Summary
Recent excitement regarding immune clearance of intracellular microorganisms has focused on two systems that maintain cellular homeostasis. One system includes cellular autophagy components that mediate degradation of pathogens in membrane-bound compartments, in a process termed xenophagy. The second system is driven by interferon– regulated GTPases that promote rupture of pathogen-containing vacuoles and microbial degradation. In the case of xenophagy, pathogen sequestration and compartmentalization suppress inflammation. In contrast, interferon-driven events can lead to exposure of pathogen-associated molecular patterns to the host cytosol with consequent inflammasome activation. Paradoxically, signals and factors involved in xenophagy also mobilize interferon-regulated GTPases, which drive the inflammatory response, indicating considerable crosstalk between these pathways. How these responses are prioritized remains to be understood. In this review, we describe mechanisms of intracellular pathogen clearance that rely on the autophagy machinery and interferon-regulated GTPases, and speculate how these pathways engage each other to balance pathogen elimination with inflammation.
Introduction
The uptake of an intracellular pathogen into a mammalian host cell initiates a battle with clear downstream consequences. A traditional view of this encounter is that the pathogen and the host are in conflict, with the winner determining if health or disease will ensue. In fact, interactions between these players are much more nuanced, with several possible consequences. At the simplest level, within a single cell, the pathogen either establishes a replication site or the host prevents infection by killing the microorganism. This binary relationship rarely captures the host-pathogen relationship. For instance, establishing a beachhead in a host cell could involve forming a latent state for the microorganism as in Mycobacterium tuberculosis or HIV infections (Cambier et al., 2014, Churchill et al., 2016). Furthermore, blocking disease progression may result in killing of microbes, but may also involve bacteriostasis or host cell death.
In this review we discuss the several host pathways that restrict intracellular microorganisms and consequences for the outcome of disease. For instance, microbial growth can be terminated by host-derived reactive oxygen or nitrogen species, routing of a membrane-bound pathogen into a lysosomal locale or subjecting cytosolic pathogens to poorly characterized lytic defense pathways, sometimes resulting in overt inflammatory responses (Martinez et al., 2015, Haldar et al., 2015). Added to this diversity of responses is a panoply of events within the host that can either block or support microbial replication. In its most extreme expression, the disease process could involve blocking of bacterial growth in a subset of cells by innate immune pathways, but eliciting an inflammatory response that supports effective pathogen replication within the animal, as is seen with Salmonella enterica during intestinal growth (Winter et al., 2010).
We will focus on restriction of intracellular pathogens by the host innate immune system, concentrating on the destruction of microorganisms while they are resident in membrane-bound compartments. The mechanism of pathogen restriction modulates the nature of the global innate immune response throughout a tissue site, which can either stimulate or prevent the production of inflammatory mediators. However, there is a critical block to progress in the field, because host components that mark invading microorganisms are poor predictors of the strategy used to destroy invaders. Therefore, specific molecular components that control each of these routes of microbial destruction need to be identified in order to better predict if an inflammation will ensue.
In the following overview, we will emphasize unresolved issues that stem from the recently discovered interface between the autophagic attack against pathogens and the action of interferon-regulated GTPases in modulating levels of released inflammatory mediators. In so doing, we acknowledge that there is also cross-regulation between the host autophagy machinery and a number of other processes important to the inflammatory response, such as regulation of autophagy by pattern recognition receptors (Travassos et al., 2010), that we will not cover here. Such cross-talk may be particularly important in maintaining intestinal homeostasis, as witnessed in human allele variants in these components that are associated with inflammatory conditions such as Crohn’s disease (Parkes et al., 2007). The reader is referred to one of a number of excellent reviews that exist on this topic (Salem et al., 2015, Lassen and Xavier, 2017).
Basic principles of xenophagy
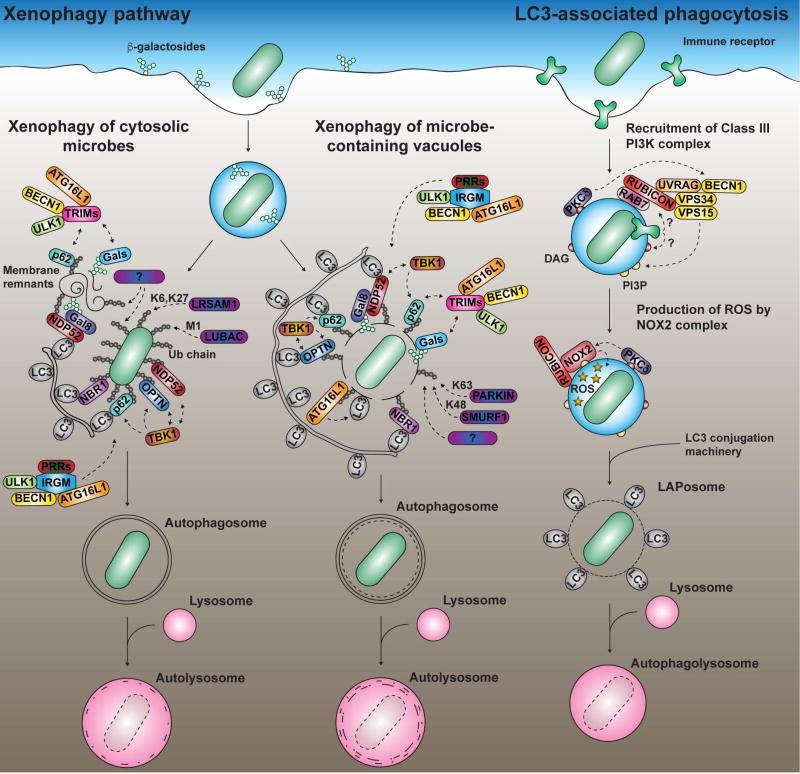
The term autophagy comprises several catabolic processes that target cytoplasmic components for lysosomal degradation (Yin et al., 2016, Galluzzi et al., 2017). These various forms of autophagy utilize overlapping sets of protein complexes that are involved in a number of cellular processes, including some unrelated to autophagic degradation (Bestebroer et al., 2013). During macroautophagy, intracellular components are sequestered into double-membrane vesicles called autophagosomes and delivered to lysosome-like compartments. As observed during starvation, macroautophagy can sequester parts of the cytoplasm, providing building blocks for housekeeping functions. In contrast, macroautophagy can also operate in a selective manner and mediates the clearance and recycling of specific components such as protein aggregates, damaged organelles and intracellular microbes. The term xenophagy is defined as selective autophagy, during which microorganisms are sequestered into autophagosomes and digested within lysosomes (Fig. 1) (Huang and Brumell, 2014, Randow and Youle, 2014). More than one pathway can lead to xenophagic degradation, and the autophagy machinery can target either cytosolic or vacuolar pathogens. The autophagy machinery also orchestrates non-canonical pathways that play roles in cell-autonomous defense and may assist other immune processes, such as phagocytosis. These xenophagy-related processes constitute a multilayered and synergistic defense network that protects virtually every subcellular compartment during the intracellular lifecycle of microbes.
FIGURE 1. Targeting of microbes by xenophagy and LC3-associated phagocytosis.
Disruption of a pathogen-containing vacuole (PVs) can lead to xenophagy. Cytosolic microbes (LEFT) and microbe-associated membranes (MIDDLE) are targeted by ubiquitin (Ub) ligases such as PARKIN, SMURF1, LRSAM1 and LUBAC (and other unknown Ub ligases as indicated by question marks) and decorated by a ubiquitin (Ub) coat of different chain linkages (M1, K6, K27, K48 and K63). These Ub chains recruit autophagy adaptors such as NBR1, NDP52, p62 and optineurin (OPTN), which bind LC3 on autophagosomal membranes. TBK1 can be recruited and phosphorylates p62 and OPTN, increasing affinity for Ub. The ATG12-ATG5-ATG16L1 complex binds ubiquitylated membranes and transfers LC3/GABARAP proteins onto PVs. Breaches in pathogen-containing vacuoles expose β-galactosides, which recruit galectins (Gals), and downstream partners. TRIM proteins bind galectins and p62, and interact with ATG16L1, Beclin-1 (BECN1) and ULK1. In humans, these autophagy regulatory proteins may also be recruited in complex with IRGM and pathogen-recognition receptors (PRRs) such as NOD2. RIGHT: TLRs, Fc receptor and CLEC7A/dectin-1 trigger LC3-associated phagocytosis (LAP). Rubicon associates with a Class III PI3K subcomplex, driving production of phosphatidylinositol 3-phosphate (PI3P). Rubicon and PI3P promotes the production of reactive oxygen species (ROS) generated by the NOX2 NADPH oxidase. Crosstalk between TLR signaling, Rubicon, the Class III PI3K subcomplex and NOX2 is likely to involve production of diacylglycerol (DAG), as well as recruitment of PKCδ and RAB7 (indicated by question marks). ROS production triggers conjugation of LC3 through an unknown mechanism. These three pathways ultimately delivered microbes to lysosome-like compartments. LC3: ATG8 orthologs (LC3/GABARAP).
