Abstract
For mammalian cells, contact-dependent regulatory controls are crucially important for controlling cellular proliferation and preventing diseases such as cancer. Candida albicans, an opportunistic fungal pathogen that normally resides within a mammalian host, also exhibits contact-dependent cellular behaviors such as invasive hyphal growth and biofilm development. Results reported here demonstrate that, in C. albicans, physical contact results in activation of the mitogen-activated protein kinase Mkc1p. This kinase is part of the fungal cell integrity pathway, a signal transduction pathway known to be activated by cell wall stress. It is demonstrated here that Mkc1p is required for invasive hyphal growth and normal biofilm development. Therefore, Mkc1p signaling contributes to contact-dependent regulation. Because responding to contact appropriately allows coordinated cellular behavior in a metazoan, commensal C. albicans cells behave like a part of the host, using contact-activated signaling to regulate fungal behavior.
Keywords: cell integrity pathway, mitogen-activated protein kinase, hyphae
The fungus Candida albicans, an opportunistic human pathogen, has evolved a repertoire of responses triggered by contact with a surface. One such response, invasive hyphal growth, occurs during infection of a host or during growth in contact with semisolid agar medium (1, 2). In these situations, hyphae and pseudohyphae, the filamentous forms of the organism, are produced and invade the tissue or medium. Formation of C. albicans biofilms, another contact-dependent response, occurs during growth on solid surfaces. This growth state is medically important because biofilms that form on implanted medical devices are notoriously difficult to eradicate, because of their characteristic high resistance to antifungal drugs (3). Sessile (attached) cells exhibit increased drug resistance soon after adherence (4).
Fungi, including C. albicans, also reorient the direction of hyphal growth in response to contact with features of the physical environment (5). This behavior, termed thigmotropism, is also observed during the growth of plant roots (6). The fungal plant pathogen Uromyces appendiculatus differentiates in response to contact with ridges of a particular height (7). These biological effects of physical contact with surfaces or features of surfaces require that organisms sense contact and process that information to produce an appropriate response. Regulatory mechanisms that control the behavior of fungal cells in response to contact are not well understood.
Production of filamentous C. albicans cells occurs in response to numerous environmental cues, and several signal transduction pathways regulate cellular morphology (8). Different pathways are needed for filamentation under different conditions. For example, a mutant strain that is extremely defective in invasive filamentation because of the absence of two transcription factors, Czf1p and Cph1p, is nevertheless able to filament under most other conditions (2). In contrast, a mutant that is defective in filamentation under many conditions, because of the absence of transcription factors Efg1p and Cph1p, retains the ability to produce invasive filaments that invade host tissue or agar medium (9, 10). Thus, there is at least one regulatory pathway, the Czf1p pathway, that specifically controls contact-dependent invasive filamentation.
The goal of the work described here was to determine how physical contact triggers cellular behaviors in C. albicans. This work was predicated on the hypothesis that growth of C. albicans in contact with a surface results in mechanical perturbation of the cell wall or plasma membrane and that this perturbation is detected by C. albicans using a signal transduction pathway known as the cell integrity pathway. The cell integrity mitogen-activated protein (MAP) kinase pathway of Saccharomyces cerevisiae contains the MAP kinase Mpk1p (also called Slt2p) and controls responses to cell wall damage or alteration in plasma membrane curvature (11, 12). Results reported here demonstrate that the C. albicans cell integrity MAP kinase, Mkc1p (13, 14), accumulates in activated form when cells are grown on a surface. In addition, C. albicans mkc1-null mutants are defective in two contact-dependent responses, invasive hyphal growth and biofilm formation, consistent with the model that the cell integrity pathway of C. albicans plays a role in contact sensing. Therefore, the C. albicans cell integrity pathway detects physical contact, allowing the organism to produce an appropriate biological response for a particular niche within the host.
Materials and Methods
Strains. All C. albicans strains were derived from CAI-4 (15) and maintained the same chromosome 1 copy number as the starting strain (trisomic). Trisomic and diploid isolates of CAI-4 exhibited similar phenotypes, demonstrating that chromosome 1 copy did not have a strong effect on contact-dependent behaviors (data not shown). MKC1-null mutants were constructed as described (13) by using plasmid pUCInt, kindly provided by F. Navarro-Garcia (Universidad Complutense de Madrid, Madrid). The following plasmids were integrated at the MKC1 locus: pCK70, a control plasmid encoding the MKC1 locus with a BglII deletion that removed most of the MKC1 gene; pCK71, encoding MKC1+ subcloned from plasmid pSN6 (a kind gift of F. Navarro-Garcia) (13); or pCK72, encoding a hemagglutinin (HA)-tagged derivative of MKC1 (MKC1-HA). S. cerevisiae Σ1278b strain L5487 (MATα ura3-52 leu2::hisG) was kindly provided by G. Fink (Whitehead Institute for Biomedical Research, Massachusetts Institute of Technology, Cambridge). pMPK1-HA, kindly provided by D. Levin (Johns Hopkins Bloomberg School of Public Health, Baltimore), was carried in S. cerevisiae S288c strain CKY338 (MATα his3Δ leu2Δ lys2Δ ura3Δ mpk1Δ::KanMX4/YEpMPK1-HA LEU2).
