Abstract
These data are presented in support of structural and evolutionary analysis of the published article entitled “The occurrence of three D-J-C clusters within the dromedary TRB locus highlights a shared evolution in Tylopoda, Ruminantia and Suina” (Antonacci et al., 2017) [1]. Here we describe the genomic structure and the gene content of the T cell receptor beta chain (TRB) locus in Camelus dromedarius. As in the other species of mammals, the general genomic organization of the dromedary TRB locus consists of a pool of TRBV genes located upstream of in tandem TRBD-J-C clusters, followed by a TRBV gene with an inverted transcriptional orientation. A peculiarity of the dromedary TRB locus structure is the presence of three TRBD-J-C clusters, which is a common feature of sheep, cattle and pig sequences.
Keywords: T cell receptor, TRB locus, Dromedary genome, Camelus dromedarius, IMGT
Specifications Table
| Subject area | Biology, genetics, genomics |
|---|---|
| More specific subject area | Genetics, Genomics and Molecular Biology |
| Type of data | Tables and figures |
| How data was acquired | A standard BLAST search (Basic Local Alignment Search Tool.http://blast.ncbi.nlm.nih.gov/Blast.cgi.) of the public dromedary genomic assembly, Long PCR on genomic DNA and cloning |
| Data format | Analyzed |
| Experimental factors | Sequence analysis and dromedary DNA extraction |
| Experimental features | Dromedary lung genomic DNA was prepared from a single healthy animal. PCRs were performed by High Fidelity DNA polymerase. The PCR products were purified and cloned into the TA-vector system. |
| Data source location | Bari and Lecce, Italy |
| Data accessibility | The whole dromedary genome shotgun sequence is available at GenBank (ID: GCA_000767585.1). Sequence data published with this article were registered in EMBL database with the Accession numberLT837971 |
Value of the data
-
•
These data insight into the genomic structure of the T cell receptor (TRB) locus in Camelus dromedaries. This results in the first, mostly complete, map of the TRB locus in a species of the Tylopoda suborder.
-
•
The dromedary TRB locus characterization can be used to increase the understanding in the evolution of Camelidae and to contribute to solving the relative placement of this species within the Artiodactyla order.
-
•
The availability of the sequence of the dromedary TRB locus allows researchers to concentrate on functional study and provides a tool to use this specie as a valuable model for immunological research.
1. Data
Data presented in the text include tables and figures giving information on the genomic structure and the gene content of the dromedary TRB locus, a mammalian species belonging to the Camelus genus. This information was obtained by integrating the sequence data deduced from the public genomic assembly [2] with sequences obtained by PCR experiments conducted in our laboratory. Table 1 describes position, classification and functionality of the TRB genes retrieved from the dromedary public genome assembly. Table 2 shows the description of the dromedary TRBV pseudogenes. Table 3 describes position, classification and functionality of the unrelated TRB genes recovered from the dromedary public genome assembly. Fig. 1 shows the deduced amino acid sequences of the dromedary TRBV genes according to IMGT unique numbering for the V-REGION [6]. Table 4 provides the list of the genomic clones of the dromedary TRBD-J-C region with the primer pairs used and the PCR conditions. Fig. 2 shows the TRBD, the TRBJ and the TRBC gene sequences.
Table 1.
Description of the TRB genes in the Camelus dromedarius genome assembly. The position of all genes and their classification and functionality are reported.