Targeting of pathogens by the canonical autophagy pathway
Several studies on xenophagy describe antimicrobial processes that resemble well-studied macroautophagic pathways although xenophagy may have unique unidentified features (Fig. 1). During macroautophagy, distinct protein complexes coordinate the initiation, nucleation, elongation, closure and recycling steps of the process (Huang and Brumell, 2014, Yin et al., 2016). The proteins ULK1/2, ATG13, FIP200 and ATG101 assemble into the ULK complex to induce the formation of an isolation membrane. The kinase activity of ULK activates the Class III PI3K complex of Beclin-1, VPS34, VPS15 and ATG14 that promotes local production of phosphatidylinositol 3-phosphate (PI3P). Domains enriched in PI3P serve as docking sites for the recruitment of effector proteins such as WIPI1/2, from which the isolation membrane nucleates. The elongation and closure of the isolation membrane are then regulated by two ubiquitin-like conjugation systems, culminating in the conjugation of members of the ATG8 protein family to phosphatidylethanolamine (PE) lipids by the ATG12-ATG5-ATG16L1 complex. There are six orthologs of ATG8 in mammals divided into the LC3 and GABARAP subfamilies that act at different stages of autophagosome formation, but also serve as docking sites for autophagy adaptors that recognize cargo (Weidberg et al., 2011, Nguyen et al., 2016). Autophagosomes ultimately mature into degradative autolysosomes following a series of fusion events with endocytic compartments.
The recognition of a specific target triggers localized autophagy (Randow and Youle, 2014), requiring the activation of the protein kinase TBK1 (Thurston et al., 2016). Recognition of pathogen-associated structures is mediated by “eat-me” signals, such as ubiquitin (Ub) chains of different linkage types, which recruit autophagy adaptors that bridge cargo with the autophagy machinery for degradation (van Wijk et al., 2017, Noad et al., 2017, Randow and Youle, 2014). These autophagy adaptors, which include NBR1, NDP52, optineurin, p62 and TAX1BP1, are important for the antibacterial response and may have functions extending beyond phagophore recruitment (Verlhac et al., 2015, Randow and Youle, 2014, Tumbarello et al., 2015). Less is known about how microorganisms are targeted by Ub chains, although E3 Ub ligases targeting cytosolic S. typhimurium (Huett et al., 2012, Noad et al., 2017, van Wijk et al., 2017) and M. tuberculosis-containing vacuoles (Manzanillo et al., 2013, Franco et al., 2017) have been identified. Interestingly, the recognition of extracellular bacterial DNA by the cGAS-STING pathway, originally identified as activating the type I interferon response, seems to be a trigger for the ubiquitylation of M. tuberculosis-containing vacuoles during infection (Watson et al., 2015).
Monitoring of pathogen vacuole (PV) integrity is likely a critical step in the process of innate immune pathogen detection (Randow and Youle, 2014). While some pathogens cause damage to membranes while in transit to the cytosol, intravacuolar pathogens may also cause membrane breaches surrounding their compartment as a consequence of their replication cycle. Both processes expose β(1,4)-linked galactosides that are recognized by cytosolic galectins. Of a dozen different galectin proteins, galectins-3, -8 and -9 have been demonstrated to detect damaged microorganism-containing vacuoles (Randow and Youle, 2014). Galectin-8 directly binds the autophagy adaptor NDP52 and mediates the engulfment of Salmonella into autophagosomes (Thurston et al., 2012), while Ub-mediated processes may amplify this response. Accordingly, tripartite motif-containing (TRIM) E3 Ub ligases bind galectins (Chauhan et al., 2016). In addition, TRIM16 binding to galectin-3 mobilizes the core autophagy components ATG16L1, ULK1 and Beclin-1 in response to damaged endomembranes, thus triggering a localized autophagy response.
Noncanonical modification of microbial-containing vacuoles by the LC3-conjugation system
In order to initiate a localized response to a microbial threat, the host has evolved mechanisms utilizing a subset of autophagy components to recognize and mark membrane structures associated with pathogens. ATG8 family members (LC3s and GABARAPs) can be directly conjugated to PVs, bypassing some of the well characterized early steps involved in selective autophagy processes (Fig. 1) (Choi et al., 2016, Kageyama et al., 2011, Lam et al., 2013, Zhao et al., 2008). Although recruitment of the autophagy machinery may dictate the formation of an isolation membrane in close proximity to the pathogen, it is important to note that the presence of ATG8 proteins directly inserted onto the membrane compartment surrounding the pathogen represents a topologically distinct process from xenophagy. By being marked with ATG8 proteins, PVs are licensed to interact with downstream components not directly associated with xenophagy, such as those of the phagolysosomal pathway (Martinez et al., 2015) and soluble antimicrobial effectors (Choi et al., 2016, Sasai et al., 2017).
During LC3-associated phagocytosis (LAP), LC3 is directly conjugated to single-membrane vacuoles shortly after cargo internalization (Fig. 1). LAP does not rely on the formation of autophagosomes nor require the ULK complex (Martinez et al., 2015). This process was initially suggested to promote phagosome maturation, but a recent study suggests that it may play other roles, so it could interface with pathogen restriction systems in a number of fashions (Cemma et al., 2016). LAP is initiated after cell surface engagement of TLRs (Martinez et al., 2015), the immunoglobulin Fc receptor FCGR2A/FCγR2A (Cemma et al., 2016) or the CLEC7A/dectin-1 receptor (Ma et al., 2012). LC3 association with enclosed phagosomes is triggered by NOX2 NADPH oxidase complex-mediated production of reactive-oxygen species (ROS) (Huang et al., 2009). Rubicon has been proposed to act as the key molecular switch that activates LAP while it interferes with canonical autophagy. In so doing, PI3P production is stimulated, promoting the assembly of the NOX2 complex (Martinez et al., 2015). During LAP, Rubicon activates a Class III PI3K subcomplex containing UVRAG (UV radiation resistance-associated gene) that is devoid of ATG14L, which usually plays an essential role in canonical autophagy. In addition, diacylglycerol (DAG)-dependent signaling contributes to LAP by recruiting PKCδ (Hubber et al., 2017, Lam et al., 2013), a kinase that targets NOX2 and the kinase JNK, triggering the release of Beclin-1 from its inhibitory interaction with Bcl-2 (Wei et al., 2008). It is tempting to speculate that the activation of PKCδ is upstream to the formation of a RAB7-Rubicon-PI3K complex (Tabata et al., 2010), the production of PI3P and the recruitment of NOX2. How the NOX2-dependent production of ROS triggers LC3 lipidation is still not understood, but may be a consequence of membrane integrity disruption (Boyle and Randow, 2015).
The ATG12-ATG5-ATG16L1 complex specifies the site of ATG8 lipidation during all autophagy-related processes (Fujita et al., 2008). Although this complex can directly bind to membranes through ATG5 in vitro, ATG12-ATG5-ATG16L1 is not recruited to membranes without a proper inducing signal within cells (Romanov et al., 2012). The targeting of membranes by the complex is often directed by ATG16L1, through at least three different interacting partners: FIP200, the PI3P-binding protein WIPI2b (WD repeat domain, phosphoinositide interacting 2b) (Dooley et al., 2014) and Ub (Fujita et al., 2013). Notably, WIPI2b binds the membrane surrounding Salmonella, promoting autophagosomal engulfment of bacteria (Dooley et al., 2014). In addition, LC3 is lipidated on the Salmonella-containing vacuole in a Ub-dependent process that is upstream of autophagosome formation (Kageyama et al., 2011, Fujita et al., 2013). Lipidation in this fashion differs from LAP, a Ub-independent process (Lam et al., 2013, Hubber et al., 2017), and may be in response to a specific pathogenic event.
