Abstract
Introduction
High-dose busulfan (busulfan) is an integral part of the majority of hematopoietic cell transplantation conditioning regimens. Intravenous (IV) busulfan doses are personalized using pharmacokinetics (PK)-based dosing where the patient’s IV busulfan clearance is calculated after the first dose and is used to personalize subsequent doses to a target plasma exposure. PK-guided dosing has improved patient outcomes and is clinically accepted but highly resource intensive.
Objective
We sought to discover endogenous plasma biomarkers predictive of IV busulfan clearance using a global pharmacometabolomics-based approach.
Methods
Using LC-QTOF, we analyzed 59 (discovery) and 88 (validation) plasma samples obtained before IV busulfan administration.
Results
In the discovery dataset, we evaluated the association of the relative abundance of 1885 ions with IV busulfan clearance and found 21 ions that were associated with IV busulfan clearance tertiles (r2 ≥ 0.3). Identified compounds were deoxycholic acid and/or chenodeoxycholic acid, and linoleic acid. We used these 21 ions to develop a parsimonious seven-ion linear predictive model that accurately predicted IV busulfan clearance in 93% (discovery) and 78% (validation) of samples.
Conclusion
IV busulfan clearance was significantly correlated with the relative abundance of 21 ions, seven of which were included in a predictive model that accurately predicted IV busulfan clearance in the majority of the validation samples. These results reinforce the potential of pharmacometabolomics as a critical tool in personalized medicine, with the potential to improve the personalized dosing of drugs with a narrow therapeutic index such as busulfan.
Keywords: pharmacometabolomics, pharmacokinetics, busulfan, biomarkers, hematopoietic cell transplantation, personalized medicine
INTRODUCTION
The goal of an allogeneic hematopoietic cell transplant (HCT) is to cure the patient – termed the host or recipient – of their underlying disease by replacing their hematopoietic cells with cells from a healthy donor (Copelan 2006). The transplantation of donor cells that are not genetically identical (i.e., allogeneic) can result in bi-directional immunologic reactions (i.e., host-versus-graft and graft-versus-host) (Copelan 2006). In HCT, grafting of cells from one individual to another provokes immunologic reactions involved in engraftment of the donor cells, graft-versus-host disease, control of a malignancy (termed graft versus tumor), the development of tolerance, and immune reconstitution (Copelan 2006). High-dose busulfan (busulfan) plays a key role in the majority of HCT conditioning regimens that do not include total body irradiation. Improving the efficacy and reducing the toxicity of intravenous (IV) busulfan is critical to avoid the devastating effects of conditioning regimens using total body irradiation (Nieder et al. 2011). Furthermore, recent data has shown improved overall survival in HCT recipients conditioned with IV busulfan as compared to total body irradiation (Copelan et al. 2013).
Busulfan has a narrow therapeutic index, and busulfan plasma exposure – typically expressed as area under the plasma concentration-time curve (AUC) – is a predictive biomarker that forecasts the likely response to IV busulfan-containing conditioning regimens (Copelan 2006; McCune and Holmberg 2009). An individual’s IV busulfan dose and clearance estimate their plasma exposure, which can be expressed as AUC or concentration at steady state (Css, defined as AUC divided by the dosing interval) (McCune and Holmberg 2009). Busulfan clearance, which is a measure of how rapidly busulfan is eliminated from the body, has moderate between-subject variability (coefficient of variation of 20.5% after IV administration) (McCune and Holmberg 2009). Low IV busulfan AUC, caused by rapid IV busulfan clearance, is associated with an increased risk of rejection or relapse, while high IV busulfan AUC is associated with an increased risk of hepatotoxicity and non-relapse mortality. Personalizing IV busulfan doses to a target plasma AUC – termed pharmacokinetics (PK)-based dosing or targeted busulfan (TBU)– improves each of these clinical outcomes (McCune and Holmberg 2009). Because of IV busulfan’s narrow therapeutic index, PK-guided IV busulfan dosing is the standard of care (Jenke et al. 2005; McCune et al. 2012; Rezvani et al. 2013). Body weight is used to estimate IV busulfan dose 1. After dose 1 administration, six to seven serial PK samples (Yeh et al. 2012) are collected and transported to the analytical facility for quantification of the plasma busulfan concentrations. The patient’s busulfan clearance is estimated using non-compartmental analysis of the busulfan concentration-time data. The busulfan clearance and target AUC for that patient are used to personalize the IV busulfan dose. Unfortunately, the current dosing method of using body weight for dose 1 IV busulfan dose rarely achieves the target AUC. Specifically, using body weight to determine dose 1 of IV busulfan results in only 24% of children (McCune et al. 2013) and 23% of adults (Yeh et al. 2012) achieving their personalized target IV busulfan AUC.
Despite the acceptance of AUC as a predictive biomarker of the patient’s response to busulfan-containing HCT regimens, identifying novel biomarkers is desirable because of the rapidly evolving trend of shorter IV busulfan courses (Tarantal et al. 2012) and because relapse and non-relapse mortality continue to be problematic even with PK-guided IV busulfan dosing (Jenke et al. 2005; Rezvani et al. 2013). McCune et al. recently developed a population pharmacokinetic model for IV busulfan from a large cohort of HCT recipients (N=1610, 92% pediatric) (McCune et al. 2014). Age and normal fat mass (NFM) were identified as patient-specific characteristics that were associated with IV busulfan clearance (McCune et al. 2014). Additionally, to date, predictors of clinical outcomes for the other common components of the HCT conditioning regimens, such as cyclophosphamide or fludarabine pharmacokinetics, have not been found (McCune et al. 2007; McCune et al. 2012). Therefore, we propose using global pharmacometabolomics profiling to identify potential biomarkers of IV busulfan clearance. Following statistical analyses, candidate ions can be selected for metabolite identification and the analysis of metabolic pathways (Nicholson et al. 2002).
In recent years, the use of pre-dose metabolite profiling to predict drug response has been evaluated for other fields.(Clayton et al. 2009; Clayton et al. 2006). With the current focus on precision medicine, metabolomics is emerging as a tool to predict both pharmacokinetic and pharmacodynamic outcomes.{Rotroff, 2016 #19023}{Kaddurah-Daouk, 2015 #19024}{Kaddurah-Daouk, 2014 #19025}{Everett, 2015 #19026} Targeted and global metabolomics have been used to identify altered metabolic pathways, thereby providing insight into the underlying causes of variability and degree of response to drug treatment. Using the metabolomics analyses of pre-dose samples, investigators have been able to predict the efficacy and the potential for toxicity for various classes of drugs.
Thus, we sought to determine whether endogenous pharmacometabolomics-based biomarkers obtained before IV busulfan administration can predict IV busulfan clearance in a HCT patient population.
METHODS
Study population
Between April 2006 to November 2012, HCT recipients aged 21.5 to 65.8 years underwent PK-guided dosing of IV busulfan (Table 1) under the auspices of their HCT treatment protocol. The samples were quantitated at the College of American Pathologists (CAP)-certified Busulfan Pharmacokinetics Laboratory. Information on the subjects’ age, sex, height, total (i.e., actual) body weight (TBW), dosing weight (calculated as previously described (Rezvani et al. 2013), and HCT conditioning regimen were available. Total body weight was used for dosing if the total body weight was less than the ideal body weight, whereas adjusted ideal body weight was used if the total body weight was greater than the ideal body weight. Subjects received antiemetics, antibiotics, and antifungals per Fred Hutch Standard Practice Guidelines, minimizing potential confounders. All subjects received phenytoin to prevent IV busulfan-induced seizures.
