 expression was further investigated in 62 glioma tissues and 10 normal brain tissues. As shown in Fig. 1B, the expression of miR-543 was markedly reduced in glioma samples compared with normal brain tissues (P<0.01). In addition, miR-543 expression in high-grade (grades III and IV) gliomas was significantly decreased compared with low-grade (grades I and II) gliomas (P<0.01; Fig. 1C). Taken together, these findings indicate that miR-543 might be a hallmark of glioma and associated with the progression of glioma.
Figure 1.
miR-543 expression is downregulated in glioma cell lines and glioma tissues analyzed by qRT-PCR. (A) The expression level of miR-543 was significantly reduced in all analyzed glioma cell lines compared with NHA (P<0.01). (B) The expression of miR-543 was markedly reduced in glioma samples compared with normal brain tissues (P<0.01). (C) miR-543 expression in high-grade (grades III and IV) gliomas was significantly reduced compared with low-grade (grades I and II) gliomas (P<0.01). *P<0.01.
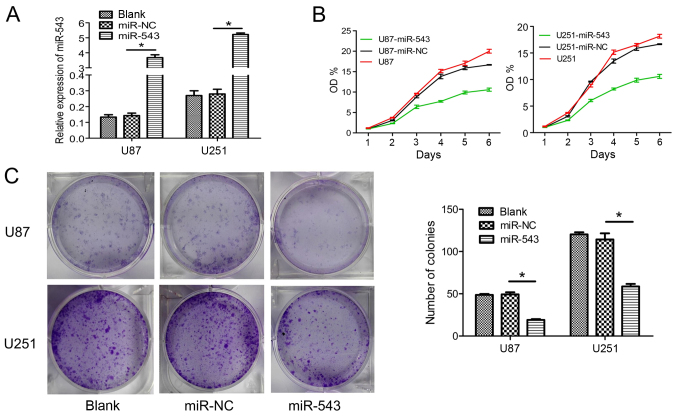
miR-543 inhibited the growth of glioma cells in vitro
To further examine the role of miR-543 in glioma cells, U87 and U251 cells were transfected with miR-543 mimic or miR-NC. For subsequent assays, U87-miR-543 and U251-miR-543 represent the glioma cells transfected with miR-543 mimic. The negative controls U87-miR-NC and U251-miR-NC represent the glioma cells transfected with miR-NC. U87 and U251 cells were used as the blank control. Increased expression of U87-miR-543 and U251-miR-543 were confirmed using qRT-PCR (Fig. 2A). Then, glioma cell growth was detected in vitro. CCK-8 assays showed that the overexpression of miR-543 significantly inhibited cell proliferation in U87 and U251 cells compared with control groups (Fig. 2B). Furthermore, the overexpression of miR-543 markedly inhibited colony formation in glioma cells (Fig. 2C). These results suggested that miR-543 contributed to the reduced proliferation of glioma cells.
Figure 2.
Overexpression of miR-543 inhibits the growth of glioma cells in vitro. (A) qRT-PCR was used to detect miR-543 expression in U87 and U251 cells transfected with the miR-543 mimic or miR-NC. The increased expression of U87-miR-543 and U251-miR-543 was confirmed. (B) Cell proliferation was measured by CCK-8 assay, and the overexpression of miR-543 significantly inhibited cell proliferation in both U87 and U251 cells compared with control groups (P<0.01). (C) Cell colony formation ability was measured by plate colony formation assay. Overexpression of miR-543 markedly inhibited colony formation in both U87 and U251 cells (P<0.01). *P<0.01.
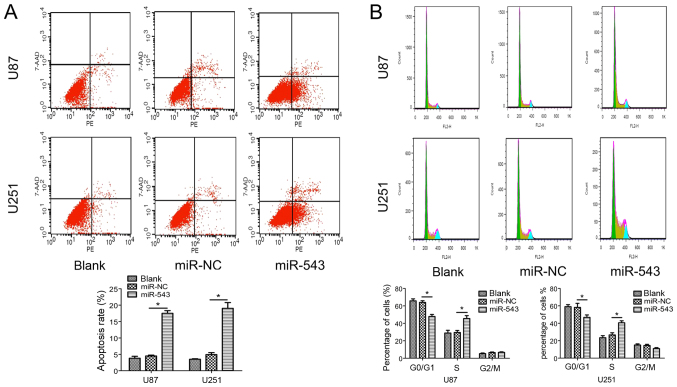
miR-543 induces apoptosis and inhibits the cell cycle
Furthermore, flow cytometry was used to examine the effects of miR-543 on cell apoptosis and cell cycle distribution. The results showed that the proportion of apoptotic cells transfected with miR-543 was significantly increased compared with control cells (Fig. 3A). In addition, as shown in Fig. 3B, miR-543 overexpression increased the percentage of cells in the S phase (P<0.01). The result showed that miR-543 overexpression in U87 and U251 cells significantly induced cell cycle arrest at the S phase. Collectively, these findings indicate that miR-543 acts as a tumor suppressor in the growth of glioma, likely via modulating cell cycle progression.