ATG8 proteins are also directly conjugated to the Toxoplasma- and Chlamydia-containing PVs in a species-specific manner through a mechanism reminiscent of LAP (Haldar et al., 2014, Choi et al., 2014). The lipidation of ATG8 proteins to these PVs does not lead unequivocally to degradation in lysosome-like compartments, but mediates the recruitment of immunity-related GTPases (Park et al., 2016, Sasai et al., 2017). This recruitment is an important link to interferon-induced clearance of intracellular pathogens as described in the following sections.
Interferon-regulated response to intracellular pathogens and regulation of inflammasome activation
Interferons (IFNs) are proteins secreted in response to infection that play a pivotal role in the immune response. IFNs are divided into three subfamilies including type I (IFN-α, IFN-β and other less characterized subtypes), type II (IFN-γ) and type III IFNs. IFN-γ, originally referred to as the macrophage-activating factor, stands out from type I and III IFNs, as the most important mediator of immunity against parasites, viruses and bacteria. IFNα/β induce an antiviral state, but, in contrast to IFN-γ, are not always protective against bacterial infections (McNab et al., 2015). Type III IFNs have been relatively recently discovered and appear to have a function similar to type I IFN, although restricted to epithelial cells (Wack et al., 2015).
Cells induce type I IFNs as a consequence of host pattern recognition receptors binding microbial products. These receptors include TLR4, which engages lipopolysaccharide (LPS), and the RNA recognition proteins TLR7 and RIG-1-like receptors (RLRs) (Wu and Chen, 2014). In addition, cyclic dinucleotides such c-di-AMP, c-di-GMP and cGAMP drive this response, either as microbial products or generated by the host protein cGAS, an enzyme that responds to microbial double stranded DNA by synthesizing 2’,3’cGAMP. Each of the cyclic dinucleotides activates the STING protein, which drives upregulation of type I IFNs (Chen et al., 2016). In addition, the release of cytokines by infected cells activates a signaling cascade that results in the secretion of IFN-γ by several types of immune cells (Schroder et al., 2004).
There are over 2000 IFN-stimulated genes, and of these, four families of GTPases are among the most abundantly expressed (Boehm et al., 1998; Pilla-Moffett et al., 2016). These include the MX viral resistance proteins (72–82 kDa), the immunity-related GTPases (IRGs; 21–47 kDa), the guanylate binding proteins (GBPs; 65–73 kDa) and the very large inducible GTPases (FLIGs/GVNs) (Pilla-Moffett et al., 2016). Bioinformatically and structurally, these subfamilies appear closest in similarity to dynamin-like GTPases, with each having an N-terminal GTPase domain linked to a C-terminal helical domain (Ghosh et al., 2004, Kim et al., 2011). That said, the biochemical behavior of these proteins does not exactly mimic dynamins or other families of GTPases, making it difficult to predict exact biochemical functions of each family member. Although the predominance of these GTPases in the IFN transcriptional response has been established for two decades, only in recent years has there been an explosion of interest in determining their functions in restricting the growth of intracellular pathogens.
For the purposes of understanding how cells respond to intravacuolar pathogens, the most important to consider are the IRGs and the GBPs, which have been linked to the disintegration of vacuole-localized microorganisms during the disease process. The IRGs consist of the GKS effectors (subdivided into the Irga, Irgb, Irgc and Irgd subgroups in the mouse) and IRGM regulatory subfamilies (Irgm1, Irgm2 and Irgm3 in the mouse; IRGM in the human). In hosts such as rodents that have both subfamilies of Irgs, the effector GKS proteins are distinguished based on the presence of a canonical catalytic GKS sequence motif in the GTPase domain, while the regulatory Irgms have the GMS substitution that defines their altered function. Although the GBPs can be grouped into those having or lacking C-terminal prenylation sites, there is no clear evidence that prenylation differentiates function among members of this subfamily.
A major shortcoming in studies related to IFN-induced GTPases is that there is no clear demonstration that the molecular details learned in the mouse can be applied to the human. In contrast to the impressive amount of information now available in the mouse, our knowledge of the roles of these GTPases in human cells lags, and there may be important unidentified players in the human response. Some differences in the interferon-regulated response between human and mouse can be attributed to massive contraction of the IRG family in humans (Bekpen et al., 2005). Divergent results between the two species can also be attributed to the fact that mouse studies are often performed in primary macrophages, while immortalized cell lines are commonly used to interrogate human responses. The high conservation of human and mouse GBPs, on the other hand, indicates that members of this protein family probably function similarly in the two species. Work on human GBPs is still rather slim compared to that of the mouse, so further work is necessary to relate function in the two species. The gap in knowledge regarding the role of GBPs in interferon-regulated restriction in humans is exemplified by the fact that GBP localization around PVs appears to be dependent on the cell type being infected by the pathogen (Johnston et al., 2016, Haldar et al., 2016). Clearly, more work needs to be performed to understand how differences in cell types control the interferon-regulated response as well as to identify evolutionarily-conserved and species-specific responses.
In the mouse, there is ample evidence that both GBPs and IRGs are involved in restricting the growth of pathogens (Fig. 2). As these family members restrict a large spectrum of pathogens, it is unlikely that there is a single mode of action for the entire family, or even that a single member plays only one role in the host innate immune response. That said, considerable evidence exists that GBPs can be recruited to the PV and promote or regulate membrane lysis of either the PV (Yamamoto et al., 2012, Meunier et al., 2014, Kravets et al., 2016) or the pathogen itself (Meunier et al., 2015, Man et al., 2015, Kravets et al., 2016). GBPs play a central role in initiating inflammatory responses to intracellular pathogens by facilitating the presentation of material from invading microorganisms to inflammasomes, which in turn activate cell death pathways executed by either caspase-1 or caspase-11 proteases. The most compelling evidence for a role in inflammasome signaling is in the mouse, in which a single deletion that eliminates five of the eleven GBPs (Gbpch3−/−) has profound defects in the activation of both caspase-1 and 11 (Pilla et al., 2014, Meunier et al., 2015, Meunier and Broz, 2015). It is important to point out that there is negative interplay between inflammasome-associated caspase activation and autophagy (Saitoh and Akira, 2016). As an example, during the IFN-regulated attack of Salmonella-containing vacuoles, inflammasome activation in mouse macrophages is increased in cells lacking ATG5 (Meunier et al., 2014). Therefore, the extent of inflammasome activation and consequent host cell death is negatively modulated by components of the autophagy machinery.
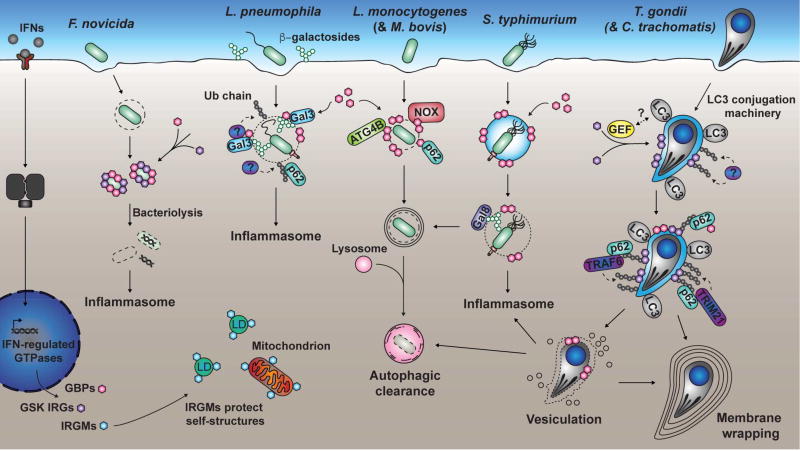
FIGURE 2. IFN-regulated response to intracellular pathogens.
