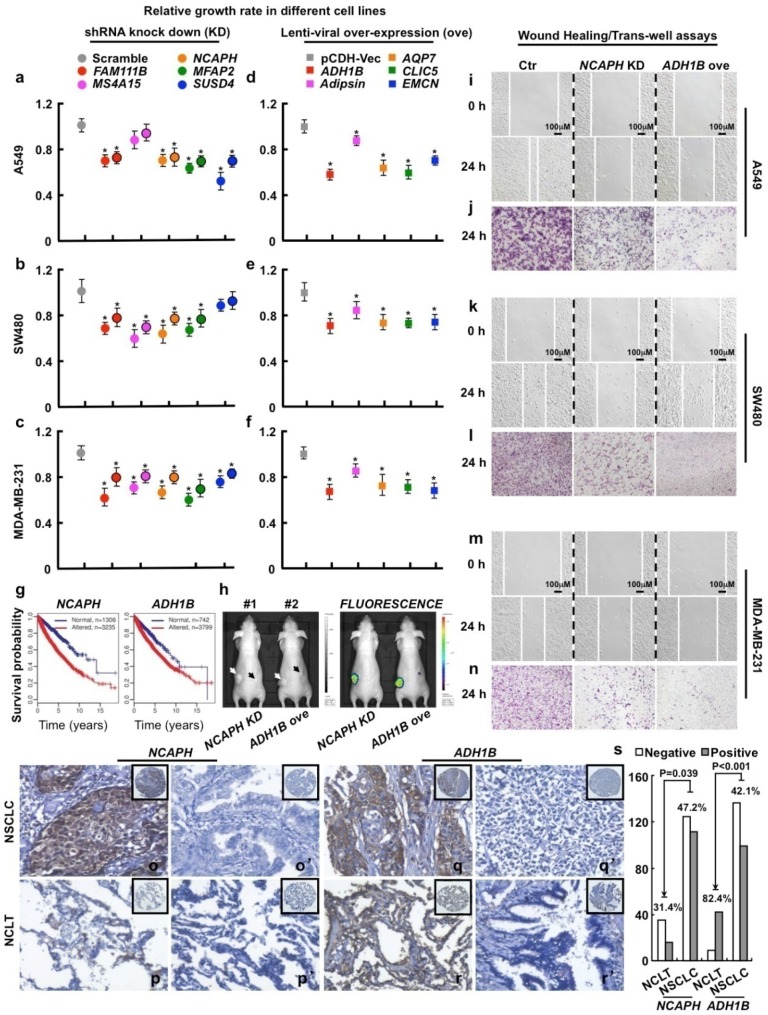
Figure 3.
Functional study of 10 picked genes from the top 500 dysregulated ones in different tumor cell lines. (a-c) 3 up-regulated genes (FAM111B, NCAPH and MFAP2) and 2 cancer-specifically dysregulated genes (MS4A15 and SUSD4) were inhibited by 2 independent shRNAs targeting to different mRNA regions (indicated by different colors with or without black circle), respectively. Here, representative day 4 cell numbers normalized to scramble shRNA cells (considered as 1) were shown, *P < 0.05. (d-f) 5 down-regulated genes (ADH1B, Adipsin, AQP7, CLIC5 and EMCN) were over-expressed, respectively. The cell numbers were counted and normalized to pCDH-Vec control cells (considered as 1), *P < 0.05. (g) Survival curves of ADH1B and NCAPH across cancer types (NCAPH, P = 2.22e-16; ADH1B, P = 6.75e-06). Normal/altered: a gene is expressed in the normal/altered levels in a sample (see Methods). (h, left) Representative photographs captured with visible light of the animals corresponding to each treatment group at day 28th after A549-luc cell injection. White arrow (#1 mouse) indicates scramble shRNA control, dark arrow (#1 mouse) indicates NCAPH KD; white arrow (#2 mouse) indicates pCDH-Vec control, dark arrow (#2 mouse) indicates ADH1B ove; total 2×106 cells for each line were injected. (h, right) Representative whole body fluorescence imaging showing a significant reduction in tumor size when NCAPH was depleted or ADH1B was over-expressed. (i, k, m) Representative wound healing assay using indicated cancer cell lines at 0 or 24 hours, respectively. (j, l, n) Representative trans-well cell migration assay using indicated cell lines at 24 hours. (o-r') IHC, DAB staining, 200X. (o-o', q-q') The positive and negative expression patterns of NCAPH or ADH1B protein, respectively, were shown in the NSCLC (p-p', r-r') The positive and negative expression patterns of NCAPH or ADH1B protein, respectively, were shown in the NCLT. (s) Quantification data for IHC. All the experiments were repeated at least 3 times, representative images were shown.