Abstract
Sensory neurons expressing members of the seven-transmembrane V1r receptor superfamily allow mice to perceive pheromones. These receptors, which exhibit no sequence homology to any known protein except a weak similarity to taste receptors, have only been found in mammals. In the mouse, the V1r repertoire contains >150 members, which are expressed by neurons of the vomeronasal organ, a structure present exclusively in some tetrapod species. Here, we report the existence of a single V1r gene in multiple species of a non-terrestrial, vomeronasal organ-lacking taxon, the teleosts. In zebrafish, this V1r gene is expressed in chemosensory neurons of the olfactory rosette with a punctate distribution, strongly suggesting a role in chemodetection. This unique receptor gene exhibits a remarkably high degree of sequence variability between fish species. It likely corresponds to the original V1r present in the common ancestor of vertebrates, which led to the large and very diverse expansion of vertebrate pheromone receptor repertoires, and suggests the presence of V1rs in multiple nonmammalian phyla.
Keywords: olfaction, sensory systems, vomeronasal
Most living species have developed chemosensory tools, allowing them to perceive the outside world. Surrounding chemical stimuli may be very diverse in their composition and carry different types of information. Thus, for example, chemoreception often allows food localization and its evaluation, danger identification, or gender discrimination. In many chordates, this latter type of information exchange, which involves pheromone perception and is necessary for the survival of the species, is mediated by the olfactory system.
Olfactory sensory neurons express different types of chemoreceptors in vertebrates. First, odorant receptors (1), which are members of the rhodopsin G protein-coupled receptor family and number >1,000 in rodents, are found in most species. Second, V2r receptors (2–4), whose numerous members are closely related to calcium-sensing and glutamate receptors, are also expressed by olfactory sensory neurons in a wide range of vertebrates, including fish (5, 6). These receptors represent chemodetectors, at least in teleosts, where one of their members has been shown to mediate the perception of basic amino acids (5), and are thought to possibly be involved in pheromone detection in mammals (although no direct evidence supports such a role). Third, V1r receptors (7), which are unrelated in sequence to the two other chemoreceptor families, have been shown to mediate pheromone perception in the mouse (8, 9). These seven-transmembrane receptors are expressed by sensory neurons located in a peculiar structure, the vomeronasal organ, which evolved in the nose of some tetrapods, and that is physically separated from the main olfactory system (10). The only complete V1r repertoires described to date pertain to two macrosmic mammals, the mouse and the rat, and contain ≈100–150 functional receptors, which are further classified into 14 very distinct families (11–13).
To date, despite the first identification of V1rs already 10 years ago, and their identification in multiple species (7, 14–19), the search for V1r receptors outside mammalian species has resulted in failure. Taking advantage of the availability of the sequence of some almost completed teleost genomes, we investigated the potential existence of V1r genes in species of this vomeronasal organ-lacking aquatic group. We identified a V1r-like gene, very surprisingly single and highly divergent between species, in the genomes of medaka [Oryzias latipes (Ol)], zebrafish [Danio rerio (Dr)], Danio malabaricus (Dm), and of two pufferfish species, fugu [Takifugu rubripes (Tr)], and tetraodon [Tetraodon nigroviridis (Tn)]. We found the zebrafish V1r-like gene exclusively expressed by olfactory sensory neurons. Based on its sequence and expression profile, this receptor very likely corresponds to the teleost version of the prototypic V1r chemoreceptor.
Materials and Methods
Identification of Fish V1r-Like Genes. Dm, Dr, Tr, and Tn V1r-like genes were sequenced to validate in silico identifications. Sequences, including the two T2r-like sequences, were deposited to GenBank with the following accession numbers: AY764271, AC1938737, AY279523, AY279524, AY764272, and AY764273. V1r-like amplicons from Cyprinus carpio(Cc) (koï variety) (Cc koï), Cc, Epalzeorhynchos erythrurus (Ee), Danio albonileatus (Da), Danio frankei (Df), and Botia macracanthus (Bm) genomes were obtained by using the following primers: forward, AAA GGC GTC TCC TTC CTG CTG CAG GCT GGT CTG; reverse, CGC TTT CAC CTT CCT GTT GGA GGA GAT GAT. The corresponding GenBank accession numbers are AY900114–AY900119.
Radiation Hybrid Mapping. Three Dr V1r-like-specific pairs of primers were independently used to screen 93 zebrafish/mouse hybrid cell DNAs from the radiation hybrid panel LN54 (20). The results of the corresponding PCRs were submitted to http://mgchd1.nichd.nih.gov:8000/zfrh/beta.cgi to identify the linked markers.