Figure 3.
Overexpression of miR-543 induces apoptosis and inhibited cell cycle progression. (A) miR-543 overexpression induced apoptosis in U87 and U251 cells. The upper panel presents representative miR-543 overexpression, which increased apoptosis of U87 and U251 cells. The lower panel presents the relative number of apoptotic cells. The proportion of apoptotic cells transfected with miR-543 was significantly increased compared with control cells (P<0.01). (B) The upper panel presents miR-543 overexpression that increased the cell population in the S phase compared with the control group. The lower panel presents quantitative data of cells in different cell cycle phases (P<0.01). *P<0.01.
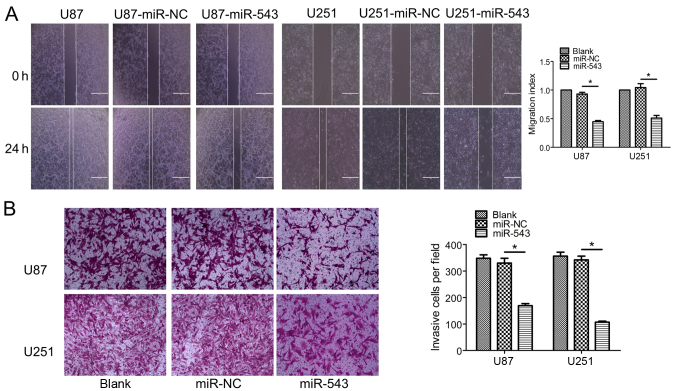
miR-543 regulates the migration and invasion of glioma cells in vitro
To further clarify the effect of miR-543 on the migration and invasion ability of U87 and U251 cells, wound healing and Transwell invasion assays were performed. As shown in Fig. 4A, a significant difference between the migration of miR-543 overexpression cells and control cells was noted (P<0.01). The Transwell invasion assay showed that the overexpression of miR-543 significantly decreased the number of glioma cells capable of invasion (Fig. 4B; P<0.01). These results suggest that miR-543 inhibits the migration and invasion of glioma cells in vitro.
Figure 4.
Overexpression of miR-543 inhibits the migration and invasion of glioma cells. (A) Cell migration was determined by a wound healing assay. A significant difference was noted between the miR-543 overexpression cells and the control cells (P<0.01). (B) The transwell invasion assay showed that the overexpression of miR-543 significantly decreased the number of glioma cells capable of invasion (P<0.01). *P<0.01.
miR-543 suppresses the growth of glioma xenografts
The in vitro study indicated that miR-543 inhibits glioma cell proliferation and invasion; therefore, the growth inhibitory effect of miR-543 in vivo was evaluated using a subcutaneously xenografted glioma model. U87 and U251 cells were chosen for the in vivo study. The glioma cells stably expressing miR-543 or miR-NC were subcutaneously inoculated into nude mice. Tumor volumes were estimated in the subcutaneous model every week after injection. Five weeks after inoculation, the mice were sacrificed and tumor tissues were extracted. Tumors formed by miR-543 overexpressing cells were visibly smaller than those formed by control cells (Fig. 5B and D). The average volume and weight of tumors formed by miR-543 overexpressing cells were significantly decreased compared with control cells (both P<0.01; Fig. 5). The results demonstrated that miR-543 could suppress the growth of glioma xenografts.
Figure 5.