Table 1.
Description of subject populationa
| Discovery dataset | Validation dataset | Entire populationb | |
|---|---|---|---|
| Number | 59 | 88 | 108 |
| Blood collection tube (BCT)c | |||
| Citrate | 59 (100%) | 0 | 59 (40.1%) |
| EDTA | 0 | 88 (100%) | 88 (59.9%) |
| Drugs present in pharmacometabolomics sample | |||
| None | 59 (100%) | 31 (35.2%) | c |
| Cyclophosphamide | 0 | 50 (56.8%) | c |
| Fludarabine | 0 | 7 (8.0%) | c |
| Age, in years | 48.7 ± 12.4 (21.6 – 64.0) | 50.8 ± 11.2 (21.6 – 65.8) | 50.4 ± 11.2 (21.6 – 65.8) |
| Dosing weight (DWT, kg)d | 70.4 ± 11.6 | 69.0 ± 11.0 | 69.4 ± 11.2 |
| Normal fat mass (NFM, kg) | 68.6 ± 14.9 | 67.3 ± 13.6 | 67.5 ± 13.9 |
| Male sex | 35 (59.3%) | 54 (61.4%) | 65 (60.2%) |
| HCT Conditioninge | |||
| Cyclophosphamide/Busulfan | 27 (45.8%) | 62 (70.5%) | 69 (63.9%) |
| Busulfan/Cyclophosphamide | 3 (5.1%) | 2 (2.3%) | 3 (2.8%) |
| Fludarabine/Busulfan | 27 (45.8%) | 17 (19.3%) | 27 (25%) |
| Fludarabine/Busulfan/Thymoglobulin | 2 (3.4%) | 7 (8%) | 9 (8.3%) |
| Busulfan dosing frequency | |||
| Every 6 hour | 11 (18.6%) | 4 (4.5%) | 11 (10.2%) |
| Every 24 hour | 48 (81.4%) | 84 (95.5%) | 97 (89.8%) |
| IV busulfan clearance (ml/min/kg NFM) | 3.17 ± 0.53 | 3.26 ± 0.56 | 3.20 ± 0.55 |
| Diagnosis | |||
| Aplastic anemia | 1 (1.7%) | 1 (1.1%) | 1 (0.9%) |
| Acute lymphoblastic leukemia | 1 (1.7%) | 1 (1.1%) | 1 (0.9%) |
| Acute myeloid leukemia | 30 (50.8%) | 35 (39.8%) | 46 (42.6%) |
| Chronic myeloid leukemia (CML) | 2 (3.4%) | 4 (4.5%) | 4 (3.7%) |
| Chronic myelomonocytic leukemia | 2 (3.4%) | 1 (1.1%) | 2 (1.9%) |
| Myelodysplastic syndrome (MDS) | 11 (18.6%) | 17 (19.3%) | 21 (19.4%) |
| MDS/CML | 1 (1.7%) | 1 (1.1%) | 1 (0.9%) |
| Myelofibrosis | 9 (15.3%) | 24 (27.3%) | 27 (25%) |
| Myeloproliferative disease | 2 (3.4%) | 4 (4.5%) | 5 (4.6%) |
Data presented as: number (%) or mean ± standard deviation (range); percentages may not total 100 because of rounding.
Number of samples (discovery and validation datasets) and number of subjects (entire population).
Of the 108 subjects, 39 subjects had samples collected on two separate occasions, once in citrate BCT (i.e., part of the discovery dataset) and once in EDTA BCT (i.e., part of the validation dataset). See “Evaluation of blood collection tubes (BCT)” section in Methods.
Dosing weight described in “IV busulfan pharmacokinetics“.
listed in administration order; all patients received PK-guided dosed IV busulfan, as described in “IV busulfan pharmacokinetics“ section.
All subjects gave written informed consent before participating in the treatment protocols. The Fred Hutch Institutional Review Board approved the treatment protocol as well as this retrospective analysis of samples to identify biomarkers of IV busulfan clearance using pharmacometabolomics.
IV busulfan pharmacokinetics
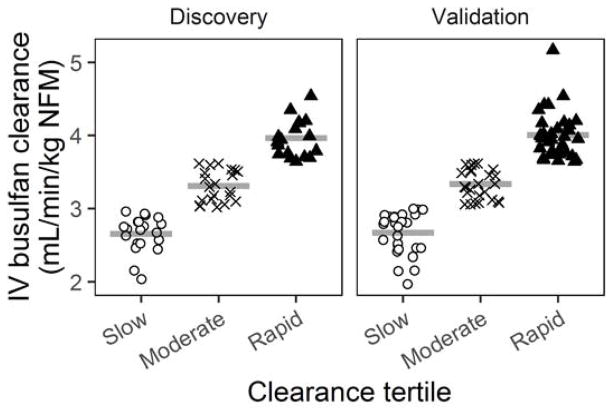
The dose 1 IV busulfan clearance was the primary outcome. The pharmacokinetic sampling schema and quantitation of plasma IV busulfan concentrations were previously described (Rezvani et al. 2013; McCune et al. 2012). In brief, for each patient, the IV busulfan concentration-time profile after dose 1 underwent noncompartmental analysis to estimate IV busulfan clearance using Phoenix WinNonlin (Certara USA, Princeton, NJ) (McCune et al. 2013). The IV busulfan clearances were expressed relative to NFM, which accounts for body size differences in children and adults so that one population pharmacokinetic model could be used for all HCT recipients. NFM was calculated as previously described (Online Resource - Methods) (McCune et al. 2014). Briefly, all PK parameters, including clearance, were scaled for body size using allometric theory and the predicted free fat mass (FFM), which was calculated from height and weight (Janmahasatian et al. 2005; Anderson and Holford 2008; Janmahasatian et al. 2008; Anderson and Holford 2009). The FFM was subsequently used with additional body metrics and IV busulfan-specific PK parameters to estimate NFM (Cortinez et al. 2010). The range of IV busulfan clearances in these patients was similar to other adult HCT populations (Online Resource Supplemental Figure 1) (McCune et al. 2014). The dataset was divided into three IV busulfan clearance tertiles (slow, moderate or rapid) (Figure 1).
Fig. 1.
IV busulfan clearance is shown in tertiles for the discovery and validation datasets. Open circles – slow clearance tertile, “X” symbols – moderate clearance tertile, black triangles – rapid clearance tertile. Horizontal lines represent the mean clearance for that group.
Metabolomic sample collection
Metabolomic profiling was conducted on plasma samples obtained before IV busulfan administration (N=147, Table 1) from 108 subjects. Plasma samples were collected in citrate or EDTA blood collection tubes (BCT) before administration of any HCT conditioning or following administration of either cyclophosphamide or fludarabine, which were administered as part of HCT conditioning.
Metabolomic sample extraction and processing
Samples for global profiling were prepared as described previously (Tay-Sontheimer et al. 2014). In brief, 200 μL of plasma were combined with 800 μL of ice-cold acetonitrile containing deuterated internal standards to monitor for shifts in retention time. Samples were vortexed for 30 seconds and centrifuged at 20,000 RCF at 4°C for 10 min. The supernatant was transferred to glass tubes and evaporated under nitrogen gas at room temperature. Samples were reconstituted by adding 25 μL of methanol and 25 μL of 0.4% acetic acid in water, vortexed, and transferred to vials for analysis.
