Abstract
The RANKL/RANK/OPG pathway plays an important role in regulating bone remodeling and bone turnover. However, the association of the genes variants with the risk of ONFH has rarely been reported. Here, we analyzed the correlation of the 10 SNPs polymorphisms of RANKL, RANK, OPG, TRAF6, and NFATC1 genes with the risk and development of ONFH in 200 ONFH patients and 177 health controls of Chinese population with using Mass ARRAY® platform. The results showed that the recessive model of NFATC1rs9518 was significantly associated with increased ONFH risk (OR:8.223, P=0.048); the proportion of stage Ⅳ patients in the rs9518TC genotype carriers was statistically higher than that of stage Ⅲ patients (P=0.03); in the T-C haplotype carriers of Naftac1, the proportion of bilateral hips lesions was also significantly enhanced than that of unilateral hip lesions(P=0.05). In addition, the proportion of idiopathic ONFH in the TT genotype carriers of OPGrs2073617 was significantly higher than that of steroid or alcohol-induced ONFH, respectively, while in the TC genotype carriers of the SNP, the proportion of idiopathic ONFH remarkably decreased compared with that of Alcohol-induced ONFH, P=0.021. Our results were first found that NFATC1rs9518 closely associated with the risk and the development of ONFH, while OPGrs2073617 statistically correlated with the etiological classification of ONFH.
Keywords: ONFH, gene polymorphism, RANKL/RANK/OPG pathway, TRAF6, NFATC1.
Introduction
Receptor activator of nuclear factor- kappa B ligand (RANKL), its receptor activator of nuclear factor-kappa B (RANK), decoy receptor osteoprotegerin (OPG), tumor necrosis factor receptor-associated factor 6 (TRAF6), and nuclear factor of activated T cells, cytoplasmic 1 (NFATC1) are five major proteins of the RANKL/RANK/OPG signaling pathway1. This pathway plays a crucial role in regulating bone remodeling, osteoclast differentiation, and a variety of pathologic conditions 2. One emerging area of RANKL/RANK/OPG pathway exerts the critical effects on the osteogenic differentiation of bone marrow mesenchymal stem cells (BMSCs), and the key genes from the signaling pathway may be expressed, regulated, and functioned in the physiology and pathology of the osteogenic differentiation of BMSCs 3. Moreover, the RANKL/RANK/ OPG pathway may mediate important and complex links between osteoblasts and osteoclast as well as the abnormal transdifferentiation between osteogenesis and adipogenesis of BMSCs, which is thought to play a central role in the molecular mechanisms of osteonecrosis of the femoral head (ONFH) 4.
Some results have been revealed that the gene variants in RANKL/ RANK/OPG signaling pathway not only associated with bone density, areal bone mineral density, and femoral neck compression strength index 5,6 but also correlated with osteoporotic fractures, rheumatoid arthritis (RA). However, it has rarely been reported that the gene variants in the pathway associate with the risk of ONFH. ONFH is a disease related to intense pain and loss of joint function, unknown molecular pathogenesis, and its incidence has been increasing during last decade 7. Although multiple gene variants have been proposed to be closely related to ONFH, no one genetic molecular mechanism has been identified as the etiology of the ONFH. In view of the important effects of the RANKL/RANK/OPG pathway on the regulation of transdifferentiation between osteogenesis and adipogenesis, we analysed the associations of 10 SNPs polymorphisms of RANKL, RANK, OPG, TRAF6, and NFATC1 genes with ONFH development in 200 ONFH patients and 177 health controls.
Materials and Methods
Subjects
A total of 200 unrelated patients with ONFH (132 males, 68 females; age: 53.46±11.48 yr) were consecutively enrolled at the Department of Orthopedics, the Second Clinical College of Jilin University, (Changchun, China) from March, 2014 to June, 2015 in the study. ONFH patients caused by direct trauma were excluded. The patients with ONFH concurrent with severe chronic diseases, such as cardiovascular diseases, congenital diseases, human immunodeficiency virus (HIV) infection, diabetes mellitus, renal dysfunction, and cancer were also excluded. ONFH were diagnosed by evidence of osteonecrosis using plain radiographs in Stages 2, 3, and 4 of the Ficat Classification system 8. On the basic of the detailed inquiry of medical history and aetiological factors, ONFH patients were classified into one of the following subgroups: alcohol-induced (71 cases (39.7%), idiopathic (64 cases (34.0%), and steroid-induced osteonecrosis (47 cases (26.3%). Steroid-induced osteonecrosis was defined by a history of taking prednisolone cumulative 2000 mg or an equivalent over 21 days. Alcohol-induced osteonecrosis was defined by the consumption of more than 900 ml of pure ethanol per week. The course of ONFH ranged from 0.5 months to 360 months, with an average of 71.75 months, and the clinical stages of ONFH consisted of 10 cases of stage II (5.6%), 54 cases of stage III (30.2%) and 115 cases (64.2%) of stage IV.
The unilateral and bilateral hips lesions were 76 cases (42.5%) and 103cases (57.5%), respectively. There were 21cases of ONFH patients who failed to undergo the clinical stages, etiological classification duo to the defect of plain radiographs or unclear aetiological factors. Moreover, 177 unrelated health control subjects (112 males, 65 females; age: 50.73 ±11.02 yr) who were age- and sex-matched for the ONFH group were consecutively enrolled at the Health Examination Center of Second Clinical College of Jilin University, (Changchun, China) from October 2014 to December 2014. Health control subjects were defined in the following manner: they had no hip pain and their fasting blood glucose, triglyceride and total cholesterol levels in serum were in normal reference range, the abdominal ultrasound examination and chest X-ray radiography were normal, and they did not suffer from cardiocerebrovascular diseases. All of the 377 participants were Han Chinese from northeast China. The study was approved by the ethics committee of the Second Clinical College of Jilin University, Changchun, China, and conducted in accordance with the World Medical Association's 2008 Declaration of Helsinki. All participants provided written informed consent for their taking part in the study.
Genomic DNA extraction and SNP selection
Approximately 2mL of venous blood was collected from all of the participants after a minimum of 10 h fasting. Genomic DNA was extracted from whole blood samples using the genomic DNA extraction kit (DP318, TianGen, BeiJing, China) following the manufacturer's protocols.