RT-PCR. Twenty rosettes were pooled for each RNA extraction. cDNA was generated by using standard protocols with an anchored oligo(dT) reverse primer [(T)23-VN]. PCR amplifications were performed by using the following primers: Dr β-actin (forward, CCC CAT TGA GCA CGG TAT T; reverse, AGC GGT TCC CAT CTC CTG), Dr OR1 (forward, CCC TCT ACG GTA CAC GAC TAT C; reverse, CAA TCA TTA TGC GGA CTT CAG), Dr OMP (forward, GAG GCC GAC GCA CAG GAG T; reverse, AAG CTA AAA ACG CCC AAG ACC ATC), and Dr V1r-like (forward, CGG CAC CGT CCC ACC ATT CAC; reverse, CTC CGC TTG CCG CTC CTG CTC TG). The following conditions were used: 5 min at 95°C, followed by 34 cycles of 1 min at 95°C, 1 min at 63°C, and 2 min at 72°C, and a final extension of 10 min at 72°C.
In Situ Hybridizations. Templates for the probes were amplified from Dr genomic DNA with the following primers: for Dr V1r-like (forward, TAT GGA CCT GTG TGT CAC; reverse, TCA TGG AAG TCC ACA TGG CAG AAG), for Dr V2r1 (forward, CCC TAA GGA AGT AGA GTT TCT G; reverse, TAT TGC CGC CAA TAG TCC AAT G), and for Dr OR1 (forward, CCC TCT ACG GTA CAC GAC TAT C; reverse, CAA TCA TTA TGC GGA CTT CAG). Digoxigenin (DIG) probes were synthesized according to the DIG RNA labeling kit supplier protocol (Roche Molecular Biochemicals). Sections were fixed in 4% paraformaldehyde for 20 min at 4°C. Hybridizations were performed overnight at 65°C by using standard protocols. Anti-DIG primary antibody coupled to alkaline phosphatase (Roche Molecular Biochemicals) and FastRed (DAKO) were used for signal detection.
Phylogenetic Trees. Amino acid sequences (and the corresponding nucleotides) were aligned from the first to the last transmembrane segments. Sequences were then aligned with all mouse V1rs with clustalx (21), followed by manual arrangement using the bioedit 6.0.5 sequence alignment software. Phylogenetic trees were generated with both DNA and amino acid alignments. Amino acid-based trees were obtained with the neighbor-joining methods (22). DNA trees were generated by using maximum likelihood (ML) methods (23) using paup* 4.0B10 (24). The modeltest program (v3.0b4) (25) was used to identify by hierachical LRT the best model for the ML analyses. The retained best-fit model was general time-reversible with a proportion of invariant sites (I = 0.0159) and a γ distribution shape parameter (G = 2.6532) calculated from the data set. Supports for branches in all trees were tested by bootstrap analyses of 1,000 replicates. The unrooted tree, including members of the human and zebrafish T2R receptor families and the mouse and fish V1Rs (Fig. 4, which is published as supporting information on the PNAS web site), was obtained from an amino acid alignment and generated by using the Fitch–Margoliash method, version 3.6a.2 (bioedit 6.0.5).
dN/dS Ratios. Phylogenetic relationships between the eight fish V1rs were reconstructed in paup* 4.0B10 (24) by using maximum likelihood methods as described above. dN/dS ratios (ω) were then estimated by using codon-based substitution site-specific models implemented in paml 3.14 (26). The following models were tested: M0, one ratio model, constant ω; M1a, nearly neutral model; M2a positive selection model allowing ω > 1; M3 discrete model, discrete classes of sites with different ω; M7, β-model, neutral with ω constrained between 0 and 1; M8 β-plus ω model, selection model adding a class of sites having ω > 1. Likelihood ratio tests were used to statistically determine the best model by comparing twice the difference of the loglikelihood values obtained for each model to the χ2 distribution with appropriate degrees of freedom. For the region-defined dN/dS ratio analyses, the k-estimator 6.0 program was used.
Results
Identification of a Single V1r-Like Sequence in Fish. Our strategy for zebrafish, medaka, and pufferfish V1r repertoires identification was initially based on homology searches of genomic and EST databases by using the tblastn and blastn algorithms. Exofish, Sanger, Human Genome Sequencing Center at Baylor College of Medicine, University of California Santa Cruz, Joint Genome Institute, National Center for Biotechnology Information, Ensembl, Mebase, National BioResource Project, Medical Research Council-Rosalind Franklin Centre for Genomics Research, and The Institute for Genomic Research databases were searched. One member of each of the 12 mouse families (11) and 14 known non-rodent mammalian V1rs were used as queries. As with many seven-transmembrane receptors in vertebrates, V1r genes have intronless coding sequences that facilitate their identification from genomic data. Sequences identified with an expected value <10–2 and with a potential coding sequence of >300 base pairs were retained, and were used as novel queries. These very relaxed criteria for inclusion of a sequence were motivated by our desire to identify not only fish V1r-like genes even distantly related to mammalian V1rs, but also potential V1r pseudogenes, because this latter category of nonfunctional sequences represents a major part of mammalian V1r repertoires (11, 27, 28).