Liquid chromatography-quadrupole time-of-flight (LC-QTOF) analysis
Global metabolomics analyses of samples were performed using an Agilent (Santa Clara, CA) 1200 LC coupled to an Agilent 6520 QTOF mass spectrometer. Samples (2 μL injection) were separated chromatographically using a 3.5 μm, 2.1 × 30 mm Agilent Zorbax SB-C8 guard column and a 1.8 μm, 2.1 × 50 mm Agilent Zorbax SB-Aq analytical column heated to 60°C. The flow rate was 0.6 mL/min with A: 0.2% acetic acid in water and B: 0.2% acetic acid in methanol (2% to 98% B in 13 min, 98% B until 19 min followed by re-equilibration for 5 min). The total run time was 24 min per sample. The MS source was maintained at 350°C with a capillary voltage of 3500 V, and the desolvation gas flow was 12 L/min. Scans were obtained between m/z 100 and 1000 at an acquisition rate of 3 spectra/sec. Data were collected using electrospray ionization (ESI) in positive and negative modes.
Data processing, filtering and normalization
The data analysis and metabolite identification are summarized in Online Resource Supplemental Figure 2. Raw data files were exported to mzData format, and the R package xcms was used to analyze raw mass spectral data (Benton et al. 2010; Smith et al. 2006; Tautenhahn et al. 2008). Feature detection was performed using the xcms “centWave” algorithm, and retention-time correction was performed using the function “peakgroups” with smoothing via a loess function. The resulting list of ion features was exported for filtering, normalization, and statistical analysis. All analyses were performed using the statistical programming language R (R Core Team 2014).
Ions were deemed uninformative and excluded when observed in fewer than 25% of all samples. A pseudo-count of one was added to every ion abundance value to allow for log-transformation, and each sample was scaled by its total ion abundance. Scaled abundances were multiplied by 1×106 for ease of presentation and then log-transformed for subsequent analyses. To remove redundant isotopic peaks, only the major peak was retained among ions that exhibited the following: (1) correlation among chromatograms greater than 0.9; (2) matching retention times; and (3) mass differences of multiples of 1.0087 Da. Finally, to ensure reproducibility, low-signal ions (mean scaled abundance below zero) were removed. The statistical analysis was based on the resulting 1885 total ions (1286 positive and 599 negative).
Selection of the discovery and validation datasets
An initial evaluation of the data using principal component analysis explored whether the global metabolome differed by the characteristics of sex, HCT conditioning regimen, diagnosis or BCT. There were no observed differences with any of these four characteristics. For statistical analyses, the samples were split into separate discovery and validation sets in a manner that maintained the greatest degree of homogeneity within each dataset while maintaining the objective evaluation of associations between ion abundance and busulfan clearance. The discovery dataset (n=59 samples) was comprised of samples collected in citrate BCT with no prior HCT conditioning administered to the subject. Samples in the validation dataset (n=88) were collected in EDTA but had greater heterogeneity with regards to conditioning, having had no conditioning (n=31), cyclophosphamide (n=50) (Rezvani et al. 2013) or fludarabine (n=7) (McCune et al. 2012) administered before sample collection.
Statistical analysis
Rather than fitting a separate univariate model for each of the 1885 ions, we allowed for correlations among ion abundances by first applying a multivariate regression model. For this, we fit a penalized least squares (ridge) regression model(Buhlmann et al. 2014) with all ion abundances using IV busulfan clearance as the outcome. This was implemented using the “refund” R package (Huang et al. 2015) in which the penalty tuning parameter is chosen automatically via a restricted maximum likelihood approach (Randolph et al. 2012). From this model, ions were selected by keeping only those whose regression coefficient had a 95% confidence interval not containing zero. Among the resulting 167 ions, this list was further reduced by retaining ions whose abundances exhibited an r2 of at least 0.3 in a (marginal) univariate linear model of association with IV busulfan clearance and a monotonic increase or decrease across subjects ordered by tertiles of IV busulfan clearance. The latter criterion served to focus on ions that would be of most predictive of IV busulfan clearance. The possible identities of these 21 ions, which are referred to as “selected” ions, were determined as described below.
In parallel, we constructed a statistical predictive model by starting with the selected ions and applying backward and forward stepwise variable-selection. The goodness of fit was assessed using the Akaike information criterion (AIC). The final linear model contained seven ions, which are referred to as “predictive” ions.
Validation dataset
To verify the ability of the seven-ion predictive model to estimate IV busulfan clearance, this model was applied to the validation dataset, and IV busulfan clearance was calculated for each subject. A predicted IV busulfan clearance was considered to be accurate for each subject if the value was within 80 to 125% of the observed IV busulfan clearance. This 80 to 125% range is used by the FDA to ascertain bioequivalence (Administration 2000) and is a reasonable metric by which to determine prediction accuracy. The model validation was completed by applying the predictive model, created using the discovery dataset, to the samples in the validation dataset.
Evaluation of blood collection tubes (BCT)
Of the 108 subjects, 39 subjects had samples collected on two separate occasions, once in citrate BCT (i.e., part of the discovery dataset) and once in EDTA BCT (i.e., part of the validation dataset). These samples allowed for a comprehensive evaluation of the within-subject variability and the effect of the BCT upon the 21 selected ions. To ascertain whether the BCT affected the ion abundance in the predictive model, the correlation was determined for each ion abundance measured in the citrate versus EDTA BCT for the 39 subjects.
Ion identification
Identification of the 21 selected ions was carried out following established methods (Xu et al. 2013) and by searching major metabolomics databases using the accurate mass (within 15 ppm) and MS/MS fragmentation spectra when available. Databases queried included METLIN (http://metlin.scripps.edu/), Massbank (http://massbank.imm.ac.cn/MassBank) and HMDB (Version 3.0 http://hmdb.ca/) using exact molecular weights. Commercially available standards were purchased from Sigma-Aldrich (St. Louis, MO) to confirm the identity of ions with a putative identification.
RESULTS
Patient characteristics
Patient pre-transplant demographics and HCT characteristics are described in Table 1. The mean age was 50.4 years (range: 21.6 to 65.8) with a slightly higher percentage of males (60.5%). The mean (± standard deviation) of the IV busulfan clearance was 3.17 ± 0.53 ml/min/kg NFM for the discovery dataset and 3.26 ± 0.56 ml/min/kg NFM for the validation dataset. The IV busulfan clearance by dosing weight and NFM are shown in Online Resources Supplemental Figures 2A and 2B.
Pharmacometabolomics
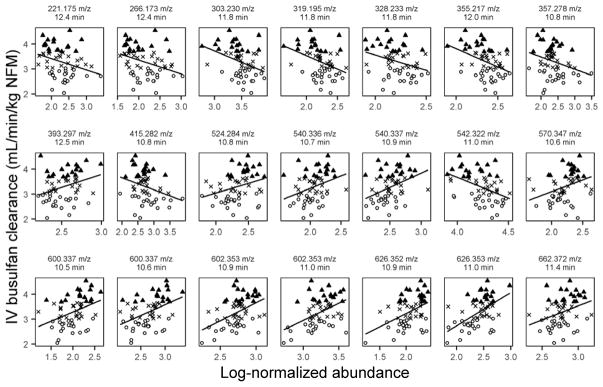
The goal of these analyses was to determine whether endogenous pharmacometabolomics-based biomarkers obtained before IV busulfan administration could be used to predict IV busulfan clearance. After aligning, filtering and normalizing the data, a total of 1885 ions were included in the analyses (Online Resource Supplemental Figure 2). The data were split into discovery and validation datasets based on the BCT. The 21 selected ions were detected within a two-minute window using our LC-QTOF conditions (range: 10.5 to 12.5 min). Of the selected ions, 11 ions were detected in negative ESI mode, and 10 ions were detected in positive ESI mode (Table 2). The ion with the strongest correlation with IV busulfan clearance (r2 = 0.57) was m/z = 626.353 with a retention time of 11.0 min and was detected in negative mode. Twelve of these selected ions were positively correlated with IV busulfan clearance, whereas the other nine ions had abundances that decreased as clearance increased (Figure 2).