DNA samples concentration and quality were detected spectrophotometrically at 260/280 nm and stored at -80°C. The HapMap database and related literature were used to select SNPs of the genes by analysing their population distribution in different countries, nationalities and regions, particularly in data obtained from an Asian population. The database: http://gvs.gs.washington.edu /GVS138/ was used to select SNPs. The search scope of the genes was from the upstream 2000bp to downstream 1000bp of RANKL, RANK, OPG, TRAF6, and NFATC1 gene, respectively, including the promoter, 3-UTR, 5-UTR, intron, and exon region. The 10SNPs from five genes, 2 SNPs of OPG, 1SNP of RANK, 3SNPs of RANKL, 2SNPs of NFATC1, and 2SNPs of TRAF6 were selected based on Linkage Disequilibrium (LD) analysis by HapMap (http://www.hapmap.org/index.html.en). The selection criteria of SNPs included in r2>0.8 or D'=1; Minority allele frequencies > 0.05. The10SNPs list of target genes is shown in Table 1.
Table 1.
Basic information of 10 SNPS in OPG, R ANK, RANKL, TRAF6, and NFATC1 genes
| Gene | Chromosome | SNP rs ID | Allele | Minor Allele | Function |
|---|---|---|---|---|---|
| OPG | 8q24 | rs2073617 | C/T | C | 5'UTR |
| rs2073618 | G/C | C | Missense | ||
| RANK | 18q22.1 | rs884205 | G/T | T | 3'UTR |
| RANKL | 13q14 | rs7984870 | C/G | C | Promoter |
| rs9525641 | C/T | C | Promoter | ||
| rs1054016 | G/T | T | Promoter | ||
| NFATC1 | 18q23 | rs754093 | G/T | G | Missense |
| rs9518 | C/T | C | 3'UTR | ||
| TRAF6 | 11p12 | rs5030411 | C/T | C | Promoter |
| rs5030416 | A/C | C | Promoter |
UTR: untranslated regions
Genotyping
Primers for polymerase chain reaction and sequencing were designed by Sequenom Assay Design 3.1 software (Sequenom, San Diego, CA, USA) according to the manufacturer's instructions, shown in Table 2. The quality inspection of the sequencing primer was completed by matrix assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF). Mass ARRAY® platform (Sequenom Analyzer 4, Inc., San Diego, CA, USA) was used to analyze the 10SNPs gene polymorphisms of 5 genes in the RANKL/RANK/OPG signaling pathway in 200 ONFH patients and 177 healthy controls. The genotyping success rates for the 10 tag SNPs were >95%, respectively.
Table 2.
List of PCR and sequencing primers of 10 SNPs
| Gene | SNP-ID | PCR primer | Product length (bp) | Sequencing primer |
Molecular weight |
|---|---|---|---|---|---|
| RUNX2 | rs3763190 | 5'ACGTTGGATGCAGAACTAGACTCCGTCAAA3' | 109 | CACGCACAACAAAAGTTAATGAGA | 7371.8 |
| 5'ACGTTGGATGAACTCAGTTTCCCTTCTCC 3' | |||||
| rs7751427 | 5'ACGTTGGATGTCTCAGCACTCTTCACCCTC 3' | 87 | CTTCACCCTCCCACCTC | 4962.2 | |
| 5'ACGTTGGATGTCTTTTTCCCTTGGGAGTCG 3' | |||||
| PPARγ | rs2920502 | 5'ACGTTGGATGCGCACAGTAGGGCCCACGG 3' | 100 | CGCAACCGGCTTTCCCGCGTCCACT | 7514.9 |
| 5'ACGTTGGATGAACGACACCAGGTAGCCTGC 3' | |||||
| IGFBP3 | rs2132572 | 5'ACGTTGGATGAGAAGACTCTCACGTGTTGC 3' | 93 | GCTACACCGCAAGTC | 4522 |
| 5'ACGTTGGATGCTTAAGGACGCATTCGCTTG 3' | |||||
| rs2854744 | 5'ACGTTGGATGTGCAGCTCGAGACTCGCCC' 3' | 105 | CCTGAACCTGCTCCTCGTGC | 6004.9 | |
| 5'ACGTTGGATGCACCTTGGTTCTTGTAGACG 3' | |||||
| rs2854746 | 5'ACGTTGGATGTCGCAGCGCACCACGGGAC 3' | 105 | GTGCCCCACGGGACCCAAGCCC | 6651.3 | |
| 5'ACGTTGGATGTGACTCTGCTGGTGCTGCTC 3' | |||||
| COL2A1 | rs1859444 | 5'ACGTTGGATGCTCTCACCTTAGCCAAACTG 3' | 100 | TTAGCCAAACTGCCTTCT | 5409.5 |
| 5'ACGTTGGATGTGGTATGAAAGGAGGCACCG 3' | |||||
| rs2070739 | 5'ACGTTGGATGGAACAGCATTGCCTATCTGG 3' | 95 | ATATCTGGACGAAGCAGCT | 5836.8 | |
| 5'ACGTTGGATGACGTCATTGGAGCCCTGGAT 3' | |||||
| rs3803183 | 5'ACGTTGGATGAGACTTACGGACATCCTGGC 3' | 119 | GGGGAGTCAGCAGCACCAGCG | 6506.2 | |
| 5'ACGTTGGATGTGAGCCATGATTCGCCTCG 3' | |||||
| rs3809322 | 5'ACGTTGGATGGGTTAAAGCCCCTCTGAGAA 3' | 112 | TCTGATATCTCTAGAACACAA | 6373.2 | |
| 5'ACGTTGGATGGCTTAGGCTATAAATGCCAG 3' | |||||
| Osteris | rs4759113 | ACGTTGGATGTCCTGATCTAGAGGCAGGTG | 124 | CCTGACTGGGGAAGCAA | 5244.4 |
| ACGTTGGATGACAAATGCACACAGTGACGC |
Statistical analysis
Shesis software (http://analysis.bio-x.cn/SHEsisMain.htm) was used to analyse the Hardy-Weinberg equilibrium and haplotypes between the ONFH and control groups and the associations of haplotypes with the clinical phenotypes of ONFH. In addition, logistical regression analyses were performed to calculate the odds ratios (OR), 95% confidence intervals (CI), and corresponding p-values of each SNP controlling for age and sex as covariates. The genetic models of dominant, recessive, and codominant were considered and the genotypes were given codes of 0, 1, and 2; 0, 1, and 1; or 0, 0, and 1 in the codominant, dominant, and recessive models, respectively. SPSS10.0 software (X2 test) was used to analyse the association of the genes polymorphisms with clinical phenotypes of ONFH. A P-value of <0.05 was considered statistically significant.