Binding of IFNs to receptors leads to upregulation of GSK IRGs, GBPs and IRGMs. IRGMs have regulatory functions and protect “self” structures from effector activity of GSK IRGs and GBPs. GSK IRGs and GBPs can lyse cytosolic F. novicida bacteria, releasing DNA and activating an inflammasome. GBPs bind to L. pneumophila PVs decorated by galectin-3 (here Gal-3), promoting the formation of a Ub coat, the recruitment of p62 and inflammasome induction. During L. monocytogenes and M. bovis infection, GBPs recruit ATG4B, NADPH oxidase components and p62, and were suggested to trigger xenophagy. GBPs disrupt S. typhimurium PVs, driving both xenophagic clearance and inflammasome induction. IRGs are recruited to T. gondii and C. trachomatis PVs downstream of LC3/GABARAP proteins and trigger formation of a Ub-associated compartment. Ub, in turn, recruits p62, which binds TRAF6, TRIM21, and GBPs. The exposed pathogens can be directly digested in the cytosol via IFN-regulated GTPases with a potential for inflammasome activation, clearance by xenophagy or wrapping in layers of membranes. A speculative interaction between an unknown guanine nucleotide exchange factor (GEF) and LC3/GABARAP proteins is indicated by a question mark. Gal8: galectin-8; LC3: ATG8 orthologs (LC3/GABARAPs); LDs: Lipid droplets.
In contrast to the role of GBP proteins in promoting cascades that lead to inflammasome activation, the action of IRG proteins can lead in multiple directions. Mouse IrgM proteins participate in critical steps that allow accurate targeting of effector proteins to the PV prior to inflammasome activation. In addition, in human cells IRG-dependent restriction of pathogen replication can lead to microbial clearance via xenophagy, which potentially bypasses inflammasome activation (Chauhan et al., 2015).
There are a number of lines of evidence indicating that IRG proteins are indispensable players in IFN-regulated restriction of intracellular pathogens, including M. tuberculosis (MacMicking et al., 2003). One of the earliest demonstrations that this family was involved in destruction of the pathogen vacuolar membrane can be traced to work with a type II Toxoplasma gondii strain in mouse cells (Martens et al., 2005). The Irga6 GKS protein (also called IIGP1) localizes around the PV, forming a “membrane attack complex” and results in PV membrane disruption and eventual destruction of T. gondii in response to IFN-γ treatment. In contrast, IRGM members localize poorly to the PV and instead accumulate on a wide variety of organelles (Haldar et al., 2013). There is considerable evidence that this localization pattern is a form of self/nonself control, in which IRGM proteins prevent GKS effectors from inappropriately acting on host cell organelles. According to this “missing-self” model, the insertion of membranolytic GKS proteins happens exclusively on compartments that lack IRGMs, such as PVs (Haldar et al., 2013). Similarly, GBP proteins mislocalize in the absence of Irgm1 and Irgm3. By marking organelles, IRGM proteins prevent inappropriate targeting of self-structures by the membranolytic IFN-regulated proteins in rodent cells.
The interface of GBP proteins with the IRG system
The model proposed for self/nonself discrimination in the rodent IRG/GBP system still raises unanswered questions. First, while providing an attractive model for how GKS and GBP proteins can be blocked from targeting host membranes, there is no clear explanation for why IRGM proteins are depleted from the PV. Second, it does not provide an explanation for what pathogen-specific signatures are being recognized on the vacuole. Finally, it does not provide an explanation for how IFN-regulated restriction of intracellular pathogens occurs in humans. In humans, IRGM is the solitary IRG family member known to play a role in pathogen restriction, and there is no evidence that it plays a regulatory role in controlling a membrane attack complex. In fact, its ability to link pathogen restriction to pattern recognition, via Beclin-1 and ATG16L1, indicates that IRGM function may have diverged greatly across species (Chauhan et al., 2015).
We propose that an evolutionarily conserved function of IRG proteins is the ability to temper inflammation. For instance, Irgm1-deficient mice suffer from hyperinflammation (Maric-Biresev et al., 2016, Schmidt et al., 2017). In human cells, IRGM appears to play a central role in xenophagy, which we envision clears pathogens without attendant inflammasome activation (Chauhan et al., 2015). GBP proteins, in contrast, drive inflammasome activation, as the disruption of a vacuolar membrane in the absence of GBP function would be predicted to increase autophagic clearance. We hypothesize that after PVs are disrupted, GBPs intervene to present pathogen pattern molecules that cause inflammasome activation, overriding autophagic clearance of the damaged compartment and driving inflammation. The fact that GBP proteins can also recruit autophagy components, such as p62, may represent a strategy to prevent out-of-control intervention by GBP family members, allowing the inflammatory response to be dampened (Kim et al., 2011, Al-Zeer et al., 2013).
Signals that allow recognition of the pathogen-containing vacuole
The initiation of IFN-regulated clearance of pathogens provides another connection to autophagy proteins. GKS-driven restriction of T. gondii is dependent on a noncanonical autophagy process that marks the PV with ATG8 proteins, resulting in recruitment of the GKS protein Irga6 (Fig. 1) (Zhao et al., 2008, Khaminets et al., 2010). Similar results were observed during C. trachomatis infection of mouse embryo fibroblasts (Haldar et al., 2014). Consistent with the model that marking of PVs is a critical step in IFN-dependent restriction in the mouse, retargeting the LC3 conjugation system to alternative target membranes results in recruitment of GKS proteins as well as GBPs following IFN-γ treatment (Park et al., 2016). This process has much in common with LAP, although the downstream consequences may be dependent on the nature of the LC3/GABARAP tagging found on the PV. For instance, post-translational modifications of ATG8 proteins could confer a pathogen-specific response that spares sterile compartments and those harboring nonpathogens (Wilkinson et al., 2015, Choi et al., 2016). It is also possible that specific orthologs of ATG8 are involved in the recruitment of interferon-regulated GTPases. Although still a matter of debate, a recent study strongly suggests a unique role for GABARAPs (especially Gabarapl2/Gate-16) in the IFN-γ-dependent response mediated by interferon-regulated GTPases (Sasai et al., 2017). Targeting by the IFN response allows other markers to tag the PV, possibly indicating a barcoding strategy. In mouse cells, the recruitment of GKS IRGs results in downstream ubiquitination of both T. gondii and C. trachomatis vacuoles, accompanied by attachment of p62 to the ubiquitinated sites (Haldar et al., 2015, Lee et al., 2015). The p62 adapter appears to amplify this signal by recruiting Ub E3 ligases such as TRAF6 (Haldar et al., 2015).
It is noteworthy that in mouse cells, Ub modification can instruct pathogen restriction processes in two different directions. First, Ub chains on the pathogen-containing vacuole can target the microorganism for xenophagic clearance. Second, ubiquitination can trigger the recruitment of GBPs that have membranolytic and bacteriolytic activity and mobilize microbial ligands for presentation to inflammasome receptors (Haldar et al., 2015). Ubiquitination of the vacuole, therefore, establishes a dynamic tension between two strategies for pathogen clearance, with one releasing inflammatory cytokines, and the other suppressing inflammation. Control of this process could be driven by pathogen-specific factors, such as insertion of pathogen-derived protein complexes that mark the vacuolar membrane as foreign or which expose galectin-binding β(1,4)-linked galactosides (Feeley et al., 2017).
What swings the immune response toward inflammation in response to intravacuolar pathogens?
Distinguishing whether intracellular microbes are degraded in membrane compartments or in the cytosol has a profound impact on the level of inflammation in response to pathogen attack. After internalization, intracellular pathogens have strategies to avoid phagolysosomal degradation, proliferating in either the cytosol or within a membrane-bound compartment. The establishment of a replication niche is a highly complex process involving the injection of numerous effectors into the host cytoplasm, potentially setting up booby traps for the pathogen that mark it for host recognition (Casson et al., 2013). Only the most highly adapted pathogens can grow within vacuoles without being detected. Once recognized as “non self,” PVs are marked for autophagic removal by molecular tags (e.g. Ub, p62 & galectin-3) that remarkably overlap with those that mark “aberrant-self” compartments, such as damaged organelles (Anding and Baehrecke, 2017). These tags also recruit IFN-regulated GTPases that coordinate attack of PVs, potentially inducing the inflammasome via GBP intervention.