Table 2.
Selected ions (n=21) and the predictive ion subset (n=7) from discovery dataset based on IV busulfan clearance.
| m/z | Retention Time (min) |
Ionization Mode |
Ion abundance by IV busulfan clearance tertile (mean ± s.d.) |
Weighted r2 |
ANOVA p value |
Number Database Matches |
Primary Database Classification/Metabolite Identifiedb |
||
|---|---|---|---|---|---|---|---|---|---|
| Slow Clearance |
Moderate Clearance |
Rapid Clearance |
|||||||
| 221.175 | 12.4 | + | 2.41 ± 0.34 | 2.27 ± 0.45 | 2.16 ± 0.34 | 0.33 | 0.1 | 27 | Isoprenoid |
| 266.173 | 12.4 | + | 2.25 ± 0.31 | 2.13 ± 0.40 | 1.99 ± 0.33 | 0.48 | 0.09 | 7 | Unknown |
| 303.230a | 11.8 | + | 3.55 ± 0.18 | 3.52 ± 0.22 | 3.36 ± 0.27 | 0.39 | 0.02 | 278 | Linoleic acid |
| 319.195 | 11.8 | + | 2.29 ± 0.16 | 2.27 ± 0.19 | 2.12 ± 0.23 | 0.31 | 0.02 | 17 | Linoleic acid |
| 328.233 | 11.8 | + | 2.25 ± 0.14 | 2.24 ± 0.14 | 2.15 ± 0.14 | 0.38 | 0.05 | 8 | Fatty Acyl |
| 355.217 | 12.0 | + | 2.40 ± 0.13 | 2.39 ± 0.14 | 2.26 ± 0.17 | 0.38 | 0.007 | 12 | Isoprenoid |
| 357.278 | 10.8 | + | 2.47 ± 0.44 | 2.37 ± 0.26 | 2.27 ± 0.28 | 0.36 | 0.2 | 46 | Chenodeoxycholic acid or Deoxycholic acid |
| 393.297a | 12.5 | + | 2.45 ± 0.16 | 2.54 ± 0.16 | 2.57 ± 0.20 | 0.40 | 0.1 | 168 | Bile Acid |
| 415.282a | 10.8 | + | 2.85 ± 0.45 | 2.73 ± 0.27 | 2.62 ± 0.29 | 0.34 | 0.1 | 121 | Chenodeoxycholic acid or Deoxycholic acid |
| 524.284 | 10.8 | - | 2.26 ± 0.22 | 2.29 ± 0.23 | 2.45 ± 0.16 | 0.33 | 0.01 | 9 | Glycerophospholipid |
| 540.336 | 10.7 | - | 1.97 ± 0.16 | 2.06 ± 0.18 | 2.13 ± 0.22 | 0.31 | 0.05 | 2 | Steroid Conjugate |
| 540.337 | 10.9 | - | 2.41 ± 0.17 | 2.55 ± 0.19 | 2.58 ± 0.22 | 0.38 | 0.02 | 2 | Steroid Conjugate |
| 542.322a | 11.0 | + | 4.28 ± 0.14 | 4.27 ± 0.14 | 4.14 ± 0.14 | 0.31 | 0.01 | 9 | Glycerophosphocholine |
| 570.347 | 10.6 | - | 2.14 ± 0.19 | 2.21 ± 0.24 | 2.30 ± 0.15 | 0.40 | 0.05 | 11 | Glycerophospholipid |
| 600.337a | 10.5 | - | 1.91 ± 0.26 | 1.98 ± 0.29 | 2.16 ± 0.31 | 0.38 | 0.03 | 20 | Glycerophospholipid |
| 600.337a | 10.6 | - | 2.55 ± 0.25 | 2.63 ± 0.27 | 2.83 ± 0.31 | 0.34 | 0.01 | 21 | Glycerophospholipid |
| 602.353 | 10.9 | - | 2.65 ± 0.19 | 2.75 ± 0.17 | 2.83 ± 0.15 | 0.31 | 0.007 | 7 | Unknown |
| 602.353 | 11.0 | - | 3.14 ± 0.23 | 3.27 ± 0.13 | 3.30 ± 0.18 | 0.32 | 0.03 | 7 | Unknown |
| 626.352 | 10.9 | - | 1.96 ± 0.27 | 2.06 ± 0.21 | 2.18 ± 0.18 | 0.34 | 0.01 | 4 | Glycerophospholipid |
| 626.353a | 11.0 | - | 2.26 ± 0.25 | 2.45 ± 0.20 | 2.54 ± 0.17 | 0.57 | 0.0006 | 4 | Glycerophospholipid |
| 662.372 | 11.4 | - | 2.73 ± 0.16 | 2.75 ± 0.17 | 2.87 ± 0.16 | 0.33 | 0.02 | 1 | Fatty Acyl |
The seven ions included in the predictive model are presented in bold text. These ions had a ridge regression coefficient whose 95% confidence interval did not contain zero, had r2 ≥ 0.3, and had a monotonic increase or decrease in abundance across busulfan clearance tertiles.
Database classification was determined from the most common classification in database hits.
Fig. 2.
Linear regressions of discovery dataset ion abundance with IV busulfan clearance of the selected 21 ions with r2 = 0.3 and monotonic change by IV busulfan clearance tertile. Open circles – slow clearance tertile, “X” symbols – moderate clearance tertile, black triangles – rapid clearance tertile. Each ion is identified by the mass-to-charge ratio (m/z) and retention time (RT in min).
A predictive model for IV busulfan clearance
A linear predictive model was subsequently built from these 21 selected ions. As described in the Statistical Analysis section, we used stepwise variable selection to construct a linear model based on seven ions as predictors of IV busulfan clearance (Table 3). The prediction of clearance was defined as accurate when the predicted IV busulfan clearance fell within 80 to 125% of the observed IV busulfan clearance.(Administration 2000) Of the 59 samples in the discovery dataset, IV busulfan clearance was accurately predicted for 55 samples (93.2%). For the 88 samples in the validation dataset, which were not used in constructing the predictive model, IV busulfan clearance was accurately predicted for 69 samples (78.4%). Figure 3 shows the observed versus predicted IV busulfan clearance for both the discovery and validation datasets.
Table 3.
Model coefficients for the seven-ion predictive model.
| Ion | Retention Time (min) | β Estimate ± Standard Error | P-value |
|---|---|---|---|
| [Intercept] | 3.15 ± 2.36 | 0.2 | |
| 600.337 | 10.5 | −2.14 ± 1.11 | 0.06 |
| 600.337 | 10.6 | 2.27 ± 1.13 | 0.05 |
| 626.353 | 11.0 | 0.80 ± 0.31 | 0.01 |
| 303.230 (Linoleic acid) | 11.8 | −0.53 ± 0.27 | 0.06 |
| 393.297 | 12.5 | 1.10 ± 0.33 | 0.002 |
| 415.282 (Chenodeoxycholic acid or deoxycholic acid) | 10.8 | −0.26 ± 0.16 | 0.1 |
| 542.522 | 11.0 | −0.89 ± 0.41 | 0.03 |
The predictive model is of form E(y)=β0 + β1 x1 + … β7 x7 where y is IV busulfan clearance and xj is the abundance of the jth ion. The coefficients, βj, were estimated using the discovery data. These values of βj were then fixed in this equation while the xj values obtained from the validation dataset were used to obtain the predicted values, y*, of the observed y.