Results
Genotypes and allele frequencies of 10 SNPs in the RANKL, RANK, OPG, TRAF6, and NFATC1 genes between ONFH and control groups
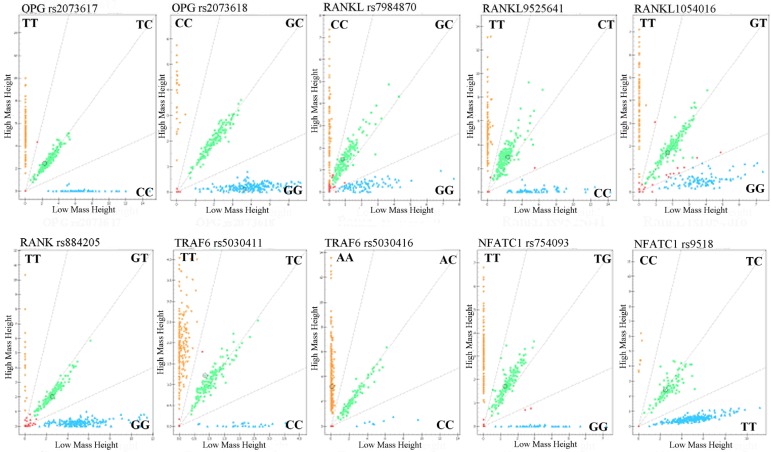
Call cluster plots genotyping 10 SNPs through Mass ARRAY® platform in 377 samples see Figure 1. Genotypes and allele frequencies of the 10 SNPs are shown in Table 3. χ2 test analysis results revealed that although the CC genotype frequency of NFATC1 rs9518CC in ONFH group was a higher tendency than that of control group (4.0% vs 0.6%, P=0.057), the genotypes and allele frequencies of the 10 SNPs were no statistical significance between ONFH and control groups. However, logistical regression analysis showed that after the adjustment of age and gender factors etc, the recessive model of NFATC1rs9518 were significantly associated with increased ONFH risk, OR (95%CI):8.223(1.015~66.651), P=0.048.
Figure 1.
Genotyping of 10SNPs in the RANKL, RANK, OPG, TRAF6, and NFATC1 genes by MALDI-ToF-MS. The green plots represents heterozygous genotypes, yellow and blue plots represent major homozygous or minor homozygous genotypes in the call cluster plot of each SNP, respectively. MALDI-ToF-MS: matrix-assisted laser desorption/ionization time-of-flight mass spectrometry.
Table 3.
Association of gene genotypes and allele frequencies in the OPG, RANK, RANKL, Traf6, and Nfatcl genes with the risk of ONFH
| Gene | dbSNP ID |
Group | Genotype(n) | MAF | HWEa | pb | Co-dominants (11 vs. 12 vs. 22) |
Dominants 12+22 vs. 11 |
Recessives 22 vs. 11+12 |
Allele 2 vs. 1 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 11 | 12 | 22 | OR (95%CI) Pc |
OR (95%CI) Pc |
OR (95%CI) Pc |
OR (95%CI) Pc |
||||||
| OPG | rs2073617 (T/C) |
TT | TC | CC | ||||||||
| Control | 59 | 85 | 33 | 0.427 | 0.807 | 0.990 | 0.989(0.738-1.325) | 0.942(0.606-1.464) | 1.052(0.618-1.791) | 0.982 (0.735-1.312) | ||
| Case | 67 | 96 | 36 | 0.422 | 0.875 | 0.940 | 0.789 | 0.852 | 0.902 | |||
| rs2073618 (G/C) |
GG | GC | CC | |||||||||
| Control | 93 | 70 | 14 | 0.277 | 0.870 | 0.726 | 0.972(0.701-1.332) | 0.997(0.658-1.512) | 0.855(0.382-1.915) | 0.966 (0.701-1.332) | ||
| Case | 104 | 84 | 12 | 0.270 | 0.355 | 0.966 | 0.990 | 0.704 | 0.833 | |||
| Rank | rs884205 (G/T) |
GG | GT | TT | ||||||||
| Control | 106 | 64 | 7 | 0.220 | 0.486 | 0.280 | 1.312(0.913-1.886) | 1.294(0.843-1.958) | 1.350(0.497-3.671) | 0.767 (0.545-1.080) | ||
| Case | 94 | 78 | 10 | 0.277 | 0.229 | 0.142 | 0.239 | 0.556 | 0.128 | |||
| RankL | rs1054016 (G/T) |
GG | GT | TT | ||||||||
| Control | 49 | 91 | 37 | 0.466 | 0.661 | 0.792 | 0.859(0.629-1.173) | 0.863(0.540P-1.380) | 0.766(0.444-1.321) | 1.105(0.823-1.485) | ||
| Case | 55 | 90 | 34 | 0.430 | 0.792 | 0.339 | 0.539 | 0.338 | 0.507 | |||
| rs7984870 (G/C) |
GG | GC | CC | |||||||||
| Control | 51 | 88 | 36 | 0.457 | 0.862 | 0.653 | 1.057(0.764-1.463) | 1.194(0.736-1.890) | 0.947(0.543-1.649) | 1.141(0.838-1.554) | ||
| Case | 37 | 79 | 34 | 0.490 | 0.510 | 0.739 | 0.490 | 0.307 | 0.403 | |||
| rs9525641 (T/C) |
TT | TC | CC | |||||||||
| Control | 50 | 90 | 36 | 0.460 | 0.689 | 0.515 | 0.978(0.715-1.336) | 1.179(0.736-1.890) | 0.765(0.442-1.293) | 1.028(0.771-1.371) | ||
| Case | 49 | 113 | 36 | 0.467 | 0.039 | 0.887 | 0.493 | 0.307 | 0.849 | |||
| Traf6 | rs5030411 (T/C) |
TT | TC | CC | ||||||||
| Control | 93 | 72 | 12 | 0.271 | 0.699 | 0.721 | 1.049(0.760-1.449) | 0.970(0.641-1.470) | 1.439(0.670-3.091) | 1.092(0.794-1.503) | ||
| Case | 102 | 79 | 18 | 0.289 | 0.632 | 0.771 | 0.887 | 0.351 | 0.588 | |||
| rs5030416 (A/C) |
AA | AC | CC | |||||||||
| Control | 126 | 48 | 3 | 0.379 | 0.515 | 0.691 | 0.920(0.609-1.390) | 0.857(0.538-1.363) | 1.591(0.371-6.813) | 1.040(0.697-1.552) | ||
| Case | 146 | 49 | 5 | 0.148 | 0.715 | 0.771 | 0.513 | 0.532 | 0.847 | |||
| Nfatc1 | rs754093 (T/G) |
TT | TG | GG | ||||||||
| Control | 69 | 82 | 26 | 0.379 | 0.838 | 0.691 | 0.907(0.669-1.230) | 0.848(0.551-1.303) | 0.944(0.519-1.715) | 0.892(0.662-1.202) | ||
| Case | 85 | 84 | 27 | 0.350 | 0.396 | 0.529 | 0.451 | 0.849 | 0.453 | |||
| rs9518 (T/C) |
TT | TC | CC | |||||||||
| Control | 123 | 55 | 1 | 0.159 | 0.061 | 0.057 | 1.163(0.780-1.735) | 1.011(0.645-1.586) | 8.223(1.015-66.651) | 1.055(0.713-1.560) | ||
| Case | 143 | 49 | 8 | 0.163 | 0.158 | 0.458 | 0.962 | 0.048 | 0.789 | |||
11: homozygotes for the major allele, 12: heterozygotes and 22: homozygotes for the minor allele.