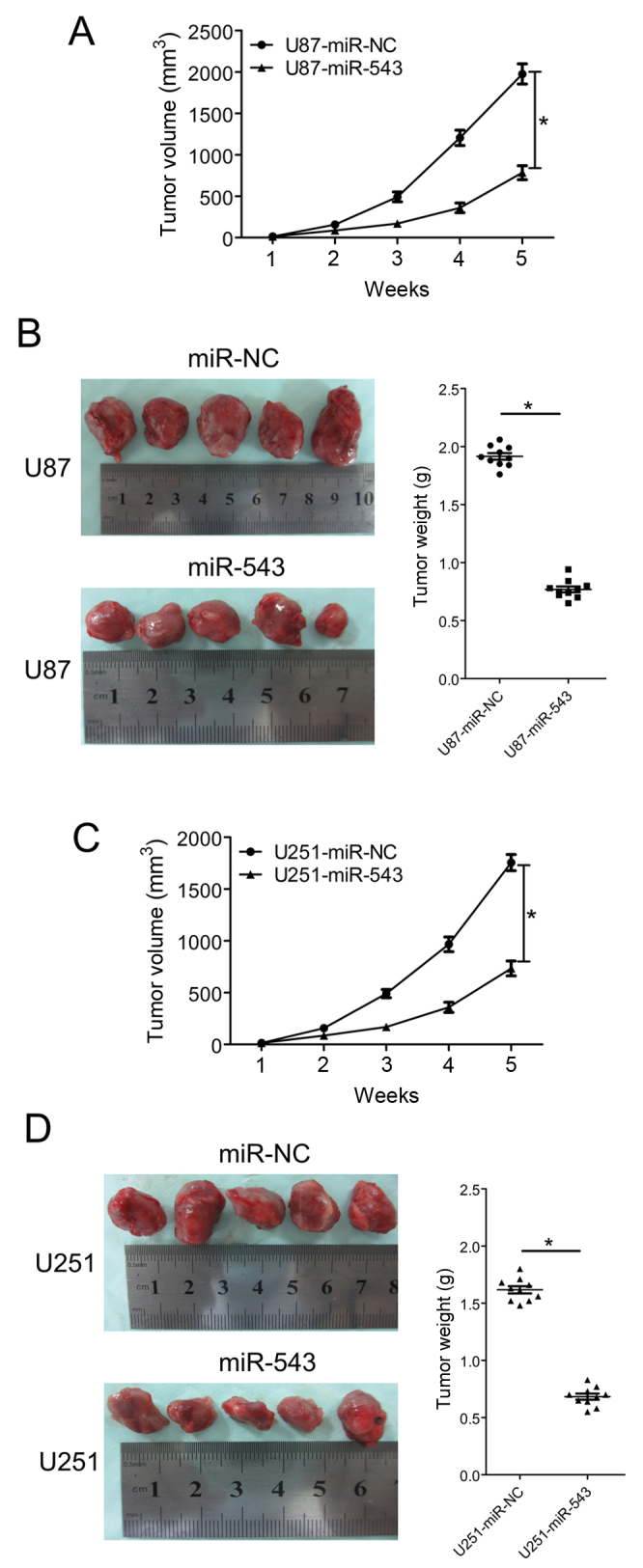
miR-543 suppresses the growth of glioma cells in vivo. (A) Tumor growth curves from the U87-miR-543 and U87-miR-NC groups. (B) Images of tumors derived from the U87-miR-543 and U87-miR-NC groups. Tumor weight in nude mice from the U87-miR-543 and U87-miR-NC groups. (C) Tumor growth curves from the U251-miR-543 and U251-miR-NC groups. (D) Images of tumors derived from the U251-miR-543 and U251-miR-NC groups. Tumor weight in the nude mice from the U251-miR-543 and U251-miR-NC groups. These images show that miR-543 inhibited glioma growth in the subcutaneous glioma nude mouse model. *P<0.01.