Fig. 3.
The performance of the seven-ion predictive model to estimate IV busulfan clearance in the discovery and validation datasets. The middle solid line represents the line of unity, and the outer dotted lines represent 80 and 125% of the observed IV busulfan clearance. Observed clearance is IV busulfan clearance (ml/min/kg NFM) estimated using noncompartmental analysis. Predicted clearance is IV busulfan clearance (ml/min/kg of NFM) estimated using the seven-ion predictive model. Open circles – slow clearance tertile, “X” symbols – moderate clearance tertile, black triangles – rapid clearance tertile.
Evaluation of BCT
For the seven predictive ions, the ion abundances were similar when plasma was collected in citrate or EDTA BCTs. A strong positive correlation (r2 from 0.50 to 0.85) was found for six of the seven ions, suggesting that the BCT did not greatly affect ion abundance (Online Resource Supplemental Figures 3). The predicted IV busulfan clearances from the citrate BCT samples were comparable to the predicted IV busulfan clearances from the EDTA BCT samples, suggesting that the BCT did not substantially affect the results of the predictive model (Online Resources, Supplemental Figures 3 and 4).
Ion identification
Four of the 21 selected ions were potentially identified. For the remaining 16 ions, the hypothesized classes as determined by METLIN is listed in Table 2. The authentic standards for both chenodeoxycholic acid and deoxycholic acid, which differ only in the position of a hydroxyl group and thus have identical molecular weights, eluted at the same time (~10.8 min) and with peaks at the same m/z as the ions with m/z of 357.279 and 415.282. These ions represent the loss of two water molecules (m/z of 357.279; i.e., [M+H - 2 H2O]+) and the sodium adduct (m/z of 415.282; i.e., [M+Na]+) of chenodeoxycholic acid and/or deoxycholic acid (monoisotopic MW = 392.293 Da). When selected samples were analyzed at a collision energy of 20 V, the fragmentation pattern of the 357.279 m/z ion was similar to that of authentic standards of both chenodeoxycholic acid and deoxycholic acid. We were not able to fragment the 415.282 m/z ion. Chenodeoxycholic acid and deoxycholic acid could not be resolved chromatographically using the stated LC-QTOF conditions.
The ions with m/z of 303.230 and 319.193 eluting at 11.8 minutes are consistent with the sodium (i.e., [M+Na]+) and potassium (i.e., [M+K]+) adducts of linoleic acid. The authentic standard for linoleic acid gave rise to peaks with m/z of 281.247 ([M+H]+), 303.230 ([M+Na]+), and 319.193 ([M+K]+) that co-eluted within 0.1 min of the peaks seen in clinical plasma samples. The putative [M+H]+ peak was also observed in clinical samples, but a slightly overlapping peak at the same m/z likely interfered with its accurate integration, and that ion was not significantly correlated with busulfan clearance. The hypothesized sodium and potassium adducts of linoleic acid were unable to be fragmented successfully, precluding confirmation of its identity based on fragmentation pattern.
DISCUSSION
These analyses provide encouraging results demonstrating the potential of pharmacometabolomics-based markers for informing IV busulfan dosing. Using a semi-quantitative, untargeted LC-MS platform to measure predose endogenous metabolite ions in plasma in citrate BCT, we built a statistical model from a small number of ions that provides a modest but reproducible capability to predict IV busulfan clearance in a separate dataset of subjects whose plasma samples were collected in a different BCT (i.e., EDTA).
Busulfan is a bi-functional alkylating agent that rapidly (i.e., within 2 minutes) damages DNA in a dose-dependent manner ex vivo (Morales-Ramirez et al. 2006). Busulfan plasma exposure has proven to be a predictive marker for outcomes, and PK-guided busulfan dosing has been shown to lower rates of rejection, nonrelapse mortality, and relapse in select HCT recipients (Deeg et al. 2006; Radich et al. 2003). Although the recent increase in PK-guided dosing of busulfan shows that this strategy is feasible, the resource intensity of pharmacokinetic sampling has been a barrier. PK-guided dosing is resource-intensive, involving the collection of multiple blood samples after IV busulfan administration, quantitation of plasma busulfan concentrations, and subsequent pharmacokinetic analysis to calculate patient-specific busulfan clearance (as clearance = dose/AUC) for personalizing future doses to the target AUC. More efficient methods to estimate IV busulfan systemic exposure and clearance are desirable because shorter (i.e., 2-day) busulfan regimens are increasingly popular. The combination of estimating busulfan doses to achieve target busulfan AUC based on a population pharmacokinetic model (McCune et al. 2014) and pre-dose pharmacometabolomics may decrease the need for resource-intensive PK-guided dosing.
After IV administration of radiolabeled busulfan, less than 50% of the administered dose is recovered in the urine (Nadkarni et al. 1959; Vodopick et al. 1969). Approximately one-third (i.e., 32.8 ± 2.2%) of busulfan is irreversibly bound to plasma proteins, primarily albumin (Ehrsson and Hassan 1984). Only a small fraction (<3%) of a busulfan dose is excreted unchanged in the urine, with negligible amounts in the feces (Hassan et al. 1989; Ehrsson et al. 1983) The primary route of elimination for busulfan is by hepatic glutathione conjugation, catalyzed by glutathione transferases (GSTs) (Hassan and Ehrsson 1987; Gibbs et al. 1996; Czerwinski et al. 1996). Of the various classes of GSTs, busulfan is predominantly conjugated by GSTA1-1 (alpha class), with GSTM1-1 (mu class) and GSTP1-1 (pi class) participating to a lesser degree (Czerwinski et al. 1996). The conjugated metabolite, γ-glutamyl-β-(S-tetrahydrothiophenium)-alanyl-glycine (THT+) (Gibbs et al. 1997; Hassan et al. 1996), is subsequently metabolized by various cytochrome P450 enzymes, specifically CYP1A1, 2B6, 2C8, 2C9, and 2C19 (El-Serafi et al. 2014).
The associations of IV busulfan clearance with various genetic polymorphisms, usually those regulating GSTA1-1 and GSTM1-1 hepatic protein expression, have been examined. Polymorphisms in GSTA1 and GSTM1 are not consistently associated with IV busulfan clearance, making constitutional genetics-based dosing infeasible (Zwaveling et al. 2008; Abbasi et al. 2011). In HCT recipients, plasma alpha GST activity was only moderately correlated with hepatic GST expression (r2=0.567) (Poonkuzhali et al. 2001) as was blood GSH with IV busulfan clearance (r2=0.45) (Almog et al. 2011). Thus, lymphocyte mRNA or protein activity will not sufficiently reflect hepatic mRNA or protein expression of IV busulfan-metabolizing enzymes, making RNA or protein techniques obtained from a blood sample infeasible biomarkers for IV busulfan PK. Alternative techniques, such as pharmacometabolomics, can be used to identify biomarkers to replace PK-guided IV busulfan dosing, with the eventual goal of improving overall survival after IV busulfan-containing HCT conditioning regimens. The work herein is encouraging proof of principle that IV busulfan clearance can be predicted by endogenous pharmacometabolomics.