Complete coding sequences were either directly extracted from the databases (T. rubripes), corrected after direct extraction (D. rerio, see Materials and Methods), assembled by using various trace sequences (O. latipes), or completed by reverse PCR amplification of the lacking segments using genomic DNA as template (D. malabaricus and T. nigroviridis). Identified sequences were then directly amplified from genomic DNA from the fish species and sequenced. A probable sequencing error leading to a frameshift was found at nucleotide position 793 (from the ATG) in the zebrafish sequence from the Ensembl database. Our own sequencing of the DrV1r-like coding sequence corrected the frameshift. Because the position of the zebrafish V1r-like gene was not assigned in the genomic databases, we determined it by radiation hybrid mapping using zebrafish/mouse somatic cell hybrids (ref. 20; see Materials and Methods), to be linked to EST fa27e09 on chromosome 22.
We identified a single sequence with significant homology to mammalian V1rs in each of the five fish species, alignment of which strongly suggests a direct orthologous relation between the five genes (Fig. 1). Surprisingly, unlike what is observed in all tested species to date, no V1r-like pseudogene was identified in any of the fish species.
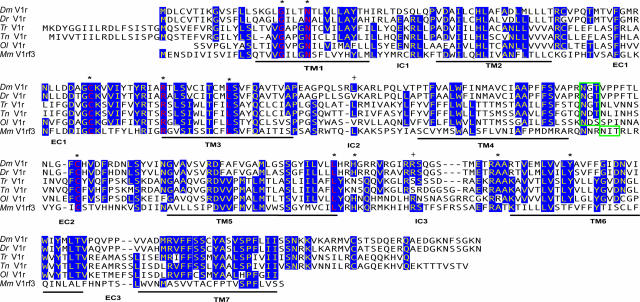
Fig. 1.
Fish V1r receptors. Alignment of the deduced amino acid of the zebrafish (Dr), giant danio (Dm), medaka (Ol), tetraodon (Tn), and fugu (Tr) V1r-like genes and the mouse (Mm) V1rf3 gene is shown. Conserved residues (at least four of six) are highlighted in blue. The 10 residues that are found in virtually all mouse V1rs are indicated by asterisks. The corresponding conserved residues in fish sequences are red. The green box indicates the position of the conserved N-linked glycosylation sites (NXS/T). +, the positions corresponding to the polymorphic variants found in the Tü, AB, and WIK Dr strains.
Taking advantage of the possibility to generate an “inferred ancestor” with the fish V1r-like genes, we used this synthetic sequence to further explore genomic databases. These searches for potential V1r homologues in nonvertebrate species, including deuterostomians (such as echinoderms) and protostomians (such as mollusks, nematodes, and arthropods), did not lead to the identification of any V1r-like sequence (data not shown), suggesting that V1rs appeared after the emergence of the first chordates.
Because mouse V1r and T2r taste receptors exhibit some (although very limited) sequence homology, and, therefore, likely evolved from a common ancestor, we investigated a possible closer relationship between these two receptor types in zebrafish. We performed a genome-wide search for zebrafish T2r sequences by using as queries members of all known human and mouse T2r subfamilies. We identified two zebrafish T2r related sequences (see Fig. 4), which, when aligned with rodent V1rs and human T2rs, clearly show a closer relationship of the fish V1rs to the rodent V1rs than to any of the T2r sequences, indicating that the divergence between V1rs and T2rs predated the sarcopterygian-actinopterygian split.
Variability of Fish V1r-Like Receptors. Mouse V1r receptors exhibit 10 highly conserved amino acids, which are present in >97% of the repertoire members (11); the five fish V1r-like receptors possess 7 of these conserved amino acids. In addition, a potential glycosylation site in extracellular loop 2 present in virtually all rodent V1rs (11), is also found in the five fish V1r-like receptors (Fig. 1). The lengths of extracellular and intracellular domains, a criterion that can be used to define classes of seven transmembrane receptors (29), are similar for mouse and fish V1rs (Fig. 1).