As a model for how these pathways are coordinated, inflammasome activation could provide a fail-safe mechanism that acts as a last resort when other cell-autonomous defenses fail at clearing an infection. In this scenario, phagolysosomal processing or autophagy-related processes can sequentially act on microbes upstream of inflammasome activation, with each path having calibrated consequences on inflammation. Several lines of evidence support the idea that the autophagy machinery blocks inflammasome activation in the response to intracellular microbes. Autophagy-related processes reduce exposure of microbial patterns to the host cytosol by intersecting with the phagolysosomal pathway to promote LAP and bacterial clearance (Martinez et al., 2015), and by mediating the repair of damaged membranes during the infection (Kreibich et al., 2015). Further, autophagy interferes with inflammasome induction by directly digesting the pathogen (Shi et al., 2012, Meunier et al., 2014) and degrading inflammasome components, which could further serve to control inflammasome activation (Shi et al., 2012).
The IFN response may intervene in this process to drive inflammatory clearance of infection by sensitizing the host cell to inflammasome activation. Most notably, IFN induction in the mouse is tightly connected to the ability of the cell to generate and present microbial molecules to inflammasomes (Casson et al., 2013, Case et al., 2013). IFN exposure is also known to regulate the expression of thousands of genes, perhaps leading to the post-translational modification (PTM) of proteins normally involved in autophagy thereby increasing their affinity for immunity-related GTPases. For example, PTM of ATG8 proteins (Wilkinson et al., 2015) may allow the recruitment of a specific subset of guanine nucleotide exchange factors (GEFs) that could activate and recruit immunity-related GTPases on PVs. Similarly, PTM of autophagy adaptors such as p62 (Pilli et al., 2012) could increase their affinity for GBPs while decreasing their interaction with conjugated LC3.
PTM of galectin also has the potential to modulate the interaction of PVs with Tripartite motif-containing (TRIM) proteins. TRIM proteins interact with autophagy regulatory proteins (e.g. Beclin-1, ULK1) that can recruit membranes to the vicinity of PVs (Kimura et al., 2016), and this could be modulated by PTM of galectin. Several TRIMs are upregulated by IFNs (Ozato et al., 2008) which could promote the conjugation of ATG8 proteins on PVs. The topology and the nature of the membrane on which ATG8 proteins are lipidated is likely to be an important determinant of the downstream outcome, especially considering that human IRGM is likely to have tropism for autophagosomes (Chauhan et al., 2015) and may not directly bind to PVs. Another strategy that could modulate these events is the Ub-like modifier ISG15, which controls the function of key immune response players through ISGylation. Interestingly, ISGylation of Beclin-1 negatively regulates canonical autophagy in response to type I IFN exposure (Xu et al., 2015). One hypothesis is that ISGylation of Beclin-1 inhibits Class III PI3K complexes involved in autophagy, but still allows the formation of PI3P on the PV through a mechanism resembling LAP (Martinez et al., 2015). This would allow immunity-related GTPases to be recruited to the PV while simultaneously blocking xenophagy.
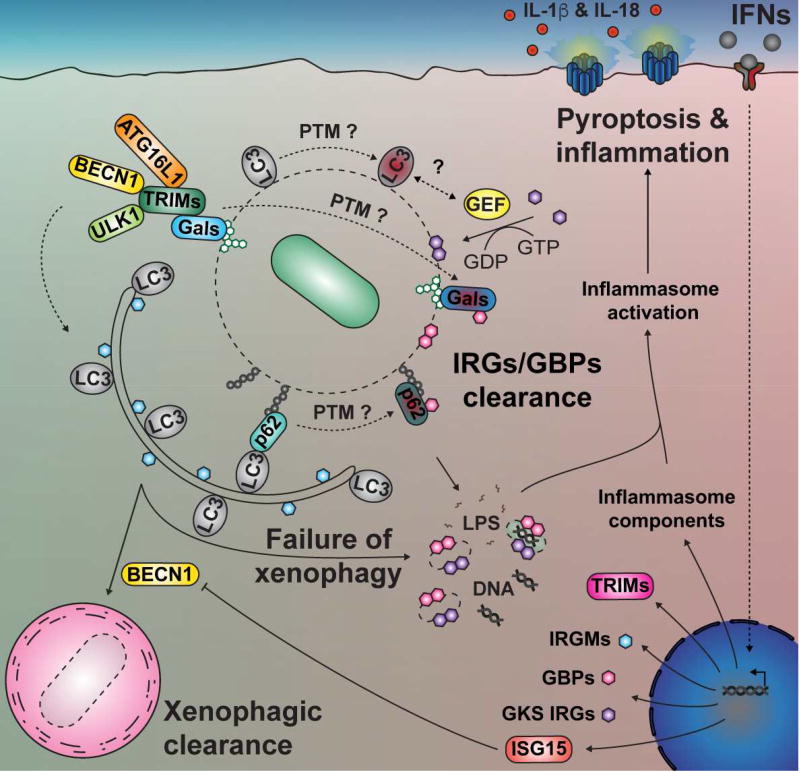
As summarized by Fig. 3, the host recognizes and decorates PVs with Ub proteins, members of the ATG8 (LC3/GABARAP) family, and autophagy adaptors such as p62. These markers are well-characterized coordinators of autophagic removal of organelles and microbes. It is now appreciated that they also have a broader function in innate immunity, allowing the recruitment of IFN-regulated GTPases, which participate in pathogen dissolution and drive inflammasome induction. Therefore, seemingly identical markers of pathogen attack can lead to multiple pathways for pathogen clearance. Future research should aim to identify microbial and host factors that push microbial clearance toward pathways that alter the dynamic tension between inhibition and stimulation of downstream inflammatory responses. We suspect that several host checkpoints will be identified that allow host cells to keep inflammation in check and remain intact as they eliminate microbes through xenophagy. In the absence of these checkpoints the trigger will be pulled, driving an inflammatory response with potentially extreme consequences for the cell.
FIGURE 3. Factors that predispose inflammasome induction.
Pathogen-containing vacuoles (PVs) are marked by Ub, galectins (Gals), autophagy adaptors (e.g. p62) and LC3/GABARAP proteins. These markers can trigger xenophagy, but also recruitment of IFN-regulated GTPases, activating inflammasomes. The IFN response sensitizes cells to the action of IRGs/GBPs and inflammasome induction by transcriptional up-regulation. Post-translational modifications (PTMs) of galectins, p62, LC3/GABARAP proteins and autophagy regulatory proteins have the potential to change their interacting partners and may participate (as indicated by question marks) in checkpoint mechanisms that distinguish a xenophagic pathway to pathways that induce cell death and inflammation. A speculative interaction between an unknown guanine nucleotide exchange factor (GEF) and LC3/GABARAP proteins is also indicated by a question mark. In addition, particular orthologs of ATG8 (e.g. Gabarapl2/Gate-16) may be more specifically involved in recruiting interferon-regulated GTPases to PVs (not illustrated here). GEF: Guanine nucleotide exchange factor; BECN1: Beclin-1; LC3: ATG8 orthologs (LC3/GABARAPs).