| Gene classification | Functionalitya | NCBI Reference Sequence | Positionb |
|---|---|---|---|
| TRBV1 | F | NW_011591622 | 861263-861886 |
| TRBV2 | F | NW_011591622 | 932263-932714 |
| TRBV3 | P | NW_011591622 | 927952-928412 |
| TRBV5S1 | F | NW_011591622 | 937384-937843 |
| TRBV5S2 | F | NW_011591622 | 940879-941358 |
| TRBV5S3 | F | NW_011591622 | 955293-955748 |
| TRBV6 | F | NW_011591622 | 944809-945237 |
| TRBV7S1 | F | NW_011591622 | 947134-947581 |
| TRBV7S2 | F | NW_011591622 | 962228-962689 |
| TRBV8 | F | NW_011591622 | 950124-950593 |
| TRBV9 | P | NW_011591622 | 965923-966346 |
| TRBV10 | F | NW_011591622 | 970368-970809 |
| TRBV11 | F | NW_011591622 | 975860-976308 |
| TRBV12S1 | P | NW_011591622 | 981727-982197 |
| TRBV12S2 | P | NW_011591622 | 992125-992569 |
| TRBV14 | P | NW_011591622 | 995472-995906 |
| TRBV15S1 | F | NW_011591622 | 997569-998023 |
| TRBV15S2 | F | NW_011591622 | 999129-999583 |
| TRBV16 | F | NW_011591622 | 1003645-1004098 |
| TRBV19 | F | NW_011591622 | 1018094-1018641 |
| TRBV20 | F | NW_011591622 | 1020910-1021565 |
| TRBV21S1 | F | NW_011591622 | 1028337-1028797 |
| TRBV21S2 | F | NW_011591151 | 70843-70731 |
| TRBV21S3 | P | NW_011591151 | 62738-62511 |
| TRBV22 | F | NW_011591151 | 46518-46381 |
| TRBV23 | P | NW_011591151 | 60590-60480 |
| TRBV24 | P | NW_011591151 | 56428-56106 |
| TRBV25 | F | NW_011591151 | 52347-52219 |
| TRBV26 | F | NW_011591151 | 66428-66297 |
| TRBV27 | F | NW_011591151 | 41158-41032 |
| TRBV28 | F | NW_011591151 | 32762-32640 |
| TRBV29 | F | NW_011591151 | 27109-26837 |
| TRBD1 | F | NW_011591151 | 9932-9943 |
| TRBJ1-1 | F | NW_011591151 | 9247-9294 |
| TRBJ1-2 | F | NW_011591151 | 9116-9159 |
| TRBJ1-3 | F | NW_011591151 | 8861-8910 |
| TRBJ1-4 | F | NW_011591151 | 8258-8308 |
| TRBJ1-5 | F | NW_011591151 | 7982-8031 |
| TRBJ1-6 | F | NW_011591151 | 7491-7543 |
| TRBC1 | F | NW_011591151 | EX1 4773-5166 |
| EX2 4311-4328 | |||
| EX3 4044-4150 | |||
| EX4 3711-3731 | |||
| TRBC3 | nd | NW_011591151 | EX2 2866-2883 |
| EX3 2599-2705 | |||
| EX4 2266-2286 | |||
| TRBJ3-1 | F | NW_011620189 | 653-702 |
| TRBJ3-1 | F | NW_011601111 | 2234-2283 |
| TRBJ3-2 | F | NW_011601111 | 2426-2476 |
| TRBJ3-3 | F | NW_011601111 | 2642-2690 |
| TRBJ3-4 | nd | NW_011601111 | 2787-2814 |
| TRBJ2-2 | F | NW_011616084 | 215-265 |
| TRBJ2-3 | nd | NW_011616084 | 2-46 |
| TRBJ2-6 | nd | NW_011607149 | 185-231 |
| TRBC2 | nd | NW_011593440 | EX1 1911-2149 |
| EX2 2622-2639 | |||
| EX3 2800-2906 | |||
| EX4 3190-3210 | |||
| TRBV30 | F | NW_011593440 | 14509-14160 |
nd: not defined (indicates that the nt sequence of the gene is incomplete and its functionality cannot be defined).
L-PART1/ V-exon for TRBV genes and coding sequence for TRBD and TRBJ.
Table 2.
Description of the Camdro TRBV pseudogenes.
| TRBV genes | Defective Leader | Frameshift | Stop codon | Defective splice sites | Defective RSS |
|---|---|---|---|---|---|
| TRBV3 | ● | ||||
| TRBV9 | ● | ● | |||
| TRBV12S1 | ● | ||||
| TRBV12S2 | ● | ||||
| TRBV14 | ● | ||||
| TRBV21S3 | ● | ● | |||
| TRBV23 | ● | ||||
| TRBV24 | ● | ● |
Table 3.