Of the 21 selected ions associated with IV busulfan clearance tertiles, deoxycholic acid and/or chenodeoxycholic acid as well as linoleic acid were identified. We could not find any published literature evaluating the effect of these bile acids upon busulfan metabolism, though bile acids have been shown to inhibit human GST activity in vitro.(Singh et al. 1988) Singh et al. showed that human GSTs are inhibited by bile acids, including chenodeoxycholic acid, with varying potency based on the GST isozyme studied.(Singh et al. 1988) Conjugation by GST is the first step in busulfan metabolism and inhibition of this step could lead to a decrease in busulfan clearance. In our study, while we did observe that individuals with slow clearance showed elevated levels of linoleic acid and deoxycholic acid and/or chenodeoxycholic acid when compared to those with rapid clearance, further work needs to be done to confirm this association.
A different bile acid – ursodeoxycholic acid – is administered to HCT recipients to lower the risk of hepatotoxicity. In HCT recipients, there were no statistically significant differences in cyclosporine pharmacokinetics between ursodeoxycholic acid and placebo but its effect upon other HCT medications – including busulfan – have not been examined. Chenodeoxycholic acid increases hepatic mRNA expression of CYP2B6, 2C8, 2C19 and 3A4 in a human hepatocyte model (Chaudhry et al. 2013), and induces CAR, FXR, and PXR in primary hepatocytes and Caco-2 cells (Martin et al. 2008). CAR induces expression of CYP1A1, and CAR and PXR both induce expression of CYP2B6, 2C9, and 2C19 (Wang et al. 2012). These CYPs are involved in the metabolism of the busulfan metabolite THT+ (El-Serafi et al. 2014). Also, GSH conjugates of chenodeoxycholic acid have been found in the bile of human infants (although not adults) (Mitamura et al. 2011), which may be relevant since the primary route of busulfan metabolism is via conjugation of GSH by GSTs. If GST-mediated glutathionylation of busulfan is lower in some patients than in others, then GST-mediated glutathionylation of bile acids could also be low in these patients, leading to relatively higher concentrations of the bile acids chenodeoxycholic acid and/or deoxycholic acid (i.e., the ions with m/z of 357.278 and 415.282 with retention time of 10.8 minutes) – and lower concentrations of the products of that reaction, GSH-bile-acid conjugates (not measured). In this analysis, chenodeoxycholic acid or deoxycholic acid were negatively correlated with IV busulfan clearance. The putative mechanism of deoxycholic acid and chenodeoxycholic acid association with IV busulfan clearance could be via their influence upon THT+ metabolism or could reflect GST activity, but in vitro studies are needed to test and to clarify these hypotheses.
There are some limitations to this work. IV busulfan plasma exposure was not the primary endpoint because it is determined by the administered IV busulfan dose (McCune et al. 2013), the administration interval, and the patient-specific IV busulfan clearance. The purpose of PK-guided dosing is to estimate the patient-specific IV busulfan clearance, which was the primary endpoint. Given the substantive number of ions identified detected with global pharmacometabolomics approaches, we focused upon a discrete and well-defined endpoint (i.e., busulfan clearance) over clinical outcomes because of the heterogeneity of the patient population (Table 1). Typical of an HCT analysis, there was considerable heterogeneity in the additional components of the HCT conditioning regimens, in the underlying diagnoses of the patient population, and in the type of allogeneic graft, all of which can affect clinical outcomes (Copelan 2006). The samples were collected in two different BCT, which fortunately did not appear to influence the ions of interest. We assigned samples to the discovery dataset because of their homogeneity (i.e., no HCT conditioning medications in the pharmacometabolomics samples). Despite the difference in HCT conditioning in the discovery and validation datasets, IV busulfan clearance was adequately estimated by the predictive model in the validation datasets (i.e., 78.4% of the validation samples were within 80% to 125% of the observed IV busulfan clearance values). More precise estimation of IV busulfan clearance is desirable to allow more accurate personalization of IV busulfan doses to achieve the desired busulfan plasma exposure. Finally, as in other pharmacometabolomics studies, identification of ions of interest is a hurdle. We identified deoxycholic acid and/or chenodeoxycholic acid and linoleic acid as part of the 21 selected ions, but only one ion as part of the seven predictive ions. An analysis of deoxycholic acid, chenodeoxycholic acid, and linoleic acid concentrations, quantitated using targeted assays, and subsequent correlation with IV busulfan clearance should be conducted. Finally, these results should be confirmed in a prospective study to determine how precisely the predictive model is able to estimate IV busulfan clearance and what other information can be incorporated (e.g., population-based PK model) (McCune et al. 2014) to improve the estimation of the IV busulfan dose for HCT patients.
OVERALL CONCLUSIONS
We evaluated the association of the relative abundance of 1885 ions with IV busulfan clearance and found 21 ions that were significantly correlated with IV busulfan clearance. The seven-ion predictive model accurately predicted IV busulfan clearance in the majority of the validation samples. These results reinforce the potential of pharmacometabolomics as a critical tool in personalized medicine, with the potential to improve the personalized dosing of drugs with a narrow therapeutic index such as busulfan.
Supplementary Material
Acknowledgments
The authors thank the study subjects, their caregivers, and the patient care staff for their support of this study. This work was supported by research funding from the National Institute of Health (CA182963, ES07033, CA18029, and CA15704)
Funding: This study was funded by the National Institute of Health (CA182963, ES07033, CA18029, and CA15704)
Footnotes
Conflict of Interest: All authors declare that they have no conflict of interest
Ethical approval: All procedures performed in studies involving human subjects were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Bibliography
- Abbasi N, Vadnais B, Knutson JA, Blough DK, Kelly EJ, O’Donnell PV, et al. Pharmacogenetics of intravenous and oral busulfan in hematopoietic cell transplant recipients. J Clin Pharmacol. 2011;51(10):1429–1438. doi: 10.1177/0091270010382915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Administration, F. a. D. [Accessed October 18 2013];Guidance for Industry: Bioavailability and Bioequivalence Studies for Orally Administered Drug Products — General Considerations. 2000 http://www.fda.gov/downloads/Drugs/DevelopmentApprovalProcess/HowDrugsareDevelopedandApproved/ApprovalApplications/AbbreviatedNewDrugApplicationANDAGenerics/UCM154838.pdf.