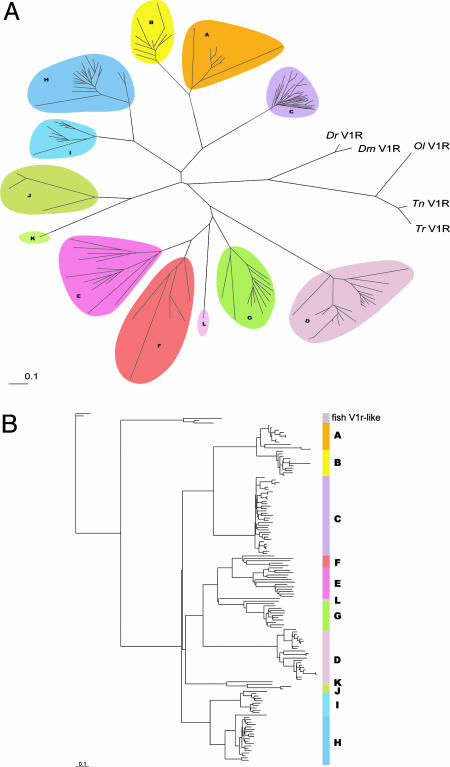
The closest mammalian relatives to the five fish V1r-like sequences are members of the V1rf and V1rc families, the closest homologue for zebrafish V1r being mouse V1rf3 with an identity of 33% at the amino acid level, and 44% at the nucleotide level. All mouse and fish V1rs were aligned and an unrooted tree was generated. This analysis was performed by using both amino acid (Fig. 2A) and nucleotide sequences (see Fig. 5, which is published as supporting information on the PNAS web site). Fish V1r-like are as distant from the 12 mouse families as these latter are from each other, and, therefore, pertain to none of the mouse families. The same alignment, using both nucleotides and amino acid sequences, was used to generate two rooted trees, using zebrafish V1r-like as an outgroup (Figs. 2B and 6, which is published as supporting information on the PNAS web site).
Fig. 2.
The fish and mouse V1r repertoires. (A) An unrooted tree representing V1r-like sequences from the five fish species together with the complete mouse V1r repertoire (the 12 families). Amino acid sequences were used for the generation of this tree. A comparable tree was generated by using nucleotide sequences (see Fig. 6). (B) Rooted tree, including all mouse V1r nucleotide sequences and the five fish sequences (Dr was used as an out-group). The corresponding tree for proteins is shown in Fig. 5.
The five fish species are phylogenetically unequally related: the separation between zebrafish and the other three fish species dates back ≈110–160 million years, whereas the medaka and pufferfish common ancestor is 60–80 million years old. The closer relatives fugu and tetraodon, probably diverged ≈18–30 million years ago (30–32). Interestingly, the fish sequences appear to be remarkably divergent between each other. We observe only 37% identity at the amino acid level between zebrafish and tetraodon, 60% between the more closely related species medaka and tetraodon, and 87% between D. malabaricus and D. rerio.
The peculiar variability between the fish V1r-like receptors prompted us to have a closer look at the evolutionary forces acting on these genes. First, we investigated the ratio (ω) of nonsynonymous (dN) vs. synonymous (dS) nucleotide substitutions per site between fish V1rs. Generally, purifying selection is inferred when dS is greater than dN, whereas positive (or diversifying) selection is inferred when the dN/dS ratio exceeds 1. Because of the too high sequence divergence between members of our dataset for such an analysis (i.e., T. nigroviridis and D. rerio), we identified V1rs from species more closely related to Danio species, i.e., pertaining to the Cyprinidae family. We amplified, by using PCR primers located outside the sequence corresponding to V1r TMI and VII, V1r sequences from the Ee, Da, Df, Cc, Cc (koï) species, and from one member of the Cyprinidae related family Cobitidae, Bm.
Pairwise dN/dS ratios using this now more homogenous dataset ranged between 0.09 (DmV1r–BmV1r) and 0.55 [CcV1r–Cc(koï)V1r], with an overall average of 0.17, providing evidence for purifying selection among fish V1rs when all sites are treated equally. We performed a more localized dN/dS analysis aimed at the separate analysis of regions with potentially different functions. Because nothing is known about the structure/function of V1rs, we considered, for each analysis, each V1r transmembrane extracellular and intracellular segment separately on each fish V1r pairs (see Fig. 7, which is published as supporting information on the PNAS web site). Apparently, different domains exhibit various levels of purifying selection when compared with each other: mean dN/dS between 0.28 and 0.29 for IC1 and IC2, and between 0.01 and 0.07 for TM3, 4, EC2, and 3 [significantly different with a paired two-tailed t test (P < 0.01), see also Fig. 7]. In agreement with our data, IC1 and TM3 were previously shown to exhibit the highest and lowest dN/dS ratios, respectively, when comparing members of the mouse a and b V1r families (33).