Acknowledgments
We thank Daniel A. Portnoy for the critical reading of this manuscript. G.M. is a postdoctoral scholar in the Portnoy laboratory, supported by a fellowship from Fonds de Recherche du Québec - Nature et technologies (FRQNT). R.R.I. is an investigator of the Howard Hughes Medical Institute (HHMI). This work was supported by National Institutes of Health grants 1P01 AI063302 (D.A.P.), 1R01 AI027655 (D.A.P.), R01AI110684 (R.R.I.), and RO1 AI113211 (R.R.I).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Al-Zeer MA, Al-Younes HM, Lauster D, Abu Lubad M, Meyer TF. Autophagy restricts Chlamydia trachomatis growth in human macrophages via IFNG-inducible guanylate binding proteins. Autophagy. 2013;9:50–62. doi: 10.4161/auto.22482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anding AL, Baehrecke EH. Cleaning House: Selective Autophagy of Organelles. Dev Cell. 2017;41:10–22. doi: 10.1016/j.devcel.2017.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekpen C, Hunn JP, Rohde C, Parvanova I, Guethlein L, Dunn DM, Glowalla E, Leptin M, Howard JC. The interferon-inducible p47 (IRG) GTPases in vertebrates: loss of the cell autonomous resistance mechanism in the human lineage. Genome Biol. 2005;6:R92. doi: 10.1186/gb-2005-6-11-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestebroer J, V’Kovski P, Mauthe M, Reggiori F. Hidden behind autophagy: the unconventional roles of ATG proteins. Traffic. 2013;14:1029–41. doi: 10.1111/tra.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm U, Guethlein L, Klamp T, Ozbek K, Schaub A, Futterer A, Pfeffer K, Howard JC. Two families of GTPases dominate the complex cellular response to IFN-gamma. J Immunol. 1998;161:6715–23. [PubMed] [Google Scholar]
- Boyle KB, Randow F. Rubicon swaps autophagy for LAP. Nat Cell Biol. 2015;17:843–5. doi: 10.1038/ncb3197. [DOI] [PubMed] [Google Scholar]
- CAmbier CJ, Falkow S, Ramakrishnan L. Host evasion and exploitation schemes of Mycobacterium tuberculosis. Cell. 2014;159:1497–509. doi: 10.1016/j.cell.2014.11.024. [DOI] [PubMed] [Google Scholar]
- Case CL, Kohler LJ, Lima JB, Strowig T, De Zoete MR, Flavell RA, Zamboni DS, Roy CR. Caspase-11 stimulates rapid flagellin-independent pyroptosis in response to Legionella pneumophila. Proc Natl Acad Sci U S A. 2013;110:1851–6. doi: 10.1073/pnas.1211521110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson CN, Copenhaver AM, Zwack EE, Nguyen HT, Strowig T, Javdan B, Bradley WP, Fung TC, Flavell RA, Brodsky IE, Shin S. Caspase-11 activation in response to bacterial secretion systems that access the host cytosol. PLoS Pathog. 2013;9:e1003400. doi: 10.1371/journal.ppat.1003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cemma M, Grinstein S, Brumell JH. Autophagy proteins are not universally required for phagosome maturation. Autophagy. 2016;12:1440–6. doi: 10.1080/15548627.2016.1191724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan S, Kumar S, Jain A, Ponpuak M, Mudd MH, Kimura T, Choi SW, Peters R, Mandell M, Bruun JA, Johansen T, Deretic V. TRIMs and Galectins Globally Cooperate and TRIM16 and Galectin-3 Co-direct Autophagy in Endomembrane Damage Homeostasis. Dev Cell. 2016;39:13–27. doi: 10.1016/j.devcel.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan S, Mandell MA, Deretic V. IRGM governs the core autophagy machinery to conduct antimicrobial defense. Mol Cell. 2015;58:507–21. doi: 10.1016/j.molcel.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Sun L, Chen ZJ. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat Immunol. 2016;17:1142–9. doi: 10.1038/ni.3558. [DOI] [PubMed] [Google Scholar]
- Choi J, Biering SB, Hwang S. Quo vadis? Interferon-inducible GTPases go to their target membranes via the LC3-conjugation system of autophagy. Small GTPases. 2016:1–9. doi: 10.1080/21541248.2016.1213090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Park S, Biering SB, Selleck E, Liu CY, Zhang X, Fujita N, Saitoh T, Akira S, Yoshimori T, Sibley LD, Hwang S, Virgin HW. The parasitophorous vacuole membrane of Toxoplasma gondii is targeted for disruption by ubiquitin-like conjugation systems of autophagy. Immunity. 2014;40:924–35. doi: 10.1016/j.immuni.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill MJ, Deeks SG, Margolis DM, Siliciano RF, Swanstrom R. HIV reservoirs: what, where and how to target them. Nat Rev Microbiol. 2016;14:55–60. doi: 10.1038/nrmicro.2015.5. [DOI] [PubMed] [Google Scholar]
- Dooley HC, Razi M, Polson HE, Girardin SE, Wilson MI, Tooze SA. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol Cell. 2014;55:238–52. doi: 10.1016/j.molcel.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeley EM, Pilla-Moffett DM, Zwack EE, Piro AS, Finethy R, Kolb JP, Martinez J, Brodsky IE, Coers J. Galectin-3 directs antimicrobial guanylate binding proteins to vacuoles furnished with bacterial secretion systems. Proc Natl Acad Sci U S A. 2017;114:E1698–e1706. doi: 10.1073/pnas.1615771114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco LH, Nair VR, Scharn CR, Xavier RJ, Torrealba JR, Shiloh MU, Levine B. The Ubiquitin Ligase Smurf1 Functions in Selective Autophagy of Mycobacterium tuberculosis and Anti-tuberculous Host Defense. Cell Host Microbe. 2017;21:59–72. doi: 10.1016/j.chom.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19:2092–100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Morita E, Itoh T, Tanaka A, Nakaoka M, Osada Y, Umemoto T, Saitoh T, Nakatogawa H, Kobayashi S, Haraguchi T, Guan JL, Iwai K, Tokunaga F, Saito K, Ishibashi K, Akira S, Fukuda M, Noda T, Yoshimori T. Recruitment of the autophagic machinery to endosomes during infection is mediated by ubiquitin. J Cell Biol. 2013;203:115–28. doi: 10.1083/jcb.201304188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Baehrecke EH, Ballabio A, Boya P, Bravo-San Pedro JM, Cecconi F, Choi AM, Chu CT, Codogno P, Colombo MI, Cuervo AM, Debnath J, Deretic V, Dikic I, Eskelinen EL, Fimia GM, Fulda S, Gewirtz DA, Green DR, Hansen M, Harper JW, Jaattela M, Johansen T, Juhasz G, Kimmelman AC, Kraft C, Ktistakis NT, Kumar S, Levine B, Lopez-Otin C, Madeo F, Martens S, Martinez J, Melendez A, Mizushima N, Munz C, Murphy LO, Penninger JM, Piacentini M, Reggiori F, Rubinsztein DC, Ryan KM, Santambrogio L, Scorrano L, Simon AK, Simon HU, Simonsen A, Tavernarakis N, Tooze SA, Yoshimori T, Yuan J, Yue Z, Zhong Q, Kroemer G. Molecular definitions of autophagy and related processes. EMBO J. 2017 doi: 10.15252/embj.201796697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Uthaiah R, Howard J, Herrmann C, Wolf E. Crystal structure of IIGP1: a paradigm for interferon-inducible p47 resistance GTPases. Mol Cell. 2004;15:727–39. doi: 10.1016/j.molcel.2004.07.017. [DOI] [PubMed] [Google Scholar]
- Haldar AK, Foltz C, Finethy R, Piro AS, Feeley EM, Pilla-Moffett DM, Komatsu M, Frickel EM, Coers J. Ubiquitin systems mark pathogen-containing vacuoles as targets for host defense by guanylate binding proteins. Proc Natl Acad Sci U S A. 2015;112:E5628–37. doi: 10.1073/pnas.1515966112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar AK, Piro AS, Finethy R, Espenschied ST, Brown HE, Giebel AM, Frickel EM, Nelson DE, Coers J. Chlamydia trachomatis Is Resistant to Inclusion Ubiquitination and Associated Host Defense in Gamma Interferon-Primed Human Epithelial Cells. MBio. 2016:7. doi: 10.1128/mBio.01417-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar AK, Piro AS, Pilla DM, Yamamoto M, Coers J. The E2-like conjugation enzyme Atg3 promotes binding of IRG and Gbp proteins to Chlamydia-and Toxoplasma-containing vacuoles and host resistance. PLoS One. 2014;9:e86684. doi: 10.1371/journal.pone.0086684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar AK, Saka HA, Piro AS, Dunn JD, Henry SC, Taylor GA, Frickel EM, Valdivia RH, Coers J. IRG and GBP host resistance factors target aberrant, “non-self” vacuoles characterized by the missing of “self” IRGM proteins. PLoS Pathog. 