- Almog S, Kurnik D, Shimoni A, Loebstein R, Hassoun E, Gopher A, et al. Linearity and stability of intravenous busulfan pharmacokinetics and the role of glutathione in busulfan elimination. Biol Blood Marrow Transplant. 2011;17(1):117–123. doi: 10.1016/j.bbmt.2010.06.017. S1083-8791(10)00277-6 [pii] [DOI] [PubMed] [Google Scholar]
- Anderson BJ, Holford NH. Mechanism-based concepts of size and maturity in pharmacokinetics. Annu Rev Pharmacol Toxicol. 2008;48:303–332. doi: 10.1146/annurev.pharmtox.48.113006.094708. [DOI] [PubMed] [Google Scholar]
- Anderson BJ, Holford NH. Mechanistic basis of using body size and maturation to predict clearance in humans. Drug Metab Pharmacokinet. 2009;24(1):25–36. doi: 10.2133/dmpk.24.25. JST.JSTAGE/dmpk/24.25 [pii] [DOI] [PubMed] [Google Scholar]
- Benton HP, Want EJ, Ebbels TM. Correction of mass calibration gaps in liquid chromatography-mass spectrometry metabolomics data. Bioinformatics. 2010;26(19):2488–2489. doi: 10.1093/bioinformatics/btq441. [DOI] [PubMed] [Google Scholar]
- Buhlmann P, Kalisch M, Meier L. High-Dimensional Statistics with a View Toward Applications in Biology. Annual Review of Statistics and Its Application. 2014;1:255–U809. doi: 10.1146/annurev-statistics-022513-115545. [DOI] [Google Scholar]
- Chaudhry AS, Thirumaran RK, Yasuda K, Yang X, Fan Y, Strom SC, et al. Genetic variation in aldo-keto reductase 1D1 (AKR1D1) affects the expression and activity of multiple cytochrome P450s. Drug Metab Dispos. 2013;41(8):1538–1547. doi: 10.1124/dmd.113.051672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton TA, Baker D, Lindon JC, Everett JR, Nicholson JK. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc Natl Acad Sci U S A. 2009;106(34):14728–14733. doi: 10.1073/pnas.0904489106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton TA, Lindon JC, Cloarec O, Antti H, Charuel C, Hanton G, et al. Pharmaco-metabonomic phenotyping and personalized drug treatment. Nature. 2006;440(7087):1073–1077. doi: 10.1038/nature04648. [DOI] [PubMed] [Google Scholar]
- Copelan EA. Hematopoietic stem-cell transplantation. N Engl J Med. 2006;354(17):1813–1826. doi: 10.1056/NEJMra052638. [DOI] [PubMed] [Google Scholar]
- Copelan EA, Hamilton BK, Avalos B, Ahn KW, Bolwell BJ, Zhu X, et al. Better leukemia-free and overall survival in AML in first remission following cyclophosphamide in combination with busulfan compared to TBI. Blood. 2013 doi: 10.1182/blood-2013-07-514448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortinez LI, Anderson BJ, Penna A, Olivares L, Munoz HR, Holford NH, et al. Influence of obesity on propofol pharmacokinetics: derivation of a pharmacokinetic model. Br J Anaesth. 2010;105(4):448–456. doi: 10.1093/bja/aeq195. [DOI] [PubMed] [Google Scholar]
- Czerwinski M, Gibbs JP, Slattery JT. Busulfan conjugation by glutathione S-transferases alpha, mu, and pi. Drug Metab Dispos. 1996;24(9):1015–1019. [PubMed] [Google Scholar]
- Deeg HJ, Storer BE, Boeckh M, Martin PJ, McCune JS, Myerson D, et al. Reduced incidence of acute and chronic graft-versus-host disease with the addition of thymoglobulin to a targeted busulfan/cyclophosphamide regimen. Biol Blood Marrow Transplant. 2006;12(5):573–584. doi: 10.1016/j.bbmt.2005.12.036. [DOI] [PubMed] [Google Scholar]
- Ehrsson H, Hassan M. Binding of busulfan to plasma proteins and blood cells. J Pharm Pharmacol. 1984;36(10):694–696. doi: 10.1111/j.2042-7158.1984.tb04847.x. [DOI] [PubMed] [Google Scholar]
- Ehrsson H, Hassan M, Ehrnebo M, Beran M. Busulfan Kinetics. Clin Pharmacol Ther. 1983;34:86–89. doi: 10.1038/clpt.1983.134. [DOI] [PubMed] [Google Scholar]
- El-Serafi I, Naughton S, Saghafian M, Abedi-Valugerdi M, Mattsson J, Hagbjörk A, et al. PhD defense. Karolinska Institute; Sep 5, 2014. The role of flavin-containing monooxygenase 3 (FMO3) in busulphan metabolism. [Google Scholar]
- Gibbs JP, Czerwinski M, Slattery JT. Busulfan-glutathione conjugation catalyzed by human liver cytosolic glutathione S-transferases. Cancer Res. 1996;56(16):3678–3681. [PubMed] [Google Scholar]
- Gibbs JP, Murray G, Risler L, Chien JY, Dev R, Slattery JT. Age-dependent tetrahydrothiophenium ion formation in young children and adults receiving high-dose busulfan. Cancer Res. 1997;57(24):5509–5516. [PubMed] [Google Scholar]
- Hassan M, Ehrsson H. Metabolism of 14C-busulfan in isolated perfused rat liver. Eur J Drug Metab Pharmacokinet. 1987;12(1):71–76. doi: 10.1007/BF03189864. [DOI] [PubMed] [Google Scholar]
- Hassan M, Fasth A, Gerritsen B, Haraldsson A, Syruckova Z, van den Berg H, et al. Busulphan kinetics and limited sampling model in children with leukemia and inherited disorders. Bone Marrow Transplant. 1996;18(5):843–850. [PubMed] [Google Scholar]
- Hassan M, Oberg G, Ehrsson H, Ehrnebo M, Wallin I, Smedmyr B, et al. Pharmacokinetic and metabolic studies of high-dose busulphan in adults. Eur J Clin Pharmacol. 1989;36(5):525–530. doi: 10.1007/BF00558081. [DOI] [PubMed] [Google Scholar]
- Huang L, Scheipl F, Goldsmith J, Gellar J, Harezlak J, McLean MW, et al. R package version 0.1-12. ed. 2015. refund: Regresson with Functional Data. [Google Scholar]
- Janmahasatian S, Duffull SB, Ash S, Ward LC, Byrne NM, Green B. Quantification of lean bodyweight. Clin Pharmacokinet. 2005;44(10):1051–1065. doi: 10.2165/00003088-200544100-00004. [DOI] [PubMed] [Google Scholar]
- Janmahasatian S, Duffull SB, Chagnac A, Kirkpatrick CM, Green B. Lean body mass normalizes the effect of obesity on renal function. Br J Clin Pharmacol. 2008;65(6):964–965. doi: 10.1111/j.1365-2125.2008.03112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenke A, Freiberg-Richter J, Johne C, Knoth H, Schleyer E, Ehninger G, et al. Targeting once-daily intravenous busulfan in combination with fludarabine before allogeneic hematopoietic cell transplantation. Bone Marrow Transplant. 2005;35(6):627–628. doi: 10.1038/sj.bmt.1704831. [DOI] [PubMed] [Google Scholar]
- Karlsson MO, Jonsson EN, Wiltse CG, Wade JR. Assumption testing in population pharmacokinetic models: illustrated with an analysis of moxonidine data from congestive heart failure patients. J Pharmacokinet Biopharm. 1998;26(2):207–246. doi: 10.1023/a:1020561807903. [DOI] [PubMed] [Google Scholar]
- Laboratory. Seattle Cancer Care Alliance Busulfan Pharmacokinetics. [Accessed February 1 2016];Sending Samples to the Busulfan Laboratory. http://www.seattlecca.org/client/documents/Req_Q6-IV_Q24-IV_Busulfex_v2.pdf.