We then tested whether positive selection could be identified in a limited number of sites in our dataset, a selection type that may have been masked in our first analysis if only a limited number of sites are under such selection and most remain under purifying selection; such situation is known to occur for multiple genes, including primate V1rs (16). We generated a phylogenetic tree that included the eight Cyprinidae/Cobitidae sequences (see Fig. 8, which is published as supporting information on the PNAS web site), and used this phylogeny to perform an analysis in which the type of selection varies among different codon sites, and that allows for statistically testing different selection models (34–36), some of which accommodate the possibility of positive selection (Fig. 8). We did not find any statistically significant evidence for a model involving positive selection for fish V1r-like genes (statistical support or lack of support for the results are obtained by using likelihood ratio tests), although we and others could, with an identical approach, clearly identify positively selected sites in a set of mouse V1r genes from the b family (see Fig. 8) and in a set of primate V1rs (16).
Second, we investigated potential sequence variations within the zebrafish species. We cloned and sequenced the V1r-like coding sequences from the zebrafish strains Tü, WIK, and AB, and identified six polymorphic sites, two of them leading to amino acid changes. Compared with our original sequence (Tü, from the Ensembl database), we found an R to H change in IC3, and a V to L substitution in IC2 of the AB and WIK strains, respectively (see Fig. 1).
Third, a closer look at V1r sequence identities at both the amino acid and nucleotide levels surprisingly often showed higher homology at the nucleotide levels (for example, tetraodon and zebrafish V1r-like genes share 50% and 37% homology at the nucleotide and protein levels, respectively).
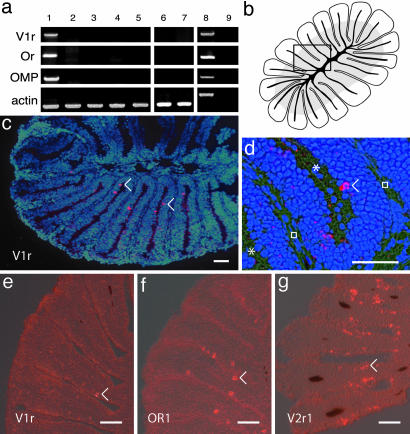
Expression of V1r-Like Genes in Fish. The existence of V1r-like sequences in teleosts, sequences that are phylogenetically clearly related to mammalian V1rs, does not necessarily imply that the corresponding proteins represent pheromone receptors. During animal evolution, gene amplification events have, at numerous times, led to neofunctionalization (37). Thus, the role of pheromone receptors played by V1rs in mammals could represent a recent use gained after an initial gene duplication. We investigated this possibility indirectly by analyzing the expression pattern of the zebrafish V1r-like genes in several tissues, including eyes, gills, olfactory bulb, brain, muscle, heart, intestine, barbels, and lips (the two latter structures are known to be chemosensory organs), and the olfactory sensory organ of the fish, the olfactory rosette. This latter structure is an external bilateral structure located on both sides of the head, which corresponds to the mammalian nostrils, although it does not connect with the throat. We first explored V1r-like expression by RT-PCR, and consistently observed its expression in the rosette (Fig. 3a). We were unable to find V1r-like transcripts in any of the other tested tissues. Inside the rosette, the numerous lamellae are covered with olfactory sensory neurons. We explored the cell type expressing the V1r-like gene by in situ hybridization of olfactory rosette sections from adult animals. Multiple olfactory sensory neurons expressing V1r-like transcripts were identified, and were apparently randomly dispersed in the neurosensory epithelium (Fig. 3 c and d). Consistent with the expression of a single V1r-like gene in the zebrafish olfactory system, a similar number of V1r-like-expressing neurons were identified at low and high stringency (data not shown). V1r-like expression was found in both male and female adult animals; analysis of serial sections indicated an average of 80 +/–11 olfactory sensory neurons expressing the V1r-like gene per rosette (n = 10). Unlike what is observed for some olfactory receptor genes which are expressed by sensory neurons from specific concentric zones in the rosette (38), and unlike the unexpected clustering of V2r-expressing sensory neurons we observed (Fig. 3 f and g), no obvious zone-specific V1r-like expression was observed in the sensory epithelium, except that most V1r-like-expressing sensory neurons were located in the middle part of the rosette z axis. In addition, when evaluating the basal/apical position of the soma of the V1r-expressing neurons in the neuroepithelium (by dividing the epithelium in three zones: apical, medial, and basal), we found a clear majority of the neurons located in the apical part (57%, 37%, and 6% respectively, n = 210). This last observation suggests that these sensory neurons are either microvillous olfactory neurons or crypt cells, the third neurosensory cell type being ciliated olfactory neurons, whose somata are mostly basally located.
Fig. 3.