ap-values of deviation from Hardy-Weinberg equilibrium between the ONFH group and control group.
b χ2 test were used for frequencies of genotype analyses
c Logistic regression analyses were used for calculations. bold: p-value < 0.05
The genotypes association of 10 SNPs in the RANKL, RANK, OPG, TRAF6, and NFATC1 genes with the clinical phenotypes of ONFH
Correlation analysis revealed that in the TT genotype carriers of OPGrs2073617, the proportion (49.2%) of idiopathic ONFH was significantly higher than those of Steroid-induced ONFH (29.8%)and Alcohol-induced ONFH (22.9%), respectively, while in the TC genotype carriers of the SNP, the proportion (36.1%) of idiopathic ONFH remarkably decreased compared with that (58.6%) of Alcohol-induced ONFH, P=0.021; in addition, the stage Ⅳ patients proportion of the TC genotype carriers revealed an increased tendency compared with the stage Ⅲ patients in spite of no statistical significance (P=0.066). However, in the TC genotype carriers of NFATC1rs9518, the proportion of the stage Ⅳ patients (29.6%) was statistically higher than that of the stage Ⅲ patients (14.8%), P=0.03, shown in Table 4.
Table 4.
The association of SNPs genotypes in OPG, RANK, RANKL, TRAF6, and NFATC1 genes with the clinical phenotypes of ONFH
| Gene ID | SNP ID | Geno-type | Gender n(%) | Age (yr) | Etiological Classification n (%) X2 text |
Involved hip lesions n(%) X2 text | Clinical stage n (%) X2 text |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| male | female | Alca | Ster | Idio | Unilateral | Bilateral | StageⅡ | Stage Ⅲ | Stage Ⅳ | ||||||||
| OPG |
rs2073617 (C/T) |
CC | 24(20.0) | 9(15.5) | 47.18±10.59 | 13(18.6) | 11(23.4) | 9(14.8) | 13(17.1) | 20(19.6) | 1(10.0) | 14(25.9) | 18(15.8) | ||||
| TT | 36(30.0) | 24(41.4) | 45.85±12.81 | 16(22.9) | 14(29.8) | 30(49.2) | 29(38.2) | 31(30.4) | 7(70.0) | 15(27.8) | 38(33.3) | ||||||
| TC | 60(50.0) | 25(43.1) | 48.95±12.31 | 41(58.6) | 22(46.8) | 22(36.1) | 34(44.7) | 51(50.0) | 2(20.0) | 25(46.3) | 58(50.9) | ||||||
| P | 0.314 | 0.317 | 0.021* | 0.555 | 0.066 | ||||||||||||
| rs2073618 (C/G) |
GG | 65(53.7) | 29(50.0) | 46.89±12.20 | 31(43.7) | 24(51.1) | 39(63.9) | 40(52.6) | 54(52.4) | 8(80.0) | 24(44.4) | 62(53.9) | |||||
| CC | 8(6.6) | 2(3.4) | 51.30±5.14 | 4(5.6) | 2(4.3) | 4(6.6) | 4(5.3) | 6(5.8) | 1(10.0) | 3(5.6) | 6(5.2) | ||||||
| GC | 48(39.7) | 27(46.6) | 48.02±12.79 | 36(50.7) | 21(44.7) | 18(29.5) | 32(42.1) | 43(41.7) | 1(10.0) | 27(50.0) | 47(40.9) | ||||||
| P | 0.536 | 0.517 | 0.166 | 0.987 | 0.212 | ||||||||||||
| RANK | rs884205 (G/T) | GG | 61(53.5) | 25(49.0) | 47.12±12.84 | 36(55.4) | 24(54.5) | 26(46.4) | 36(54.5) | 50(50.5) | 4(40.0) | 26(52.0) | 56(53.3) | ||||
| TT | 7(6.1) | 3(5.9) | 45.35±9.16 | 3(4.6) | 2(4.5) | 5(8.9) | 1(1.5) | 9(9.1) | 1(10.0) | 2(4.0) | 7(6.7) | ||||||
| GT | 46(40.4) | 23(45.1) | 48.26±12.23 | 26(40.0) | 18(40.9) | 25(44.6) | 29(43.9) | 40(40.4) | 5(50.0) | 22(44.0) | 42(40.0) | ||||||
| P | 0.848 | 0.725 | 0.764 | 0.136 | 0.864 | ||||||||||||
| RANKL | rs1054016 (G/T) |
GG | 31(29.0) | 18(34.0) | 46.60±11.75 | 17(26.6) | 15(36.6) | 17(30.9) | 21(31.8) | 28(29.8) | 4(40.0) | 17(35.4) | 28(27.5) | ||||
| TT | 18(16.8) | 9(17.0) | 44.15±12.30 | 15(23.4) | 5(12.2) | 7(12.7) | 9(13.6) | 18(19.1) | 1(10.0) | 7(14.6) | 19(18.6) | ||||||
| GT | 58(54.2) | 26(49.1) | 48.38±12.49 | 32(50.0) | 21(51.2) | 31(56.4) | 36(54.5) | 48(51.1) | 5(50.0) | 24(50.0) | 55(53.9) | ||||||
| P | 0.791 | 0.28 | 0.435 | 0.657 | 0.790 | ||||||||||||
| rs79848709 | GG | 20(22.0) | 15(33.3) | 46.52±12.22 | 10(18.9) | 14(38.9) | 11(23.4) | 14(25.0) | 21(26.3) | 3(33.3) | 12(32.4) | 20(22.2) | |||||
| (C/G) | CC | 18(19.8) | 8(17.8) | 45.52±12.63 | 14(26.4) | 3(8.3) | 9(19.1) | 10(17.9) | 16(20.0) | 1(11.1) | 3(8.1) | 22(24.4) | |||||
| GC | 53(58.2) | 22(48.9) | 47.94±13.19 | 29(54.7) | 19(52.8) | 27(57.4) | 32(17.9) | 43(53.8) | 5(55.6) | 22(59.5) | 48(53.3) | ||||||
| P | 0.359 | 0.677 | 0.124 | 0.919 | 0.245 | ||||||||||||