Identification of miR-543-regulated proteins using a quantitative proteomic approach
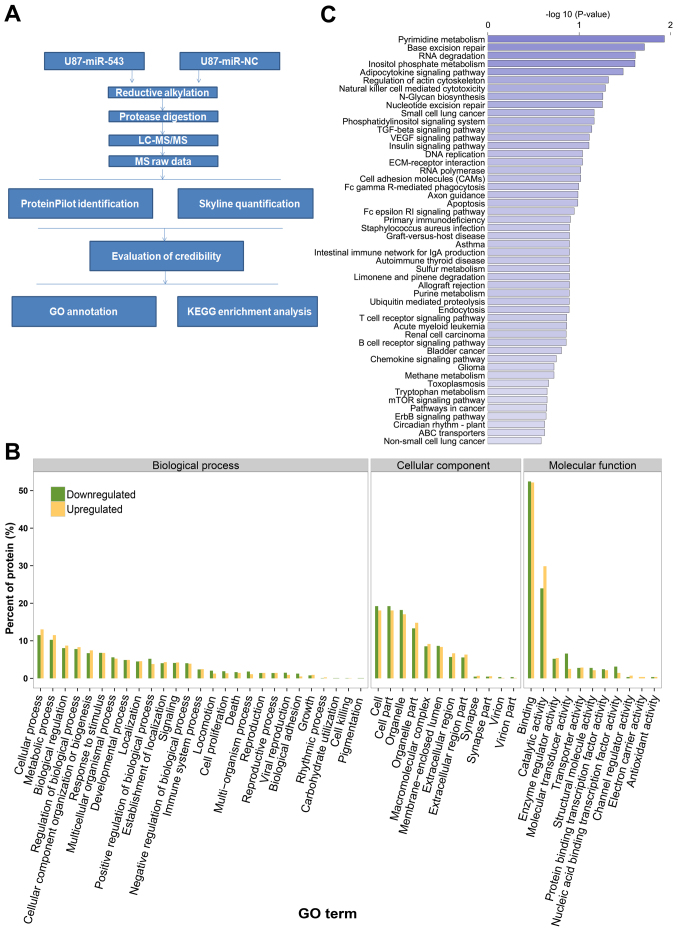
Given that miR-543 was downregulated in U87 cells, we hypothesized that the upregulation of miR-543 alters the expression of its target genes. A label-free quantitative proteomics approach was used to identify dysregulated proteins in U87 glioma cells with or without miR-543 overexpression. The workflow in the quantitative proteomics approach is presented in Fig. 6A. A total of 2,446 proteins were quantified according to the criteria described in Materials and methods. Among all the quantified proteins, 339 proteins were differentially expressed after miR-543 overexpression. Of these, 165 were upregulated and 174 were downregulated. The identified differentially expressed proteins included CKAP5, SYNJ2, POMP, CSN5, NOL7 and GOLM1. Table I presents the previously noted 20 upregulated and downregulated proteins.
Figure 6.
Quantitative proteomics identification of miR-543-regulated proteins and pathways. (A) The workflow in the label-free quantitative proteomics approach in this study is presented. The label-free quantitative proteomics approach was used to identify dysregulated proteins in U87 glioma cells with or without miR-543 overexpression. (B) The miR-543-regulated proteins were classified by their GO cellular components, biological process and molecular function. (C) Enrichment analysis of KEGG pathways among miR-543 regulated proteins. The horizontal axis presents the value of -log10 (P-value), whereas the vertical axis presents the pathway name. The larger value of the horizontal axis suggests a more significantly enriched pathway.
Table I.
Proteins differentially expressed after miR-543 was overexpressed in U87 glioma cells.
| Protein | miR-543:miR-NC |
|---|---|
| Upregulated (20) | |
| Q5LJA5 | 34.14326014 |
| A0A024RDE8 | 26.47960363 |
| GSLG1 | 20.15439479 |
| B7Z410 | 19.53041636 |
| ARK72 | 16.30160722 |
| AN32B | 10.8127484 |
| B4DEX5 | 10.1040579 |
| CKAP5 | 10.04365302 |
| Q6IAX1 | 8.927064395 |
| A0A024QZN8 | 7.504923236 |
| F8W8R3 | 7.432445004 |
| SYNJ2 | 7.16088658 |
| POMP | 6.727857068 |
| IF4B | 6.688933707 |
| A0A0C4DFT3 | 6.599762499 |
| EXOS3 | 6.591721953 |
| B7ZM73 | 5.992988916 |
| SSRD | 5.988039064 |
| Q53HS1 | 5.814460789 |
| MPPA | 5.634894759 |
| Downregulated (20) | |
| H32 | 0.036087 |
| B3KVB3 | 0.057414 |
| CUL4A | 0.05955 |
| GOLM1 | 0.062021 |
| NUP53 | 0.074681 |
| D6R905 | 0.07473 |
| Q5U091 | 0.079087 |
| Q53G69 | 0.090318 |
| A0A024R7M6 | 0.092072 |
| RUSD2 | 0.096349 |
| OVCA2 | 0.103314 |
| RBM15 | 0.121837 |
| C9K0W8 | 0.133651 |
| A0A024R7D5 | 0.145933 |
| ARI3C | 0.1481 |
| NOL7 | 0.154437 |
| A8K4H7 | 0.163771 |
| CE051 | 0.174426 |
| B4DKM0 | 0.176571 |
| PP2AA | 0.178847 |
Bioinformatic analysis of miR-543-regulated proteins
Based on the results of the proteomic analysis, the miR-543-regulated proteins were classified by their GO cellular components, biological process and molecular function to achieve an overview of the GO distribution (Fig. 6B). GO cellular component classification results revealed that many miR-543-regulated proteins were assigned to cells, cell parts, organelle and organelle parts. GO biological process analysis revealed that the miR-543-regulated proteins were involved in diverse biological processes, such as cellular process, metabolic process, biological regulation, regulation of biological process and cellular component organization or biogenesis. In the GO molecular function category, binding, catalytic activity, enzyme regulator activity and molecular transducer activity were noted as the main classifications. As a whole, this bioinformatic information indicated that miR-543 may act as an important regulator of different cellular functions across extensive biological processes.