2013;9:e1003414. doi: 10.1371/journal.ppat.1003414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Brumell JH. Bacteria-autophagy interplay: a battle for survival. Nat Rev Microbiol. 2014;12:101–14. doi: 10.1038/nrmicro3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Canadien V, Lam GY, Steinberg BE, Dinauer MC, Magalhaes MA, Glogauer M, Grinstein S, Brumell JH. Activation of antibacterial autophagy by NADPH oxidases. Proc Natl Acad Sci U S A. 2009;106:6226–31. doi: 10.1073/pnas.0811045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubber A, Kubori T, Coban C, Matsuzawa T, Ogawa M, Kawabata T, Yoshimori T, Nagai H. Bacterial secretion system skews the fate of Legionella-containing vacuoles towards LC3-associated phagocytosis. Sci Rep. 2017;7:44795. doi: 10.1038/srep44795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huett A, Heath RJ, Begun J, Sassi SO, Baxt LA, Vyas JM, Goldberg MB, Xavier RJ. The LRR and RING domain protein LRSAM1 is an E3 ligase crucial for ubiquitin-dependent autophagy of intracellular Salmonella Typhimurium. Cell Host Microbe. 2012;12:778–90. doi: 10.1016/j.chom.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston AC, Piro A, Clough B, Siew M, Virreira Winter S, Coers J, Frickel EM. Human GBP1 does not localize to pathogen vacuoles but restricts Toxoplasma gondii. Cell Microbiol. 2016;18:1056–64. doi: 10.1111/cmi.12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama S, Omori H, Saitoh T, Sone T, Guan JL, Akira S, Imamoto F, Noda T, Yoshimori T. The LC3 recruitment mechanism is separate from Atg9L1-dependent membrane formation in the autophagic response against Salmonella. Mol Biol Cell. 2011;22:2290–300. doi: 10.1091/mbc.E10-11-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaminets A, Hunn JP, Konen-Waisman S, Zhao YO, Preukschat D, Coers J, Boyle JP, Ong YC, Boothroyd JC, Reichmann G, Howard JC. Coordinated loading of IRG resistance GTPases on to the Toxoplasma gondii parasitophorous vacuole. Cell Microbiol. 2010;12:939–61. doi: 10.1111/j.1462-5822.2010.01443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BH, Chee JD, Bradfield CJ, Park ES, Kumar P, Macmicking JD. Interferon-induced guanylate-binding proteins in inflammasome activation and host defense. Nat Immunol. 2016;17:481–9. doi: 10.1038/ni.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BH, Shenoy AR, Kumar P, Das R, Tiwari S, Macmicking JD. A family of IFN-gamma-inducible 65-kD GTPases protects against bacterial infection. Science. 2011;332:717–21. doi: 10.1126/science.1201711. [DOI] [PubMed] [Google Scholar]
- Kimura T, Mandell M, Deretic V. Precision autophagy directed by receptor regulators - emerging examples within the TRIM family. J Cell Sci. 2016;129:881–91. doi: 10.1242/jcs.163758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravets E, Degrandi D, Ma Q, Peulen TO, Klumpers V, Felekyan S, Kuhnemuth R, Weidtkamp-Peters S, Seidel CA, Pfeffer K. Guanylate binding proteins directly attack Toxoplasma gondii via supramolecular complexes. Elife. 2016:5. doi: 10.7554/eLife.11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreibich S, Emmenlauer M, Fredlund J, Ramo P, Munz C, Dehio C, Enninga J, Hardt WD. Autophagy Proteins Promote Repair of Endosomal Membranes Damaged by the Salmonella Type Three Secretion System 1. Cell Host Microbe. 2015;18:527–37. doi: 10.1016/j.chom.2015.10.015. [DOI] [PubMed] [Google Scholar]
- Lam GY, Cemma M, Muise AM, Higgins DE, Brumell JH. Host and bacterial factors that regulate LC3 recruitment to Listeria monocytogenes during the early stages of macrophage infection. Autophagy. 2013;9:985–95. doi: 10.4161/auto.24406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassen KG, Xavier RJ. Genetic control of autophagy underlies pathogenesis of inflammatory bowel disease. Mucosal Immunol. 2017;10:589–597. doi: 10.1038/mi.2017.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Sasai M, Ma JS, Sakaguchi N, Ohshima J, Bando H, Saitoh T, Akira S, Yamamoto M. p62 Plays a Specific Role in Interferon-gamma-Induced Presentation of a Toxoplasma Vacuolar Antigen. Cell Rep. 2015;13:223–33. doi: 10.1016/j.celrep.2015.09.005. [DOI] [PubMed] [Google Scholar]
- Ma J, Becker C, Lowell CA, Underhill DM. Dectin-1-triggered recruitment of light chain 3 protein to phagosomes facilitates major histocompatibility complex class II presentation of fungal-derived antigens. J Biol Chem. 2012;287:34149–56. doi: 10.1074/jbc.M112.382812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macmicking JD, Taylor GA, Mckinney JD. Immune control of tuberculosis by IFN-gamma-inducible LRG-47. Science. 2003;302:654–9. doi: 10.1126/science.1088063. [DOI] [PubMed] [Google Scholar]
- Man SM, Karki R, Malireddi RK, Neale G, Vogel P, Yamamoto M, Lamkanfi M, Kanneganti TD. The transcription factor IRF1 and guanylate-binding proteins target activation of the AIM2 inflammasome by Francisella infection. Nat Immunol. 2015;16:467–75. doi: 10.1038/ni.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzanillo PS, Ayres JS, Watson RO, Collins AC, Souza G, Rae CS, Schneider DS, Nakamura K, Shiloh MU, Cox JS. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature. 2013;501:512–6. doi: 10.1038/nature12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maric-Biresev J, Hunn JP, Krut O, Helms JB, Martens S, Howard JC. Loss of the interferon-gamma-inducible regulatory immunity-related GTPase (IRG), Irgm1, causes activation of effector IRG proteins on lysosomes, damaging lysosomal function and predicting the dramatic susceptibility of Irgm1-deficient mice to infection. BMC Biol. 2016;14:33. doi: 10.1186/s12915-016-0255-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens S, Parvanova I, Zerrahn J, Griffiths G, Schell G, Reichmann G, Howard JC. Disruption of Toxoplasma gondii parasitophorous vacuoles by the mouse p47-resistance GTPases. PLoS Pathog. 2005;1:e24. doi: 10.1371/journal.ppat.0010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Malireddi RK, Lu Q, Cunha LD, Pelletier S, Gingras S, Orchard R, Guan JL, Tan H, Peng J, Kanneganti TD, Virgin HW, Green DR. Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat Cell Biol. 2015;17:893–906. doi: 10.1038/ncb3192. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Mcnab F, Mayer-Barber K, Sher A, Wack A, O’Garra A. Type I interferons in infectious disease. Nat Rev Immunol. 2015;15:87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier E, Broz P. Interferon-induced guanylate-binding proteins promote cytosolic lipopolysaccharide detection by caspase-11. DNA Cell Biol. 2015;34:1–5. doi: 10.1089/dna.2014.2701. [DOI] [PubMed] [Google Scholar]
- Meunier E, Dick MS, Dreier RF, Schurmann N, Kenzelmann Broz D, Warming S, Roose-Girma M, Bumann D, Kayagaki N, Takeda K, Yamamoto M, Broz P. Caspase-11 activation requires lysis of pathogen-containing vacuoles by IFN-induced GTPases. Nature. 2014;509:366–70. doi: 10.1038/nature13157. [DOI] [PubMed] [Google Scholar]