| rs9525641 | CC | 21(17.6) | 8(13.8) | 45.03±12.01 | 16(23.2) | 5(10.6) | 8(13.1) | 10(13.2) | 19(18.8) | 1(10.0) | 7(13.0) | 21(18.6) | |||||
| (C/T) | TT | 28(23.5) | 16(27.6) | 47.78±11.29 | 14(20.3) | 15(31.9) | 15(24.6) | 17(22.4) | 27(26.7) | 3(30.0) | 16(29.6) | 25(22.1) | |||||
| TC | 70(58.8) | 34(58.6) | 48.20±12.68 | 39(56.5) | 27(57.4) | 38(62.3) | 49(64.5) | 55(54.5) | 6(60.0) | 31(57.4) | 67(59.3) | ||||||
| P | 0.736 | 0.467 | 0.285 | 0.383 | 0.748 | ||||||||||||
| TRAF6 | rs5030411 | CC | 11(9.2) | 6(10.3) | 49.65±11.68 | 6(8.6) | 4(8.5) | 7(11.5) | 5(6.6) | 12(11.8) | 2(20.0) | 8(14.8) | 7(6.1) | ||||
| (C/T) | TT | 63(52.5) | 30(51.7) | 47.88±11.52 | 36(51.4) | 28(59.6) | 29(47.5) | 44(57.9) | 49(48.0) | 6(60.0) | 27(50.0) | 60(52.6) | |||||
| TC | 46(38.3) | 22(37.9) | 46.63±13.28 | 28(40.0) | 15(31.9) | 25(41.0) | 27(35.5) | 41(40.2) | 2(20.0) | 19(35.2) | 47(41.2) | ||||||
| P | 0.969 | 0.622 | 0.776 | 0.319 | 0.236 | ||||||||||||
| rs5030416 | CC | 5(4.1) | 0(0.0) | 46.40±15.13 | 4(5.6) | 1(2.1) | 0(0.0) | 2(2.6) | 3(2.9) | 0(0.0) | 3(5.6) | 2(1.7) | |||||
| (A/C) | AA | 87(71.9) | 45(77.6) | 47.80±12.18 | 53(74.6) | 36(76.6) | 43(70.5) | 62(81.6) | 70(68.0) | 7(70.0) | 39(72.2) | 86(74.8) | |||||
| CA | 29(24.0) | 13(22.4) | 47.16±12.11 | 14(19.7) | 10(21.3) | 18(29.5) | 12(15.8) | 30(29.1) | 3(30.0) | 12(22.2) | 27(23.5) | ||||||
| P | 0.271 | 0.934 | 0.244 | 0.109 | 0.648 | ||||||||||||
| NFATC1 | rS754093 | GG | 14(11.9) | 9(15.8) | 44.4±12.9 | 9(12.9) | 7(15.6) | 7(11.7) | 8(10.7) | 15(15.0) | 1(10.0) | 7(15.1) | 14(12.5) | ||||
| (T/G) | GT | 58(49.2) | 18(31.6) | 47.0±11.6 | 28(40.0) | 24(53.3) | 24(40.0) | 34(45.3) | 42(42.0) | 5(50.0) | 26(49.1) | 45(40.2) | |||||
| TT | 46(39.0) | 30(52.6) | 48.6±12.4 | 33(47.1) | 14(31.1) | 29(48.3) | 33(44.0) | 43(43.0) | 4(40.0) | 19(35.8) | 56(47.3) | ||||||
| P | 0.086 | 0.324 | 0.434 | 0.694 | 0.710 | ||||||||||||
| Rs9518 (T/C) |
TT | 86(71.1) | 39(67.2) | 47.4±12.7 | 51(71.8) | 31(66.0) | 43(70.5) | 58(76.4) | 67(71.9) | 6(60.0) | 41(75.9) | 78(67.8) | |||||
| TC | 28(23.1) | 18(31.0) | 48.7±11.0 | 17(23.9) | 11(23.4) | 18(29.5) | 15(19.7) | 31(26.5) | 4(40.0) | 8(14.8) | 34(29.6) | ||||||
| CC | 7(5.8) | 1(1.7) | 44.4±12.7 | 3(4.2) | 5(10.6) | 0(0.0) | 3(3.9) | 5(4.9) | 0(0.0) | 5(9.3) | 3(2.6) | ||||||
| P | 0.289 | 0.461 | 0.116 | 0.277 | 0.073(0.030)* | ||||||||||||
a:Alc : Alcohol-induced; Ster: steroid- induced; Idio: idiopathic; * stageⅡvs stageⅢvs stageⅣ:P=0.073: stageⅢ vs stageⅣP=0.030
The haplotypes of OPG, RANKL, TRAF6, and NFATC1 genes and their association with the clinical phenotypes of ONFH
We calculated the Linkage disequilibrium (LD) coefficients (|D′| and r2) among SNPs in the different genes using Shesis software (http://analysis. bio-x.cn/myAnalysis.php) on the basic of the genotypes of the SNPs. There are the 4 haplotypes of C-C, C-G, T-C, and T-G between rs2073617(C/T) and rs2073618 (G/C) of OPG gene, the 6 haplotypes of G-C-C, G-G-T, T-C-C, T-G-C, T-G-T, and G-C-T among rs1054016(G/T), rs7984870(G/C), and rs9525641(T/C)of RANKL gene, the 4 haplotypes of C-A, C-C, T-C, and T-G between rs5030411(C/T) and rs5030416(C/A) of TRAF6 gene, and the 4 haplotypes of G-C, G-T, T-C, and T-T between rs754093(G/T)and rs9518(C/T) of Naftac1 gene, respectively. The haplotypes frequencies were no statistical significance between the ONFH and control groups(data not shown) but correlation analysis between the haplotypes and clinical phenotypes of ONFH showed that in the T-C haplotype carriers of Naftac1 gene, the proportion of bilateral hips lesions (13.3%)was significantly increased compared with that (6.9%) of unilateral hip lesions, OR(95%CI): 2.063 (0.975~4.367), P=0.05; in the C-C haplotype carriers of TRAF6 gene, the proportion of bilateral hips lesions revealed higher tendency than that of unilateral hips lesions despite no statistical significance, OR(95%CI):1.821 (0.969~3.423), P=0.06, shown in Table 5. The correlation analysis between the haplotypes and clinical stages of ONFH did not reveal statistical significance (data not shown).