To gain insights into the function of miR-543, the enrichment analysis of KEGG pathways among miR-543 regulated proteins was performed. Fig. 6C showed the pathways associated with miR-543 regulated proteins with the bioinformatic analysis. Moreover, according to the P-values obtained with hypergeometric tests, we identified pathways markedly enriched and likely involved in miR-543-mediated tumorigenesis. The enriched pathways included pyrimidine metabolism, base excision repair, RNA degradation, inositol phosphate metabolism, adipocytokine signaling pathway and regulation of the actin cytoskeleton. These results may indicate the involvement of miR-543 in intricate physiological and pathological systems.
Discussion
It was believed that the exploration of the molecular mechanisms involved in tumor initiation and progression affords the possibility to identify molecules that may serve as new targets for the effective therapy of glioma. There is accumulating evidence demonstrating that miRNAs play crucial roles in tumorigenic processes, such as cell proliferation, apoptosis, angiogenesis and invasion (17–19). miRNAs have been associated with tumor suppressor and oncogenic activities in glioma (20–22). Therefore, it has been proposed that miRNAs may act as potential targets for the gene treatment of glioma.
Some studies have demonstrated the association between miR-543 and several human cancers. For instance, Yu et al (12) demonstrated that miR-543 was dramatically overexpressed in hepatocellular carcinoma (HCC), and the overexpression of miR-543 promoted the proliferative and invasive potential of the HCC cell line HepG2, and miR-543 acted as an oncogene by targeting PAQR3 in HCC. Moreover, miR-543 promoted gastric cancer cell proliferation by targeting SIRT1 (23). On the contrary, Fan et al (13) reported that miR-543 expression was significantly downregulated in tumors from patients with colorectal cancer (CRC), and miR-543 inhibited the growth and metastasis of CRC cells in vitro and in vivo by targeting KRAS, MTA1 and HMGA2. Another similar study found that miR-543 suppressed endometrial cancer oncogenicity via targeting FAK and TWIST1 expression (24). The different roles of miR-543 in different tumors suggested that miR-543 may exert different functions in different cell types. To the best of our knowledge, the role of miR-543 in glioma has not been reported. In the present study, the expression level of miR-543 was extensively downregulated in glioma cell lines and tissues. Furthermore, the expression level of miR-543 was negatively associated with high-grade glioma. A functional study demonstrated that miR-543 in glioma cells induced apoptosis and inhibited cell growth, the cell cycle, migration and invasion. In addition, the in vivo study showed that miR-543 suppressed the tumorigenicity of glioma cells. According to all these results, miR-543 may function as a tumor suppressor in glioma and may serve as a potential biomarker for glioma.