- Martin P, Riley R, Back DJ, Owen A. Comparison of the induction profile for drug disposition proteins by typical nuclear receptor activators in human hepatic and intestinal cells. Br J Pharmacol. 2008;153(4):805–819. doi: 10.1038/sj.bjp.0707601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCune JS, Baker KS, Blough DK, Gamis A, Bemer MJ, Kelton-Rehkopf MC, et al. Variation in Prescribing Patterns and Therapeutic Drug Monitoring of Intravenous Busulfan in Pediatric Hematopoietic Cell Transplant Recipients. J Clin Pharmacol. 2013;53(3):264–275. doi: 10.1177/0091270012447196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCune JS, Batchelder A, Deeg HJ, Gooley T, Cole S, Phillips B, et al. Cyclophosphamide following Targeted Oral Busulfan as Conditioning for Hematopoietic Cell Transplantation: Pharmacokinetics, Liver Toxicity, and Mortality. Biol Blood Marrow Transplant. 2007;13(7):853–862. doi: 10.1016/j.bbmt.2007.03.012. [DOI] [PubMed] [Google Scholar]
- McCune JS, Bemer MJ, Barrett JS, Scott Baker K, Gamis AS, Holford NH. Busulfan in infant to adult hematopoietic cell transplant recipients: a population pharmacokinetic model for initial and bayesian dose personalization. Clin Cancer Res. 2014;20(3):754–763. doi: 10.1158/1078-0432.CCR-13-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCune JS, Holmberg LA. Busulfan in hematopoietic stem cell transplant setting. Expert Opin Drug Metab Toxicol. 2009;5(8):957–969. doi: 10.1517/17425250903107764. [DOI] [PubMed] [Google Scholar]
- McCune JS, Woodahl EL, Furlong T, Storer B, Wang J, Heimfeld S, et al. A pilot pharmacologic biomarker study of busulfan and fludarabine in hematopoietic cell transplant recipients. Cancer Chemother Pharmacol. 2012;69(1):263–272. doi: 10.1007/s00280-011-1736-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitamura K, Hori N, Iida T, Suzuki M, Shimizu T, Nittono H, et al. Identification of S-acyl glutathione conjugates of bile acids in human bile by means of LC/ESI-MS. Steroids. 2011;76(14):1609–1614. doi: 10.1016/j.steroids.2011.10.001. [DOI] [PubMed] [Google Scholar]
- Morales-Ramirez P, Miranda-Pasaye S, Cruz-Vallejo VL, Vallarino-Kelly T, Mendiola-Cruz MT. Kinetic of genotoxic expression in the pharmacodynamics of busulfan. Arch Med Res. 2006;37(3):316–321. doi: 10.1016/j.arcmed.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Mould DR, Holford NH, Schellens JH, Beijnen JH, Hutson PR, Rosing H, et al. Population pharmacokinetic and adverse event analysis of topotecan in patients with solid tumors. Clin Pharmacol Ther. 2002;71(5):334–348. doi: 10.1067/mcp.2002.123553. [DOI] [PubMed] [Google Scholar]
- Nadkarni MV, Trams EG, Smith PK. Preliminary Studies on the Distribution and Fate of TEM, TEPA, and Myeleran in the Human. Cancer Res. 1959;19:713–718. [PubMed] [Google Scholar]
- Nicholson JK, Connelly J, Lindon JC, Holmes E. Metabonomics: a platform for studying drug toxicity and gene function. Nat Rev Drug Discov. 2002;1(2):153–161. doi: 10.1038/nrd728. [DOI] [PubMed] [Google Scholar]
- Nieder ML, McDonald GB, Kida A, Hingorani S, Armenian SH, Cooke KR, et al. National Cancer Institute-National Heart, Lung and Blood Institute/pediatric Blood and Marrow Transplant Consortium First International Consensus Conference on late effects after pediatric hematopoietic cell transplantation: long-term organ damage and dysfunction. Biol Blood Marrow Transplant. 2011;17(11):1573–1584. doi: 10.1016/j.bbmt.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poonkuzhali B, Chandy M, Srivastava A, Dennison D, Krishnamoorthy R. Glutathione S-transferase activity influences busulfan pharmacokinetics in patients with beta thalassemia major undergoing bone marrow transplantation. Drug Metab Dispos. 2001;29(3):264–267. [PubMed] [Google Scholar]
- R Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2014. [Google Scholar]
- Radich JP, Gooley T, Bensinger W, Chauncey T, Clift R, Flowers M, et al. HLA-matched related hematopoietic cell transplantation for chronic-phase CML using a targeted busulfan and cyclophosphamide preparative regimen. Blood. 2003;102(1):31–35. doi: 10.1182/blood-2002-08-2619. [DOI] [PubMed] [Google Scholar]
- Randolph TW, Harezlak J, Feng Z. Structured penalties for functional linear models-partially empirical eigenvectors for regression. Electron J Stat. 2012;6:323–353. doi: 10.1214/12-ejs676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezvani AR, McCune JS, Storer BE, Batchelder A, Kida A, Deeg HJ, et al. Cyclophosphamide followed by Intravenous Targeted Busulfan for Allogeneic Hematopoietic Cell Transplantation: Pharmacokinetics and Clinical Outcomes. Biol Blood Marrow Transplant. 2013;19(7):1033–1039. doi: 10.1016/j.bbmt.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinger DH, Vicini P, Blough DK, O’Donnell PV, Pawlikowski MA, McCune JS. Development of a Population Pharmacokinetics-Based Sampling Schedule to Target Daily Intravenous Busulfan for Outpatient Clinic Administration. J Clin Pharmacol. 2010;50(11):1292–1300. doi: 10.1177/0091270009357430. 0091270009357430 [pii] [DOI] [PubMed] [Google Scholar]
- Singh SV, Leal T, Awasthi YC. Inhibition of human glutathione S-transferases by bile acids. Toxicol Appl Pharmacol. 1988;95(2):248–254. doi: 10.1016/0041-008x(88)90161-5. [DOI] [PubMed] [Google Scholar]
- Slattery JT, Risler LJ. Therapeutic monitoring of busulfan in hematopoietic stem cell transplantation. Ther Drug Monit. 1998;20(5):543–549. doi: 10.1097/00007691-199810000-00017. [DOI] [PubMed] [Google Scholar]
- Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem. 2006;78(3):779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- Tarantal AF, Giannoni F, Lee CC, Wherley J, Sumiyoshi T, Martinez M, et al. Nonmyeloablative conditioning regimen to increase engraftment of gene-modified hematopoietic stem cells in young rhesus monkeys. Mol Ther. 2012;20(5):1033–1045. doi: 10.1038/mt.2011.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautenhahn R, Bottcher C, Neumann S. Highly sensitive feature detection for high resolution LC/MS. BMC Bioinformatics. 2008;9:504. doi: 10.1186/1471-2105-9-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay-Sontheimer J, Shireman LM, Beyer RP, Senn T, Witten D, Pearce RE, et al. Detection of an endogenous urinary biomarker associated with CYP2D6 activity using global metabolomics. Pharmacogenomics. 2014;15(16):1947–1962. doi: 10.2217/pgs.14.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodopick H, Hamilton HE, Jackson HL, Peng CT, Sheets RF. Metabolic fate of tritiated busulfan in man. J Lab Clin Med. 1969;73(2):266–276. [PubMed] [Google Scholar]
- Wang YM, Ong SS, Chai SC, Chen T. Role of CAR and PXR in xenobiotic sensing and metabolism. Expert Opin Drug Metab Toxicol. 2012;8(7):803–817. doi: 10.1517/17425255.2012.685237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Chen Y, Zhang R, Song Y, Cao J, Bi N, et al. Global and targeted metabolomics of esophageal squamous cell carcinoma discovers potential diagnostic and therapeutic biomarkers. Mol Cell Proteomics. 2013;12(5):1306–1318. doi: 10.1074/mcp.M112.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh RF, Pawlikowski MA, Blough DK, McDonald GB, O’Donnell PV, Rezvani A, et al. Accurate targeting of daily intravenous busulfan with 8-hour blood sampling in hospitalized adult hematopoietic cell transplant recipients. Biol Blood Marrow Transplant. 2012;18(2):265–272. doi: 10.1016/j.bbmt.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaveling J, Press RR, Bredius RG, van Derstraaten TR, den Hartigh J, Bartelink IH, et al. Glutathione S-transferase polymorphisms are not associated with population pharmacokinetic parameters of busulfan in pediatric patients. Ther Drug Monit. 2008;30(4):504–510. doi: 10.1097/FTD.0b013e3181817428. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.