Description of the unrelated TRB genes in the Camelus dromedarius genome assembly. The position of all genes and their classification and functionality are reported.
| Gene classification | Functionalitya | NCBI reference sequence | Position |
|---|---|---|---|
| MOXD2 | F | NW_011591622 | 850155-856730 |
| TRY1 | F | NW_011591622 | 870036-876394 |
| TRY2 | F | NW_011591622 | 882909-888072 |
| TRY3 | nd | NW_011623391 | 1-2387 |
| TRY4 | F | NW_011591151 | 13974-17714 |
| EPBH6 | F | NW_011593440 | 46466-60647 |
nd: not defined (indicates that the nt sequence of the gene is incomplete and its functionality cannot be defined).
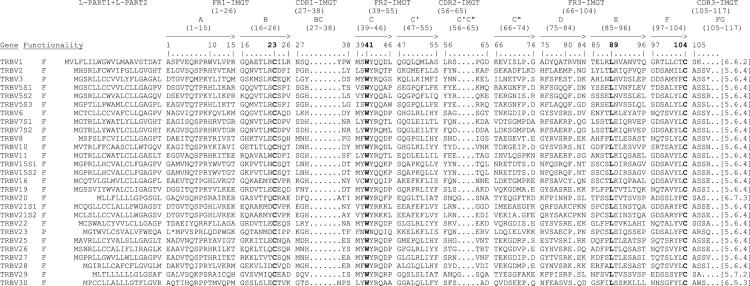
Fig. 1.
The IMGT Protein display of the dromedary TRBV genes. Only functional genes and in-frame pseudogenes are shown. The description of the strands and loops and of the FR-IMGT and CDR-IMGT is according to the IMGT unique numbering for V-REGION [6]. The amino acid length of the CDR-IMGT AA is also indicated in square brackets.
Table 4.
Camelus dromedarius D-J-C region genomic clones. The primer sequences, the PCR conditions and the size of each clone are reported.
| Clone | Primer pairs sequence (5′-3′) | Primer location | T annealing | Product length (bp) |
|---|---|---|---|---|
| pSCBJ11 | JB11U: CTTTGGAGAAGGCACCAG | TRBJ1-1 gene | 55/58 | 4396 |
| CB2L: TGGTTGCGGGGGTTGTGC | TRBC gene exon 1 | |||
| pSCJ22KN | CB2U: GCACAACCCCCGCAACCA | TRBC gene exon 1 | 53/55 | 5000 |
| JB34L: GCCAAAGTACTGAGTGTT | TRBJ3-4 gene | |||
| pSCBJ27U | JB34U: AACACTCAGTACTTTGGC | TRBJ3-4 gene | 56/58 | 4077 |
| CB2L: TGGTTGCGGGGGTTGTGC | TRBC gene exon 1 | |||
| pSCBD3 | CB2U: GCACAACCCCCGCAACCA | TRBC gene exon 1 | 55/56 | 4848 |
| JB23L: CCGCCGAAAAACAGTGTC | TRBJ2-3 gene | |||
| pSCMG1 | JB23U: GACACTGTTTTTCGGCGG | TRBJ2-3 gene | 55/58 | 3160 |
| CB2L: TGGTTGCGGGGGTTGTGC | TRBC gene exon 1 | |||
| pSCB2C8 | CB2U: GCACAACCCCCGCAACCA | TRBC gene exon 1 | 62 | 1331 |
| 3UTR:GTTGAGCTCACTTTGCAGGG | TRBC2 gene 3UTR |
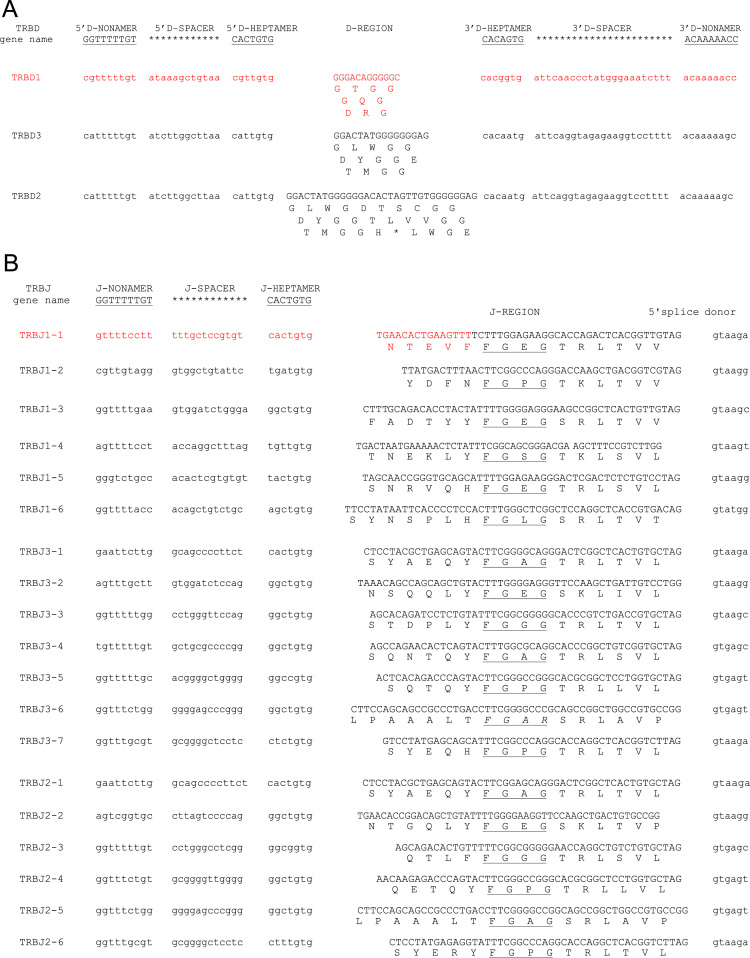
Fig. 2.
Nucleotide and deduced amino acid sequences of the dromedary TRBD (a), TRBJ (b) and TRDC (c) genes. The consensus sequence of the heptamer and nonamer are provided at the top of the figure and underlined. The numbering adopted for the gene classification is reported on the left of each gene. The gene sequence retrieved from the Ca_dromedarius_V1.0 genomic assembly is highlighted