Table 5.
Association of SNPs haplotypes in the OPG, TRAF6, RANKL, and NFATC1 genes with unilateral or bilateral hip lesions of ONFH
| SNPs | Haplotype | Bil▲n(freq) | Unil▼n(freq) | χ2 | P | OR(95%CI) |
|---|---|---|---|---|---|---|
| OPG: rs2073618(G/C) -rs2073617(C/T) |
C-C | 55.00(0.270) | 37.66(0.248) | 0.144 | 0.70 | 1.098 (0.678~1.777) |
| G-C | 36.00(0.176) | 22.34(0.147) | 0.465 | 0.49 | 1.222 (0.687~2.173) | |
| G-T | 113.00(0.554) | 89.66(0.590) | 0.720 | 0.39 | 0.831(0.542~1.275) | |
| TRAF6 : rs5030411(C/T), -rs5030416(C/A) |
C-A | 29.00(0.14) | 21.00(0.13) | 0.012 | 0.91 | 1.034 (0.564~1.894) |
| C-C | 36.00(0.176) | 16.00(0.105) | 3.540 | 0.06 | 1.821 (0.969~3.423) | |
| T-A | 139.00(0.681) | 115.00(0.75) | 2.410 | 0.12 | 0.688(0.429~1.104) | |
| RANKL: rs7984870(G/C) -rs9525641(T/C) -rs1054016(G/T) |
C-C-G | 3.04(0.020) | 5.12(0.047) | 1.661 | 0.19 | 0.399 (0.095~1.687) |
| C-C-T | 68.96(0.442) | 42.88(0.390) | 0.536 | 0.46 | 1.206 (0.730~1.993) | |
| C-T-G | 0.00(0.000) | 2.00(0.018) | _ | _ | _ | |
| G-T-G | 80.96(0.519) | 57.88(0.526) | 0.076 | 0.78 | 0.933 (0.567~1.533) | |
| G-T-T | 2.04(0.013) | 2.12(0.019) | _ | _ | _ | |
| G-C-T | 1.00(0.006) | 0.00(0.000) | _ | _ | _ | |
| Nfatc1: rs754093(G/T), -rs9518(T/C) |
G-C | 14.31(0.072) | 10.58(0.071) | 0.001 | 0.97 | 1.015 (0.445~2.316) |
| G-T | 57.69(0.288) | 39.42(0.263) | 0.282 | 0.59 | 1.137 (0.707~1.829) | |
| T-C | 26.69(0.133) | 10.42(0.069) | 3.703 | 0.05 | 2.063 (0.975~4.367) | |
| T-T | 101.31(0.507) | 89.58(0.597) | 2.842 | 0.09 | 0.692(0.451~1.062) |
▲bilateral hips lesions; ▼unilateral hip lesions
Discussion
ONFH is a disorder caused by the interactions with complex genetic and environmental factors 9. A series of genes polymorphisms have been showed to associate with ONFH risk 10,11,12 while the effects of genes polymorphisms in RANKL/RANK/OPG signaling pathway on the development of ONFH have been remained unclear. RANK/RANKL/OPG pathway plays an important role in the pathogenesis of RA and osteoporosis due to the crucial molecular pathway for coupling between osteoblasts and osteoclasts. It was early proved that there was the transdifferentiation disorder between osteogenesis and adipogenesis of BMSCs related to this pathway during the development of ONFH 13. In order to explore the effects of genes polymorphisms in the RANK/RANKL/OPG pathway on the development of ONFH, we genotyped the 10SNPs of RANKL, RANK, OPG, TRAF6, and NFATC1 genes. Our results showed that the gene polymorphisms of NFATC1rs9518 significantly associated with the development of ONFH as well as the clinical phenotypes of ONFH.
NFATC1gene is located in chromosome18q23 and its protein is a component of the nuclear factor of activated T cells DNA-binding transcription complex. NFATC1 play a central role in inducible gene transcription during immune response and as the master regulator of osteoclast differentiation, it is specifically induced by RANKL and dependent on both the TRAF6-NF-κB and c-Fos pathways 14 and exerts the essential and sufficient role in osteoclastogenesis 15. In view of the key effects of NFATC1 on the regulation of steogenesis and osteoclastogenesis, its gene polymorphisms were more focused on the correlation with bone metabolism and bone disorders. A Genome-wide association study (GWAS) genotyping 4608 tagging and potentially functional SNPs in 383 bone metabolism candidate genes identified novel genetic variants of NFATC1 related to cortical volumetric bone mineral density (vBMD) in older men with the osteoporotic fractures 16. Excessive osteoclast activity is a major pathological mechanism of metabolic bone disorders, such as osteopenia, RA, and osteoporosis while NFATC1 plays molecular switch roles in osteoclast pathway. It has previously been shown that embryonic stem cells with NFATC1 deficiency are not capable of differentiating into functional osteoclasts, and the retrovirus-mediated overexpression of NFATC1 induces normal osteoclast differentiation even in the absence of RANKL 15.