- Meunier E, Wallet P, Dreier RF, Costanzo S, Anton L, Ruhl S, Dussurgey S, Dick MS, Kistner A, Rigard M, Degrandi D, Pfeffer K, Yamamoto M, Henry T, Broz P. Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida. Nat Immunol. 2015;16:476–84. doi: 10.1038/ni.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TN, Padman BS, Usher J, Oorschot V, Ramm G, Lazarou M. Atg8 family LC3/GABARAP proteins are crucial for autophagosome-lysosome fusion but not autophagosome formation during PINK1/Parkin mitophagy and starvation. J Cell Biol. 2016;215:857–874. doi: 10.1083/jcb.201607039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noad J, Von Der Malsburg A, Pathe C, Michel MA, Komander D, Randow F. LUBAC-synthesized linear ubiquitin chains restrict cytosol-invading bacteria by activating autophagy and NF-kappaB. Nat Microbiol. 2017;2:17063. doi: 10.1038/nmicrobiol.2017.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozato K, Shin DM, Chang TH, Morse HC., 3rd TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol. 2008;8:849–60. doi: 10.1038/nri2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Choi J, Biering SB, Dominici E, Williams LE, Hwang S. Targeting by AutophaGy proteins (TAG): Targeting of IFNG-inducible GTPases to membranes by the LC3 conjugation system of autophagy. Autophagy. 2016;12:1153–67. doi: 10.1080/15548627.2016.1178447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkes M, Barrett JC, Prescott NJ, Tremelling M, Anderson CA, Fisher SA, Roberts RG, Nimmo ER, Cummings FR, Soars D, Drummond H, Lees CW, Khawaja SA, Bagnall R, Burke DA, Todhunter CE, Ahmad T, Onnie CM, Mcardle W, Strachan D, Bethel G, Bryan C, Lewis CM, Deloukas P, Forbes A, Sanderson J, Jewell DP, Satsangi J, Mansfield JC, Wellcome Trust Case Control C. Cardon L, Mathew CG. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat Genet. 2007;39:830–2. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilla-Moffett D, Barber MF, Taylor GA, Coers J. Interferon-Inducible GTPases in Host Resistance, Inflammation and Disease. J Mol Biol. 2016;428:3495–513. doi: 10.1016/j.jmb.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilla DM, Hagar JA, Haldar AK, Mason AK, Degrandi D, Pfeffer K, Ernst RK, Yamamoto M, Miao EA, Coers J. Guanylate binding proteins promote caspase-11-dependent pyroptosis in response to cytoplasmic LPS. Proc Natl Acad Sci U S A. 2014;111:6046–51. doi: 10.1073/pnas.1321700111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilli M, Arko-Mensah J, Ponpuak M, Roberts E, Master S, Mandell MA, Dupont N, Ornatowski W, Jiang S, Bradfute SB, Bruun JA, Hansen TE, Johansen T, Deretic V. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity. 2012;37:223–34. doi: 10.1016/j.immuni.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randow F, Youle RJ. Self and nonself: how autophagy targets mitochondria and bacteria. Cell Host Microbe. 2014;15:403–11. doi: 10.1016/j.chom.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanov J, Walczak M, Ibiricu I, Schuchner S, Ogris E, Kraft C, Martens S. Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. Embo j. 2012;31:4304–17. doi: 10.1038/emboj.2012.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Akira S. Regulation of inflammasomes by autophagy. J Allergy Clin Immunol. 2016;138:28–36. doi: 10.1016/j.jaci.2016.05.009. [DOI] [PubMed] [Google Scholar]
- Salem M, Ammitzboell M, Nys K, Seidelin JB, Nielsen OH. ATG16L1: A multifunctional susceptibility factor in Crohn disease. Autophagy. 2015;11:585–94. doi: 10.1080/15548627.2015.1017187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai M, Sakaguchi N, Ma JS, Nakamura S, Kawabata T, Bando H, Lee Y, Saitoh T, Akira S, Iwasaki A, Standley DM, Yoshimori T, Yamamoto M. Essential role for GABARAP autophagy proteins in interferon-inducible GTPase-mediated host defense. Nat Immunol. 2017 doi: 10.1038/ni.3767. [DOI] [PubMed] [Google Scholar]
- Schmidt EA, Fee BE, Henry SC, Nichols AG, Shinohara ML, Rathmell JC, Maciver NJ, Coers J, Ilkayeva OR, Koves TR, Taylor GA. Metabolic Alterations Contribute to Enhanced Inflammatory Cytokine Production in Irgm1-deficient Macrophages. J Biol Chem. 2017;292:4651–4662. doi: 10.1074/jbc.M116.770735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol. 2004;75:163–89. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- Shi CS, Shenderov K, Huang NN, Kabat J, Abu-Asab M, Fitzgerald KA, Sher A, Kehrl JH. Activation of autophagy by inflammatory signals limits IL-1beta production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol. 2012;13:255–63. doi: 10.1038/ni.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata K, Matsunaga K, Sakane A, Sasaki T, Noda T, Yoshimori T. Rubicon and PLEKHM1 negatively regulate the endocytic/autophagic pathway via a novel Rab7-binding domain. Mol Biol Cell. 2010;21:4162–72. doi: 10.1091/mbc.E10-06-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurston TL, Boyle KB, Allen M, Ravenhill BJ, Karpiyevich M, Bloor S, Kaul A, Noad J, Foeglein A, Matthews SA, Komander D, Bycroft M, Randow F. Recruitment of TBK1 to cytosol-invading Salmonella induces WIPI2-dependent antibacterial autophagy. Embo j. 2016;35:1779–92. doi: 10.15252/embj.201694491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurston TL, Wandel MP, Von Muhlinen N, Foeglein A, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482:414–8. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG, Yuan L, Soares F, Chea E, Le Bourhis L, Boneca IG, Allaoui A, Jones NL, Nunez G, Girardin SE, Philpott DJ. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- Tumbarello DA, Manna PT, Allen M, Bycroft M, Arden SD, Kendrick-Jones J, Buss F. The Autophagy Receptor TAX1BP1 and the Molecular Motor Myosin VI Are Required for Clearance of Salmonella Typhimurium by Autophagy. PLoS Pathog. 2015;11:e1005174. doi: 10.1371/journal.ppat.1005174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wijk SJL, Fricke F, Herhaus L, Gupta J, Hotte K, Pampaloni F, Grumati P, Kaulich M, Sou YS, Komatsu M, Greten FR, Fulda S, Heilemann M, Dikic I. Linear ubiquitination of cytosolic Salmonella Typhimurium activates NF-kappaB and restricts bacterial proliferation. Nat Microbiol. 2017;2:17066. doi: 10.1038/nmicrobiol.2017.66. [DOI] [PubMed] [Google Scholar]
- Verlhac P, Gregoire IP, Azocar O, Petkova DS, Baguet J, Viret C, Faure M. Autophagy receptor NDP52 regulates pathogen-containing autophagosome maturation. Cell Host Microbe. 2015;17:515–25. doi: 10.1016/j.chom.2015.02.008. [DOI] [PubMed] [Google Scholar]
- Wack A, Terczynska-Dyla E, Hartmann R. Guarding the frontiers: the biology of type III interferons. Nat Immunol. 2015;16:802–9. doi: 10.1038/ni.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson RO, Bell SL, Macduff DA, Kimmey JM, Diner EJ, Olivas J, Vance RE, Stallings CL, Virgin HW, Cox JS. The Cytosolic Sensor cGAS Detects Mycobacterium tuberculosis DNA to Induce Type I Interferons and Activate Autophagy. Cell Host Microbe. 2015;17:811–9. doi: 10.1016/j.chom.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–88. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H, Shvets E, Elazar Z. Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem. 2011;80:125–56. doi: 10.1146/annurev-biochem-052709-094552. [DOI] [PubMed] [Google Scholar]
- Wilkinson DS, Jariwala JS, Anderson E, Mitra K, Meisenhelder J, Chang JT, Ideker T, Hunter T, Nizet V, Dillin A, Hansen M. Phosphorylation of LC3 by the Hippo kinases STK3/STK4 is essential for autophagy. Mol Cell. 2015;57:55–68. doi: 10.1016/j.molcel.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, Roth JR, Baumler AJ. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–9. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Chen ZJ. Innate immune sensing and signaling of cytosolic nucleic acids. Annu Rev Immunol. 2014;32:461–88. doi: 10.1146/annurev-immunol-032713-120156. [DOI] [PubMed] [Google Scholar]
- Xu D, Zhang T, Xiao J, Zhu K, Wei R, Wu Z, Meng H, Li Y, Yuan J. Modification of BECN1 by ISG15 plays a crucial role in autophagy regulation by type I IFN/interferon. Autophagy. 2015;11:617–28. doi: 10.1080/15548627.2015.1023982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Okuyama M, Ma JS, Kimura T, Kamiyama N, Saiga H, Ohshima J, Sasai M, Kayama H, Okamoto T, Huang DC, Soldati-Favre D, Horie K, Takeda J, Takeda K. A cluster of interferon-gamma-inducible p65 GTPases plays a critical role in host defense against Toxoplasma gondii. Immunity. 2012;37:302–13. doi: 10.1016/j.immuni.2012.06.009. [DOI] [PubMed] [Google Scholar]
- Yin Z, Pascual C, Klionsky DJ. Autophagy: machinery and regulation. Microb Cell. 2016;3:588–596. doi: 10.15698/mic2016.12.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Fux B, Goodwin M, Dunay IR, Strong D, Miller BC, Cadwell K, Delgado MA, Ponpuak M, Green KG, Schmidt RE, Mizushima N, Deretic V, Sibley LD, Virgin HW. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe. 2008;4:458–69. doi: 10.1016/j.chom.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]