miRNAs generally demonstrate their biological activities by regulating their target genes through mRNA degradation and translational repression. In the latter mechanism, the mRNA of the target remains unchanged. Unlike most studies using microarray and RNA-Seq to measure dysregulated mRNAs, we used a proteomic approach to reveal protein changes after miR-543 overexpression (25,26). Studies have demonstrated that one miRNA can regulate the levels of hundreds of proteins (4); thus, it is valuable and efficient to conduct a high-throughput protein discovery for further exploration. Proteomic profiling is the most frequently used approaches for high-throughput protein detection, and proteomic pathway analyses are emerging as a pivotal method for comprehending molecular networks (27,28). Although numerous studies of miRNA function in tumors have been reported, little information is available on the landscape of differentially expressed proteins associated with miRNAs that mediate the progression of tumors. In the present study, we investigated for the first time the proteomic profile of U87-miR-543 and the corresponding negative control U87-miR-NC. The application of the proteomic profile to identify proteins should be a prerequisite before further exploration to elucidate the key molecules and molecular pathways involved in miR-543-mediated tumorigenesis. In the present study, 339 proteins were identified as dysregulated after miR-543 overexpression. Among these, 165 were upregulated and 174 were downregulated. Among the identified differentially expressed proteins, some molecules may be involved in miR-543-mediated in tumorigenesis. For example, CKAP5 (cytoskeleton-associated protein 5), also known as colonic hepatic tumor overexpressed gene (ch-TOG), played a critical role in centrosome clustering in cancer cells (29) and is essential for tumor cell survival in non-small cell lung cancer (NSCLC) and head and neck squamous cell carcinoma (HNSCC) (30). POMP, the proteasome maturation protein, was targeted by miR-101 to impair proteasome assembly and activity, resulting in the accumulation of p53 and cyclin-dependent kinase inhibitors, cell cycle arrest and apoptosis (31). As a member of the COP9 signalosome complex, CSN5 (also known as Jab1) is overexpressed in numerous human cancers and affects multiple pathways associated with cell proliferation and apoptosis (32). Moreover, multiple pathways are significantly enriched and likely involved in miR-543-mediated tumorigenesis. For instance, members of the inositol phosphate metabolism pathway regulate cell proliferation, migration and phosphatidylinositol-3-kinase (PI3K)/Akt signaling and are frequently dysregulated in cancer (33). Consequently, the present study provided deep viewpoint into the key molecules and molecular pathways involved for further exploration.
Despite the exciting potential as therapeutic targets in human cancers, there are some scientific challenges. The major challenge to miRNA-based therapeutics is an effective mode of delivery of miRNA-targeting agents. Limitations that may be overcome by delivery include, but are not limited to, poor in vivo stability, inappropriate biodistribution, disruption and saturation of endogenous RNA machinery, and untoward side effects (34). However, miRNA-based therapies, either restoring or repressing miRNAs expression and activity, hold great promise.
In conclusion, this study first found that miR-543 expression was downregulated in glioma and that its expression was negatively associated with advanced tumor stage, and miR-543 may functioned as a novel tumor suppressor in glioma. Proteomic analysis shows the landscape of differentially expressed proteins associated with miR-543, providing comprehensive and deep insight into the potential molecular mechanisms. Taken together, our data indicate that miR-543 may be a future therapeutic target in glioma therapy.
Acknowledgements
The present study was supported by the Foundation Program of Suzhou Science and Technology Project (SYSD2014092), the Research and Innovation Project for College Graduates of Jiangsu Province (KYLX_1263), and the Foundation of Young Member of the Second Affiliated Hospital of Soochow University (SDFEYQN1409).