in red. In (a), the inferred amino acid sequence of the TRBD genes in the three coding frames are reported. In (b), the donor splice site for each TRBJ is shown. The canonical FGXG amino acid motifs are underlined. The unusual TRBJ3.6 gene motif is in italics. In (c), IMGT Protein display of the dromedary TRBC genes. Descriptions of the strands and loops were collected according to the IMGT unique numbering for C-DOMAIN [7].
2. Experimental design, materials and methods
2.1. Analysis of the dromedary TRB locus retrieved from the genome assembly: identification of the related and unrelated TRB genes
We employed the recent submission to NCBI (BioProject PRJNA234474) of a draft genome sequence from the Arabian camel [2] to identify the TRB locus in this species. A standard BLAST search (Basic Local Alignment Search Tool. http://blast.ncbi.nlm.nih.gov/Blast.cgi.) of the dromedary genomic resource was then performed by using human and sheep TRB gene sequences to assess their physical location in the dromedary genome. We directly retrieved a sequence of 457871 pb (gaps included) from the PRJNA234474_Ca_dromedarius_V1.0 assembly that corresponds to eight distinct unplaced and not continuous scaffolds (Fig. 1 in [1]). The sequence comprises the MOXD2 and the EPHB6 genes that flank the 5′ and 3′ ends, respectively, of all mammalian TRB loci studied to date. All dromedary TRB genes have been recognized and annotated while taking into account both the human sequence and the sheep genomic D-J-C region as a reference [3], [4], [5] (Table 1). The functionality of V, J and C genes was predicted through the manual alignment of sequences adopting the following parameters: (a) identification of the leader sequence at the 5′ of the TRBV genes; (b) determination of proper recombination signal (RS) sequences located at 3′ of the TRBV, 5′ of the TRBJ, and 3′ and 5′ ends of the TRBD genes, respectively; (c) determination of correct acceptor and donor splicing sites; (d) estimation of the expected length of the coding regions; (e) absence of frameshifts and stop signals in the coding regions of the genes. We annotated 33 TRBV germline genes (twenty-five functional genes and eight pseudogenes) (Table 2), one TRBD, 13 TRBJ and two complete and one incomplete TRBC genes. The analysis of the 3′ part of the locus revealed the potential presence of three D-J-C clusters similar to clusters found in sheep [4], [5].