Our results first revealed that the minor homozygote (CC) and the heterozygosis (TC) of NFATC1rs9518 were a genotype of ONFH risk and a risk genotype of the advanced hip lesion of ONFH, respectively. Moreover, the T-C haplotype of NFATC1rs754093(G/T)- rs9518(T/C) also involved in bilateral hips lesions of ONFH. These results indicate that NFATC1rs9518 may be a potential molecular target associated with the risk and the development of ONFH. The rs9518 (T/C) is located in 3'UTR region of NFATC1 gene. Generally, the SNPs in 3'UTR region may correlate with up-regulation and down-regulation of gene expression 17. The NFATC1rs9518(T/C) polymorphisms might involve in abnormal gene expression and function during development of ONFH, and the detailed mechanisms remain to be further explored. In addition, our results also confirmed the association of OPGrs2073617 genotypes with the etiological classification of ONFH. Idiopathic ONFH often occurs in young and middle-aged patients, and most of the radiographically larger lesions progress to collapse, resulting in the rapid destruction of the hip joint 18. Excess corticosteroid and alcohol abuse have been identified as environmental risk factors for the development of steroid- or alcohol-induced ONFH, respectively 19, but the risk factors of idiopathic ONFH has been remained unclear.
A meta-analysis to assess the association of the 4b/a, G894T, and T786C polymorphisms of endothelial nitric oxide synthase with ONFH risk showed a significant correlation between allele a of the 4b/a polymorphism and idiopathic ONFH 20. As chondrogenesis-specific transcription factor, collagen type II alpha-1 gene (COL2A1) mutations, including c.3508G > A, have been reported to be involved in idiopathic ONFH etiology 21. OPG, as protein encoded by tumor necrosis factor superfamily11B (TNFRSF11B), is an osteoblast-secreted decoy receptor that functions as a negative regulator of bone resorption. OPG specifically binds to its ligand, OPG ligand, both of which are key extracellular regulators of osteoclast development. A study to evaluate the association of three polymorphisms in OPG gene promoter with the osteoporosis in RA demonstrated that the C allele of the C950T was associated with RA 22. The C950T SNP is located in the promoter region, near the TGF-β response area and 129 bp upstream the TATA box 23, therefore, the C950T polymorphism affected the gene expression of OPG 24. The rs2073617analyzed by our study is located in 5'UTR region of OPG gene, and the SNPs in 5'UTR region usually affects gene expression or protein function. Therefore, the effects of rs2073617 polymorphisms on OPG gene expression or its protein function in the development of ONFH needs to be further investigated.
Except for NFATC1 and OPG genes, our results failed to show the significant association of 6 SNPs polymorphisms in the RANK, RANKL, and TRAT6 genes with the risk or clinical phenotypes of ONFH in spite of the proportion of bilateral hips lesions in the C-C haplotype carriers of TRAF6rs5030411(C/T) -rs5030416(C/A) was higher tendency than that of unilateral hips lesions. However, the polymorphisms of these genes have been proved to be associated with the bone erosion in the RA patients synovium at the site of bone resorption 25. Therefore, it is necessary to further investigate the association of the genes polymorphisms with the development of ONFH in expanded samples.
The limitation of this study is that 200 ONFH patients and 177 health controls attribute to smaller sample system that may limit our statistical power to detect small differences between groups, especially for the subgroups analysis between the genotypes and clinical phenotypes. Thus, larger sample studies are necessary to further assess the variants distribution between ONFH and control groups. In addition, the correlation analysis between the SNPs polymorphisms and clinical phenotypes involve in the two-two comparison in multiple-test. Generally, multiple comparisons may increase false positive 26. Considering this study only included in the two-two comparison among 3 genotype subgroups, instead of the adjustment of the significance threshold, we adopted the single factor variance analysis with the least significant difference (LSD) effectively to control the fairway-wise error rate. However, the effects of RANK/RANKL/OPG pathway genes variants on the risk and development of ONFH remains to be further explored on the basis of the adjustment of significance threshold.
In conclusion, we completed the analysis of the genotypes, allele frequencies, and hypotypes of the 10 SNPs in the RANKL, RANK, OPG, TRAF6, and NFATC1 genes and their associations with the risk and development of ONFH in 200 ONFH patients and 177 health controls, respectively. Our results were first found that the minor homozygote (CC) of NFATC1rs9518 was a higher risk genotype of ONFH, the heterozygosis (TC) of NFATC1 rs9518 was a risk genotype related to the advanced hip lesion of ONFH, and the T-C haplotype of Naftac1rs754093(G/T) -rs9518(T/C) might be a risk haplotype involved in bilateral hips lesions of ONFH while the genotypes of OPGrs2073617 closely associated with the etiological classification of ONFH. These results indicate that NFATC1rs9518 and OPGrs2073617 are potential molecular targets associated with the occurrence and the development of ONFH.
Acknowledgments
This work was supported by the Project of Health Management Department of Jilin Province, China (Grant No. 20132003), the Project of Bethune Youth Foundation of Jilin University, China (Grant No. 2015409), the Department of Science and Technology of Jilin Province, China (Grant No.20140311006YY, 20150312022ZG), and the Development and Reform Commission of Jilin Province, China [Grant No. 2014G073, the project of application demonstration center of precision medicine for molecular diagnosis in Jilin Province (2016-2018)].