References
- 1.Ohgaki H. Epidemiology of brain tumors. Methods Mol Biol. 2009;472:323–342. doi: 10.1007/978-1-60327-492-0_14. [DOI] [PubMed] [Google Scholar]
- 2.Huse JT, Holland E, DeAngelis LM. Glioblastoma: Molecular analysis and clinical implications. Annu Rev Med. 2013;64:59–70. doi: 10.1146/annurev-med-100711-143028. [DOI] [PubMed] [Google Scholar]
- 3.Orang Valinezhad A, Safaralizadeh R, Kazemzadeh-Bavili M. Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-specific upregulation. Int J Genomics. 2014;2014:970607. doi: 10.1155/2014/970607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei Y, Schober A, Weber C. Pathogenic arterial remodeling: The good and bad of microRNAs. Am J Physiol Heart Circ Physiol. 2013;304:H1050–H1059. doi: 10.1152/ajpheart.00267.2012. [DOI] [PubMed] [Google Scholar]
- 5.Zhang S, Hao J, Xie F, Hu X, Liu C, Tong J, Zhou J, Wu J, Shao C. Downregulation of miR-132 by promoter methylation contributes to pancreatic cancer development. Carcinogenesis. 2011;32:1183–1189. doi: 10.1093/carcin/bgr105. [DOI] [PubMed] [Google Scholar]
- 6.Wang W, Zhou X, Wei M. MicroRNA-144 suppresses osteosarcoma growth and metastasis by targeting ROCK1 and ROCK2. Oncotarget. 2015;6:10297–10308. doi: 10.18632/oncotarget.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiner-Gorzel K, Dempsey E, Milewska M, McGoldrick A, Toh V, Walsh A, Lindsay S, Gubbins L, Cannon A, Sharpe D, et al. Overexpression of the microRNA miR-433 promotes resistance to paclitaxel through the induction of cellular senescence in ovarian cancer cells. Cancer Med. 2015;4:745–758. doi: 10.1002/cam4.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu Y, Zhao H, Feng L, Xu S. MicroRNA-217 inhibits cell proliferation and invasion by targeting Runx2 in human glioma. Am J Transl Res. 2016;8:1482–1491. [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng Z, Wang HZ, Li X, Wu Z, Han Y, Li Y, Chen G, Xie X, Huang Y, Du Z, et al. MicroRNA-184 inhibits cell proliferation and invasion, and specifically targets TNFAIP2 in Glioma. J Exp Clin Cancer Res. 2015;34:27. doi: 10.1186/s13046-015-0142-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H, Lu Y, Luo L, Li W, Liang C, He H, Ba Y. MicroRNA-195 inhibits the proliferation of human glioma cells by directly targeting cyclin D1 and cyclin E1. PLoS One. 2013;8:e54932. doi: 10.1371/journal.pone.0054932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haga CL, Phinney DG. MicroRNAs in the imprinted DLK1-DIO3 region repress the epithelial-to-mesenchymal transition by targeting the TWIST1 protein signaling network. J Biol Chem. 2012;287:42695–42707. doi: 10.1074/jbc.M112.387761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu L, Zhou L, Cheng Y, Sun L, Fan J, Liang J, Guo M, Liu N, Zhu L. MicroRNA-543 acts as an oncogene by targeting PAQR3 in hepatocellular carcinoma. Am J Cancer Res. 2014;4:897–906. [PMC free article] [PubMed] [Google Scholar]
- 13.Fan C, Lin Y, Mao Y, Huang Z, Liu AY, Ma H, Yu D, Maitikabili A, Xiao H, Zhang C, et al. MicroRNA-543 suppresses colorectal cancer growth and metastasis by targeting KRAS, MTA1 and HMGA2. Oncotarget. 2016;7:21825–21839. doi: 10.18632/oncotarget.7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 15.Shilov IV, Seymour SL, Patel AA, Loboda A, Tang WH, Keating SP, Hunter CL, Nuwaysir LM, Schaeffer DA. The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol Cell Proteomics. 2007;6:1638–1655. doi: 10.1074/mcp.T600050-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Conesa A, Götz S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics. 2008;2008:619832. doi: 10.1155/2008/619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu WG, Shang YL, Cong XR, Bian X, Yuan Z. MicroRNA-135b promotes proliferation, invasion and migration of osteosarcoma cells by degrading myocardin. Int J Oncol. 2014;45:2024–2032. doi: 10.3892/ijo.2014.2641. [DOI] [PubMed] [Google Scholar]
- 18.Yu SH, Zhang CL, Dong FS, Zhang YM. miR-99a suppresses the metastasis of human non-small cell lung cancer cells by targeting AKT1 signaling pathway. J Cell Biochem. 2015;116:268–276. doi: 10.1002/jcb.24965. [DOI] [PubMed] [Google Scholar]