We also identified and annotated four trypsin-like serine protease (TRY) genes (Table 3). In this context, downstream of the TRBV1 gene, proceeding from 5′ to 3′, we found as in humans two protease genes that we recognized tentatively, according to their genomic position, as TRY1 (alias PRSS58 or TRYX3) and TRY2 (alias TRY2P), respectively. A third TRY gene, named TRY3, was homologous to a gene located after the TRY2P gene in humans that was found within the NW_011623391 unplaced scaffold. Extrapolation of the synteny with the human sequence predicts that the NW_011623391 scaffold should be juxtaposed within the dromedary TRB locus, upstream of the TRBV3 gene (Fig. 1 in [1]). An additional TRY gene, classified as TRY4, was found before the D-J-C region. Thus, unlike humans, only one TRY gene encompasses the array of the TRBV genes. All dromedary TRY genes appear putatively functional with the presence of correct acceptor and donor splicing site and an absence of frameshifts and stop codon in their coding regions. The genomic structure of the MOXD2 and EPHB6 genes, which delimit the TRB locus, was also defined (Table 3).
2.2. Protein display of the dromedary TRBV genes
The deduced amino acid sequences of the germline TRBV genes were manually aligned according to IMGT unique numbering for the V-REGION [6] to maximize the percentage of identity (Fig. 1). Only potential functional genes and in-frame pseudogenes are shown. All sequences exhibit the typical framework regions (FR) and complementarity determining regions (CDR) as well as the four amino acids: cysteine 23 (1st-CYS) in FR1-IMGT, tryptophan 41 (CONSERVED-TRP) in FR2-IMGT, hydrophobic amino acid 89, and cysteine 104 (2nd-CYS) in FR3-IMGT [6]. Conversely, CDR-IMGT varies in amino acid composition and length. It should be noted that the TRBV21 genes show a difference in length of one amino acid in the FR3 that corresponds to a C′′ strand that is shorter and has a diverse amino acid sequence for TRBV21S2 compared to the TRBV21S1 gene.
2.3. Isolation of the dromedary TRBD-J-C region and analysis of the gene content
To isolate the entire TRBD-J-C region, we set up six different PCRs to produce six consecutive amplicons that cover the region between the first TRBJ and the last TRBC gene. Mostly, for each amplification, we used a primer pair, a gene-specific primer designed on the sequence of the TRBJ genes identified within the cDNA clones (see [1]), and a conserved primer constructed on the first exon of the TRBC genes. For the isolation of the TRBC2 gene, a 3'UTR lower primer derived from the sequence of the genomic assembly was used. Amplification consisted of an initial denaturation step at 93 °C for 2 min followed by 10 amplification cycles that each comprised a denaturation step at 93 °C for 10 s, an annealing step with a low temperature (53–56 °C, according to the melting temperature of the primers) for 30 s, an extension step at 68 °C for 7 min, followed by 25 cycles with a higher annealing temperature (55–58 °C, according to the melting temperature of the primers) and a gradually increasing extension time of 20 s as well as a final incubation at 68 °C for 7 min. A 30-deoxyadenosine overhang was added to blunt-ended amplicons by incubation with 1.0 unit of Platinum Taq DNA Polymerase (Invitrogen) at 72 °C for 10 min. These products were purified and cloned into the StrataClone TA-vector per the manufacturer's instructions. For each sample, 6 to 10 colonies were propagated and bi-directionally sequenced using M13 and T7 vector-specific primers. All plasmid sequence data were manually analysed. For the list of the clones with the primer pairs used and the PCR conditions see Table 4. All the obtained amplicons were sequenced (Acc. no. LT837971). The sequenced region is schematically illustrated in Fig. 3 in [1].
The nucleotide and deduced amino acid sequences of the TRBD, TRBJ and TRBC genes classified according to the similarity to the sheep sequence are shown in Fig. 2.
Acknowledgements
The “Bilateral agreement of scientific cooperation between CNR and ASRT” for the years 2009–10 is gratefully acknowledged as well as the Italian Ministry of Foreign Affairs and Egyptian Academia of Science for supporting the “Programme of scientific and technological cooperation between Italy and Egypt”. The financial support of the University of Bari “Aldo Moro” (ex 60% delivered to RA) and University of Salento is gratefully acknowledged.
Footnotes
Transparency document associated with this article can be found in the online version at 10.1016/j.dib.2017.08.002.
Contributor Information
Rachele Antonacci, Email: rachele.antonacci@uniba.it.
Mariagrazia Bellini, Email: mg1192@hotmail.it.