Expression of V1r-like transcripts in the zebrafish olfactory system. (a) Expression of Dr V1r mRNA. cDNA from different adult tissues was amplified by PCR with specific primers for DrV1r, odorant receptor 1 (OR), olfactory marker protein (OMP), and actin. Lane 1, olfactory rosette; lane 2, gills; lane 3, olfactory bulb; lane 4, brain; lane 5, heart; lane 6, barbels; lane 7, lips; lane 8, genomic DNA; lane 9, olfactory rosette minus reverse transcriptase. Because no introns are found in the coding sequence of V1r and OMP, the sizes of the cDNA amplicons correspond to the size of the genomic amplicons. The amplicons corresponding to actin amplified from genomic material contain an intron and are therefore larger than cDNA amplicons. Exclusive V1r expression is observed in the olfactory rosette. (b) Drawing of a horizontal section of an olfactory rosette (lamellae are cut perpendicular to their flat face). The gray zone in the lamellae, close to the center, indicates the location of the sensory neuroepithelium. The black lines correspond to the cartilage. The rectangle indicates the area shown in d. (c and d) In situ hybridizations of horizontal sections through the olfactory rosette, with an antisense DrV1r-like RNA probe. Neurons expressing V1r-like mRNA appear as fluorescent red. The blue color corresponds to DAPI staining of the nuclei. The asterisks and squares indicate the lumen between the lamellae and the extracellular matrix, respectively. (e–g) In situ hybridizations with V1r, OR1, and V2r1 probes, respectively. The numerous neurons positive for V2r1 likely reflect crosshybridization with multiple V2r sequences. (Scale bar, 30 μm.) Arrowheads indicate some of the labeled neurons.
Discussion
Here, we report the identification of a single, highly variable vomeronasal receptor-type gene in teleosts, the largest group of vertebrate species. This receptor is a member of the V1r gene superfamily, whose presence was only described in mammalian species, whose expression was thought to be restricted to vomeronasal sensory neurons, and which is known to mediate the perception of pheromones in the mouse. Transcription of this gene is apparently restricted, in zebrafish, to a subpopulation of olfactory sensory neurons.
Our observations indicate that, unlike what is known in mammals where V1r receptor genes may number >150, fishes apparently make use of a single V1r-like gene. This finding is surprising for two reasons. First, one may have expected other sequences related to the fish V1r-like gene, at least in one of the genomes surveyed, because one or two major genome duplication events probably affected teleost species (31). The existence of a single fish V1r-like gene may indicate that paralogous sequences never existed but does not exclude the possibility that such sequences may be today unrecognizable because of very ancient pseudogenizations. Second, large expansions of olfactory chemoreceptor families are usually the rule in vertebrates, a situation that is understandable because most of these receptors help to make sense of an unknown and diverse outside chemical world by using a combinatorial activation strategy, involving the stimulation of multiple different receptors by a single agonist. This suggests that the V1r-like receptor may be devoted to the recognition of a single or of a very limited number of molecules, for which the emergence of multiple V1r-like receptors is of no selective advantage.
The identification of a single V1r-like gene in fish, and not in protostomians, has implications related to the existence of V1rs in other animal species. It first suggests that the emergence of these genes has taken place in some primitive vertebrate. It also suggests that because the common ancestor between fish and mammals lived ≈450 million years ago, and because mammalian and fish V1r-type genes emerged very unlikely independently, genomes of species that share this ancestor should contain V1r genes, or remnants of these latter. These species include all reptiles, birds, and amphibians, species in which no V1r has been reported. Our preliminary investigations of the Xenopus laevis genome support this view because we found a few intact V1r-like genes in this species (L. Capello and I.R., unpublished data).
By applying the term V1r to teleost sequences (an acronym that stands for vomeronasal receptor type I), we face a terminology problem because our finding indicates that the emergence of V1r receptors preceded the one of the vomeronasal system, and that their presence is even independent of the existence of the structure in some currently living species. This is the reason why we adopted the term “V1r-like.” An adequate way to refer to these receptors would be to relate them to their natural agonists or to the role played by these latter. We still ignore which are these agonists in fish, but because the V1r-like expression profile in zebrafish (punctate in an olfactory chemosensory neuroepithelium) is particularly reminiscent of the one of odorant and vomeronasal receptors in mammals, we may quite safely argue that they represent chemodetectors.