References
- 1.Baud'huin M, Duplomb L, Ruiz VC. et al. Key roles of the OPG-RANK-RANKL system in bone oncology. Expert Rev Anticancer Ther. 2007;7:221–32. doi: 10.1586/14737140.7.2.221. [DOI] [PubMed] [Google Scholar]
- 2.Boyce BF, Xing L. Biology of RANK, RANKL, and osteoprotegerin. Arthritis Res Ther. 2007;9(Suppl 1):S1. doi: 10.1186/ar2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang Y, Zhang Y, Chen W. et al. Achyranthes bidentata extract exerts osteoprotective effects on steroid-induced osteonecrosis of the femoral head in rats by regulating RANKL/RANK/OPG signaling. J Transl Med. 2014;12:334.. doi: 10.1186/s12967-014-0334-7. doi: 10.1186/s12967-014-0334-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mont MA, Cherian JJ, Sierra RJ. et al. Nontraumatic osteonecrosis of the femoral Head: where do we stand today? a ten-year update. J Bone Joint Surg Am. 2015;97:1604–27. doi: 10.2106/JBJS.O.00071. [DOI] [PubMed] [Google Scholar]
- 5.Paternoster L, Lorentzon M, Vandenput L. et al. Genome-wide association Meta-Analysis of cortical bone mineral density unravels allelic heterogeneity at the RANKL locus and potential pleiotropic effects on bone. PLoS Genet. 2010;6:e1001217.. doi: 10.1371/journal.pgen.1001217. doi: 10.1371/journal.pgen. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong SS, Liu XG, Chen Y. et al. Association analyses of RANKL/RANK/OPG gene polymorphisms with femoral neck compression strength index variation in Caucasians. Calcif Tissue Int. 2009;85:104–12. doi: 10.1007/s00223-009-9255-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Russo S, Sadile F, Esposito R. et al. Italian experience on use of E.S.W. therapy for avascular necrosis of femoral head. Int J Surg. 2015;24:188–90. doi: 10.1016/j.ijsu.2015.11.038. [DOI] [PubMed] [Google Scholar]
- 8.Ficat RP. Idiopathic bone necrosis of the femoral head. Early diagnosis and treatment. J Bone Joint Surg Br. 1985;67:3–9. doi: 10.1302/0301-620X.67B1.3155745. [DOI] [PubMed] [Google Scholar]
- 9.Moya-Angeler J, Gianakos AL, Villa JC. et al. Current concepts on osteonecrosis of the femoral head. World J Orthop. 2015;6:590–601. doi: 10.5312/wjo.v6.i8.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng KT, Huang KC, Huang TW. et al. Single nucleotide polymorphisms other than factor V Leiden are associated with coagulopathy and osteonecrosis of the femoral head in chinese patients. PLoS ONE. 2014;9:e104461.doi. doi: 10.1371/journal.pone.0104461. 10.1371/journal.pone. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim H, Cho C, Cho Y. et al. Significant associations of PAI-1 genetic polymorphisms with osteonecrosis of the femoral head. BMC Musculoskelet Disord. 2011;12:160.doi. doi: 10.1186/1471-2474-12-160. 10.1186/1471-2474-12-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z, Zhang Y, Kong X. et al. Association of a polymorphism in PON-1 gene with steroid-induced osteonecrosis of femoral head in Chinese Han population. Diagn Pathol. 2013;8:186–91. doi: 10.1186/1746-1596-8-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang YS, Wang YH, Zhao GQ. et al. Osteogenic potential of human calcitonin gene-related peptide alpha gene-modified bone marrow mesenchymal stem cells. Chin Med J(Engl) 2011;124:3976–81. [PubMed] [Google Scholar]
- 14.Takayanagi H, Kim S, Koga T. et al. Induction and activation of the transcription factor NFATc1(NFAT2) integrate RANKL signaling for terminal differentiation of osteoclasts. Dev Cell. 2002;3:889–901. doi: 10.1016/s1534-5807(02)00369-6. [DOI] [PubMed] [Google Scholar]
- 15.Asagiri M, Sato K, Usami T. et al. Autoamplification of NFATc1 expression determines its essential role in bone homeostasis. J Exp Med. 2005;202:1261–9. doi: 10.1084/jem.20051150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yerges LM, Klei L, Cauley JA. et al. Candidate gene analysis of femoral neck trabecular and cortical volumetric bone mineral density in older men. J Bone Miner Res. 2010;25:330–8. doi: 10.1359/jbmr.090729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leal MF, Cirilo PD, Mazzotti TK. et al. Prohibitin expression deregulation in gastric cancer is associated with the 3'untranslated region 1630 C>T polymorphism and copy number variation. PLoS One. 2014;9:e98583.. doi: 10.1371/journal.pone.0098583. doi: 10.1371/journal.pone. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukui K, Kaneuji A, Matsumoto T. Arthroscopic correction for concomitant cam impingement in a patient with idiopathic osteonecrosis of the femoral head: A case report. Int J Surg Case Rep. 2016;19:154–8. doi: 10.1016/j.ijscr.2015.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Assouline-Dayan Y, Chang C, Greenspan A. et al. Pathogenesis and natural history of osteonecrosis. Semin Arthritis Rheum. 2002;32:94–124. [PubMed] [Google Scholar]
- 20.Song GG, Lee YH. Association of eNOS polymorphisms with susceptibility to osteonecrosis of the femur head: A meta-analysis. Z Rheumatol; 2016. [Epub ahead of print]. doi:10.1007/s00393-016-0093-3. [DOI] [PubMed] [Google Scholar]
- 21.Sakamoto Y, Yamamoto T, Miyake N, Screening of the COL2A1 mutation in idiopathic osteonecrosis of the femoral head. J Orthop Res; 2016. [Epub ahead of print]. doi:10.1002/jor.23300. [DOI] [PubMed] [Google Scholar]
- 22.Zavala-Cerna MG, Moran-Moguel MC, Cornejo-Toledo JA. et al. Osteoprotegerin polymorphisms in a Mexican population with rheumatoid arthritis and generalized osteoporosis: A preliminary report. J Immunol Res. 2015:376197. doi: 10.1155/2015/376197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thirunavukkarasu K, Miles RR, Halladay DL. et al. Stimulation of osteoprotegerin (OPG) gene expression by transforming growth factor-β (TGF-β).mapping of the OPG promoter region that mediates TGF-βeffects. J Biol Chem. 2001;276:36241–50. doi: 10.1074/jbc.M104319200. [DOI] [PubMed] [Google Scholar]
- 24.Vidal C, Formosa R, Xuereb-Anastasi AC. Functional polymorphisms within the TNFRSF11B (osteoprotegerin) gene increase the risk for low bone mineral density. J Mol Endocrinol. 2011;47:327–33. doi: 10.1530/JME-11-0067. [DOI] [PubMed] [Google Scholar]
- 25.Pettit AR, Walsh NC, Manning C. et al. RANKL protein is expressed at the pannus-bone interface at sites of articular bone erosion in rheumatoid arthritis. Rheumatology. 2006;45:1068–76. doi: 10.1093/rheumatology/kel045. [DOI] [PubMed] [Google Scholar]
- 26.William SN. How does multiple testing correction work? Nat Biotechnol. 2009;27:1135–37. doi: 10.1038/nbt1209-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]