Vito Castelli, Email: vtcast@gmail.com.
Salvatrice Ciccarese, Email: salvatricemaria.ciccarese@uniba.it.
Serafina Massari, Email: sara.massari@unisalento.it.
Transparency document. Supplementary material
Transparency document
.
References
- 1.Antonacci R., Bellini M., Pala A., Mineccia M., Hassanane M.S., Ciccarese S., Massari S. The occurrence of three D-J-C clusters within the dromedary TRB locus highlights a shared evolution in Tylopoda, Ruminantia and Suina. Dev. Comp. Immunol. 2017;76:105–119. doi: 10.1016/j.dci.2017.05.021. [DOI] [PubMed] [Google Scholar]
- 2.(a) Wu H., Guang X., Al-Fageeh M.B., Cao J., Pan S., Zhou H., Zhang L., Abutarboush M.H., Xing Y., Xie Z., Alshanqeeti A.S., Zhang Y., Yao Q., Al-Shomrani B.M., Zhang D., Li J., Manee M.M., Yang Z., Yang L., Liu Y., Zhang J., Altammami M.A., Wang S., Yu L., Zhang W., Liu S., Ba L., Liu C., Yang X., Meng F., Wang S., Li L., Li E., Li X., Wu K., Zhang S., Wang J., Yin Y., Yang H., Al-Swailem A.M., Wang J. Camelid genomes reveal evolution and adaptation to desert environments. Nat. Commun. 2014;5:5188. doi: 10.1038/ncomms6188. [DOI] [PubMed] [Google Scholar]; (b) Wu H., Guang X., Al-Fageeh M.B., Cao J., Pan S., Zhou H., Zhang L., Abutarboush M.H., Xing Y., Xie Z., Alshanqeeti A.S., Zhang Y., Yao Q., Al-Shomrani B.M., Zhang D., Li J., Manee M.M., Yang Z., Yang L., Liu Y., Zhang J., Altammami M.A., Wang S., Yu L., Zhang W., Liu S., Ba L., Liu C., Yang X., Meng F., Wang S., Li L., Li E., Li X., Wu K., Zhang S., Wang J., Yin Y., Yang H., Al-Swailem A.M., Wang J. Erratum in: Nat. Commun. 2015;6:6107. doi: 10.1038/ncomms7107. [DOI] [PubMed] [Google Scholar]
- 3.Lefranc M.-P., Giudicelli V., Duroux P., Jabado-Michaloud J., Folch G., Aouinti S., Carillon E., Duvergey H., Houles A., Paysan-Lafosse T., Hadi-Saljoqi S., Sasorith S., Lefranc G., Kossida S. IMGT®, the international ImMunoGeneTics information system® 25 years on. Nucleic Acids Res. 2015;43:D413–D422. doi: 10.1093/nar/gku1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di Tommaso S., Antonacci R., Ciccarese S., Massari S. Extensive analysis of D-J-C arrangements allows the identification of different mechanisms enhancing the diversity in sheep T cell receptor beta-chain repertoire. BMC Genom. 2010;11:3. doi: 10.1186/1471-2164-11-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antonacci R., Di Tommaso S., Lanave C., Cribiu E.P., Ciccarese S., Massari S. Organization, structure and evolution of 41 Kb of genomic DNA spanning the D-J-C region of the sheep TRB locus. Mol. Immunol. 2008;45:493–509. doi: 10.1016/j.molimm.2007.05.023. [DOI] [PubMed] [Google Scholar]
- 6.Lefranc M.-P., Pommié C., Ruiz M., Giudicelli V., Foulquier E., Truong L., Thouvenin-Contet V., Lefranc G. IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Dev. Comp. Immunol. 2003;27:55–77. doi: 10.1016/s0145-305x(02)00039-3. [DOI] [PubMed] [Google Scholar]
- 7.Lefranc M.-P., Pommié C., Kaas Q., Duprat E., Bosc N., Guiraudou D., Jean C., Ruiz M., Da Piédade I., Rouard M., Foulquier E., Thouvenin V., Lefranc G. IMGT unique numbering for immunoglobulin and T cell receptor constant domains and Ig superfamily C-like domains. Dev. Comp. Immunol. 2005;29:185–203. doi: 10.1016/j.dci.2004.07.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transparency document