In the mouse, V1rs, which represent pheromone receptors, are particularly sensitive and are very narrowly tuned (39), apparently unlike odorant receptors. An observation, in addition to the specific expression profile of the fish V1r-like genes, may indicate that they encode chemodetectors involved in species-specific interactions: the comparison of the polypeptide sequences corresponding to the fish V1r-like genes shows a strikingly low degree of homology for a member of a monogenic receptor family. This quite weak similarity naturally suggests a correlated divergence of natural ligands, and a corresponding inability of the V1r-like receptor of a given species to identify the agonists recognized by the V1r-like receptor of another. It is tempting to speculate that these natural agonists are represented by pheromones, but this possibility remains to be shown. The remark concerning the interspecies sequence variability is also valid for odorant receptors but has different implications in this case. The odorant receptor family is multigenic, and uses a combinatorial activation strategy. Modifications of a single odorant receptor, or even of multiple odorant receptors, thus, does not likely impair the ability of the system to extract information from most molecules.
Given the variability of fish V1r-like genes, we looked for possible signs of positive Darwinian selection. Such type of selection is observed in some genes involved in intercellular or interindividual interactions, such as immunity-related proteins (MHC, IgVH,...), male/female-specific proteins involved in fertilization (ZP2, bindin,...) (40), but also in mammalian V1rs (16, 28). We report clear purifying selection pressure, apparently unequal between different regions of the fish V1r genes, but no evidence for positive selection. This observation could reflect reality, and may indicate that the function played in fishes by this receptor, despite its variability, is different from the one played in mammals (this argument is, however, quite weak, because positive selection of V1rs has only been observed in mice and primates, and has not been linked to any specific role). It could also simply be the result of the limited size of our dataset possibly not allowing to identify residues under weak positive selection, although a dataset with identical size allows the identification of positive selection in mouse V1rb genes.
Because fishes only possess a single V1r-like gene, when did the vertebrate V1r repertoire expand so to reach >100 members in rodents? Possible species-specific ecological niches, group size, breeding habits, or presence of a vomeronasal organ, may correlate with specific sizes or variability of V1r repertoires. Future investigation of different vertebrate V1r repertoires will likely help to better understand the pressures that led to the large expansions (or nonexpansions), and maintenance, of V1r families in a subset of vertebrate species.
Supplementary Material
Acknowledgments
We thank Carlos Galan for expert technical assistance; Pierre Vassalli and members of the I.R. laboratory for useful comments on the manuscript; Roland Dosch, Xavier Pochon, Benoît Stadelmann, and Juan Montoya for advice; and Marc Ekker (Loeb Health Research, Ottawa, ON, Canada) for the radiation hybrid panel LN54. This work was supported by the Swiss National Foundation for Research, the European Molecular Biology Organization Young Investigator Programme, and the Schmidheiny and Schlumberger Foundations.
Author contributions: P.P. and I.R. designed research; P.P. and I.R. performed research; P.P. and I.R. analyzed data; and I.R. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY764271, AC1938737, AY279523, AY279524, AY764272, AY764273, and AY900114–AY900119).
References
- 1.Buck, L. & Axel, R. (1991) Cell 65, 175–187. [DOI] [PubMed] [Google Scholar]
- 2.Ryba, N. J. & Tirindelli, R. (1997) Neuron 19, 371–379. [DOI] [PubMed] [Google Scholar]
- 3.Matsunami, H. & Buck, L. B. (1997) Cell 90, 775–784. [DOI] [PubMed] [Google Scholar]
- 4.Herrada, G. & Dulac, C. (1997) Cell 90, 763–773. [DOI] [PubMed] [Google Scholar]
- 5.Speca, D. J., Lin, D. M., Sorensen, P. W., Isacoff, E. Y., Ngai, J. & Dittman, A. H. (1999) Neuron 23, 487–498. [DOI] [PubMed] [Google Scholar]