- 19.Tsai MM, Wang CS, Tsai CY, Chen CY, Chi HC, Tseng YH, Chung PJ, Lin YH, Chung IH, Chen CY, et al. MicroRNA-196a/−196b promote cell metastasis via negative regulation of radixin in human gastric cancer. Cancer Lett. 2014;351:222–231. doi: 10.1016/j.canlet.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 20.Zhang QQ, Xu H, Huang MB, Ma LM, Huang QJ, Yao Q, Zhou H, Qu LH. MicroRNA-195 plays a tumor-suppressor role in human glioblastoma cells by targeting signaling pathways involved in cellular proliferation and invasion. Neuro Oncol. 2012;14:278–287. doi: 10.1093/neuonc/nor216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao X, Jin W. The emerging role of tumor-suppressive microRNA-218 in targeting glioblastoma stemness. Cancer Lett. 2014;353:25–31. doi: 10.1016/j.canlet.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 22.Che S, Sun T, Wang J, Jiao Y, Wang C, Meng Q, Qi W, Yan Z. miR-30 overexpression promotes glioma stem cells by regulating Jak/STAT3 signaling pathway. Tumour Biol. 2015;36:6805–6811. doi: 10.1007/s13277-015-3400-8. [DOI] [PubMed] [Google Scholar]
- 23.Li J, Dong G, Wang B, Gao W, Yang Q. miR-543 promotes gastric cancer cell proliferation by targeting SIRT1. Biochem Biophys Res Commun. 2016;469:15–21. doi: 10.1016/j.bbrc.2015.11.062. [DOI] [PubMed] [Google Scholar]
- 24.Bing L, Hong C, Li-Xin S, Wei G. MicroRNA-543 suppresses endometrial cancer oncogenicity via targeting FAK and TWIST1 expression. Arch Gynecol Obstet. 2014;290:533–541. doi: 10.1007/s00404-014-3219-3. [DOI] [PubMed] [Google Scholar]
- 25.Wang W, Luo J, Sheng W, Xue J, Li M, Ji J, Liu P, Zhang X, Cao J, Zhang S. Proteomic profiling of radiation-induced skin fibrosis in rats: Targeting the ubiquitin-proteasome system. Int J Radiat Oncol Biol Phys. 2016;95:751–760. doi: 10.1016/j.ijrobp.2016.01.021. [DOI] [PubMed] [Google Scholar]
- 26.Zhang S, Wang W, Gu Q, Xue J, Cao H, Tang Y, Xu X, Cao J, Zhou J, Wu J, et al. Protein and miRNA profiling of radiation-induced skin injury in rats: The protective role of peroxiredoxin-6 against ionizing radiation. Free Radic Biol Med. 2014;69:96–107. doi: 10.1016/j.freeradbiomed.2014.01.019. [DOI] [PubMed] [Google Scholar]
- 27.Yan W, Chen SS. Mass spectrometry-based quantitative proteomic profiling. Brief Funct Genomics Proteomics. 2005;4:27–38. doi: 10.1093/bfgp/4.1.27. [DOI] [PubMed] [Google Scholar]
- 28.Hanash S. Disease proteomics. Nature. 2003;422:226–232. doi: 10.1038/nature01514. [DOI] [PubMed] [Google Scholar]
- 29.Fielding AB, Lim S, Montgomery K, Dobreva I, Dedhar S. A critical role of integrin-linked kinase, ch-TOG and TACC3 in centrosome clustering in cancer cells. Oncogene. 2011;30:521–534. doi: 10.1038/onc.2010.431. [DOI] [PubMed] [Google Scholar]
- 30.Martens-de Kemp SR, Nagel R, Stigter-van Walsum M, van der Meulen IH, van Beusechem VW, Braakhuis BJ, Brakenhoff RH. Functional genetic screens identify genes essential for tumor cell survival in head and neck and lung cancer. Clin Cancer Res. 2013;19:1994–2003. doi: 10.1158/1078-0432.CCR-12-2539. [DOI] [PubMed] [Google Scholar]
- 31.Zhang X, Schulz R, Edmunds S, Krüger E, Markert E, Gaedcke J, Cormet-Boyaka E, Ghadimi M, Beissbarth T, Levine AJ, et al. MicroRNA-101 suppresses tumor cell proliferation by acting as an endogenous proteasome inhibitor via targeting the proteasome assembly factor POMP. Mol Cell. 2015;59:243–257. doi: 10.1016/j.molcel.2015.05.036. [DOI] [PubMed] [Google Scholar]
- 32.Sang MM, Du WQ, Zhang RY, Zheng JN, Pei DS. Suppression of CSN5 promotes the apoptosis of gastric cancer cells through regulating p53-related apoptotic pathways. Bioorg Med Chem Lett. 2015;25:2897–2901. doi: 10.1016/j.bmcl.2015.05.057. [DOI] [PubMed] [Google Scholar]
- 33.Tan J, Yu CY, Wang ZH, Chen HY, Guan J, Chen YX, Fang JY. Genetic variants in the inositol phosphate metabolism pathway and risk of different types of cancer. Sci Rep. 2015;5:8473. doi: 10.1038/srep08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Wang Z, Gemeinhart RA. Progress in microRNA delivery. J Control Release. 2013;172:962–974. doi: 10.1016/j.jconrel.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]