- 6.Naito, T., Saito, Y., Yamamoto, J., Nozaki, Y., Tomura, K., Hazama, M., Nakanishi, S. & Brenner, S. (1998) Proc. Natl. Acad. Sci. USA 95, 5178–5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dulac, C. & Axel, R. (1995) Cell 83, 195–206. [DOI] [PubMed] [Google Scholar]
- 8.Boschat, C., Pelofi, C., Randin, O., Roppolo, D., Luscher, C., Broillet, M. C. & Rodriguez, I. (2002) Nat. Neurosci. 5, 1261–1262. [DOI] [PubMed] [Google Scholar]
- 9.Del Punta, K., Leinders-Zufall, T., Rodriguez, I., Jukam, D., Wysocki, C. J., Ogawa, S., Zufall, F. & Mombaerts, P. (2002) Nature 419, 70–74. [DOI] [PubMed] [Google Scholar]
- 10.Halpern, M. & Martinez-Marcos, A. (2003) Prog. Neurobiol. 70, 245–318. [DOI] [PubMed] [Google Scholar]
- 11.Rodriguez, I., Del Punta, K., Rothman, A., Ishii, T. & Mombaerts, P. (2002) Nat. Neurosci. 5, 134–140. [DOI] [PubMed] [Google Scholar]
- 12.Lane, R. P., Young, J., Newman, T. & Trask, B. J. (2004) Genome Res. 14, 603–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grus, W. E. & Zhang, J. (2004) Gene 340, 303–312. [DOI] [PubMed] [Google Scholar]
- 14.Rodriguez, I., Greer, C. A., Mok, M. Y. & Mombaerts, P. (2000) Nat. Genet 26, 18–19. [DOI] [PubMed] [Google Scholar]
- 15.Giorgi, D. & Rouquier, S. (2002) Chem. Senses 27, 529–537. [DOI] [PubMed] [Google Scholar]
- 16.Mundy, N. I. & Cook, S. (2003) Mol. Biol. Evol. 20, 1805–1810. [DOI] [PubMed] [Google Scholar]
- 17.Belluscio, L., Koentges, G., Axel, R. & Dulac, C. (1999) Cell 97, 209–220. [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez, I., Feinstein, P. & Mombaerts, P. (1999) Cell 97, 199–208. [DOI] [PubMed] [Google Scholar]
- 19.Wakabayashi, Y., Mori, Y., Ichikawa, M., Yazaki, K. & Hagino-Yamagishi, K. (2002) Chem. Senses 27, 207–213. [DOI] [PubMed] [Google Scholar]
- 20.Hukriede, N., Fisher, D., Epstein, J., Joly, L., Tellis, P., Zhou, Y., Barbazuk, B., Cox, K., Fenton-Noriega, L., Hersey, C., et al.. (2001) Genome Res. 11, 2127–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saitou, N. & Nei, M. (1987) Mol. Biol. Evol. 4, 406–425. [DOI] [PubMed] [Google Scholar]
- 23.Felsenstein, J. (1981) J. Mol. Evol. 17, 368–376. [DOI] [PubMed] [Google Scholar]
- 24.Swofford, D. (1999) PAUP* 4.0b10: Phylogenetic Analysis Using Parsimony (*and Other Methods) (Sinauer, Sunderland, MA).
- 25.Posada, D. & Crandall, K. A. (1998) Bioinformatics 14, 817–818. [DOI] [PubMed] [Google Scholar]
- 26.Yang, Z. (1997) Comput. Appl. Biosci. 13, 555–556. [DOI] [PubMed] [Google Scholar]
- 27.Rodriguez, I. & Mombaerts, P. (2002) Curr. Biol. 12, R409–R411. [DOI] [PubMed] [Google Scholar]
- 28.Zhang, X., Rodriguez, I., Mombaerts, P. & Firestein, S. (2004) Genomics 83, 802–811. [DOI] [PubMed] [Google Scholar]
- 29.Otaki, J. M. & Firestein, S. (2001) J. Theor. Biol. 211, 77–100. [DOI] [PubMed] [Google Scholar]
- 30.Wittbrodt, J., Shima, A. & Schartl, M. (2002) Nat. Rev. Genet. 3, 53–64. [DOI] [PubMed] [Google Scholar]
- 31.Chen, W. J., Orti, G. & Meyer, A. (2004) Trends Genet. 20, 424–431. [DOI] [PubMed] [Google Scholar]
- 32.Crnogorac-Jurcevic, T., Brown, J. R., Lehrach, H. & Schalkwyk, L. C. (1997) Genomics 41, 177–184. [DOI] [PubMed] [Google Scholar]
- 33.Lane, R. P., Cutforth, T., Axel, R., Hood, L. & Trask, B. J. (2002) Proc. Natl. Acad. Sci. USA 99, 291–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang, Z. & Swanson, W. J. (2002) Mol. Biol. Evol. 19, 49–57. [DOI] [PubMed] [Google Scholar]
- 35.Yang, Z., Nielsen, R., Goldman, N. & Pedersen, A. M. (2000) Genetics 155, 431–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong, W. S., Yang, Z., Goldman, N. & Nielsen, R. (2004) Genetics 168, 1041–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meyer, A. & Schartl, M. (1999) Curr. Opin. Cell Biol. 11, 699–704. [DOI] [PubMed] [Google Scholar]
- 38.Weth, F., Nadler, W. & Korsching, S. (1996) Proc. Natl. Acad. Sci. USA 93, 13321–13326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leinders-Zufall, T., Lane, A. P., Puche, A. C., Ma, W., Novotny, M. V., Shipley, M. T. & Zufall, F. (2000) Nature 405, 792–796. [DOI] [PubMed] [Google Scholar]
- 40.Wolfe, K. H. & Li, W. H. (2003) Nat. Genet. 33, Suppl., 255–265. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.