Construction of Plasmids pCK70, pCK71, and pCK72. To construct an integrating plasmid with C. albicans URA3 as the selectable marker, the MKC1-encoding fragment from pSN6 (a kind gift of F. Navarro-Garcia) (13) was released by digestion with Ecl36I and PstI and cloned into PstI- and SmaI-digested pRC3915 (16), generating pCK71. For construction of C-terminally HA-tagged MKC1, the HA tag and selectable marker were amplified from plasmid pFA6a-3HA-HIS3MX6 (17) by using the primers MKCHAF (5′AAAGCTAGAGGAAGAGCTTGGGTTTGGAT TAGAT TGGTGCTATGT TCGGATCCCCGGGT TAATTAACA-3′) and MKCHAR (5′ACGTGGTTGTGTGTTTCAACTACTGGTGGTCGTTACAGTAGTTG TTATGGCGCGCCGAATTCGAGCTCGTTTAAACTG-3′). The introduced AscI site in MKCHAR is italicized. Plasmid pSN6 was cotransformed into S. cerevisiae cells with the HA-HIS3MX6 PCR product. Recombinant plasmids in which the HA-HIS fragment had recombined into the MKC1 gene of pSN6 were isolated. After digestion with AscI and religation, a plasmid encoding HA-tagged MKC1 with its native 3′ end was constructed. The insert from this plasmid was subcloned into pRC3915, as described above, generating pCK72. To construct a control plasmid that would integrate at MKC1 but did not encode intact MKC1, the 3.8-Kb XbaI fragment from plasmid pSN6 was subcloned into vector pNUB1, a derivative of pNEB193 (New England Biolabs) that contains the XbaI-to-ScaI C. albicans URA3 fragment cloned at the NdeI site. To delete most of the MKC1 gene, this plasmid was digested with BglII and religated, generating plasmid pCK70.
Growth Media. Yeast extract/peptone/dextrose (YPD)-rich and yeast extract/peptone/sucrose (YPS)-rich media and synthetic defined (SD) and complete medium (CM) defined media were as described (2, 18). RPMI medium 1640 was from Sigma. In some experiments, uridine was added to 60 μg/ml. For surface growth and agar invasion, medium containing 1% agar (Difco), 0.7% SeaPlaque low-melt agarose (FMC), or 5% gelatin (Sigma) was used. Escherichia coli strains were cultured in Luria (L) broth or on L plates (19) containing ampicillin (100 μg/ml).
Cell Extraction and Detection of Phospho-MAP Kinase. Cells of strains CKY357 (mkc1-null), CKY358 (mkc1/mkc1/MKC1), or CKY359 (mkc1/mkc1/MKC1-HA) were grown under the following conditions: (i) in SD liquid medium, at 37°C, with or without 3.1 μg/ml Nikkomycin Z for 4 h, (ii) in CM-uracil liquid medium at 25°C with or without chlorpromazine (0.5 mM) for 1 h, (iii) in YPS liquid medium at 25°C in logarithmic (log) phase, (iv) in YPS liquid medium or on the surface of YPS-agarose or YPS-gelatin plates for 3 days at 25°C, (v) on the surface of YPS-agarose medium followed by growth in YPS liquid medium for 1 h at 25°C, and (vi) in RPMI medium 1640 liquid medium or plated in polystyrene dishes in RPMI medium 1640 at 37°C for 4 h. S. cerevisiae cells were grown in YPD or CM-leu liquid medium at 25°C in log or stationary phase and shifted to 42°C for 1 h in liquid medium or grown on the surface of YPD or CM-leu medium.
Cells were collected over ice, washed with ice-cold PBS, and immediately extracted in lysis buffer essentially as described (20), except that the buffer contained Complete protease inhibitor mixture (Boehringer Mannheim). The cells were vortex mixed in a Tomy (Tokyo) mixer with 0.5-mm zirconium-glass beads for 30 sec, followed by chilling in ice for a total of six cycles. Extracts were clarified by centrifugation in a microcentrifuge (Eppendorf) at maximum speed for 10 min at 4°C. Total protein was determined by using the Pierce Micro BCA assay as described by the manufacturer. Eighty micrograms of total protein was loaded per lane on a full-size 8.5% polyacrylamide SDS gel, fractionated by electrophoresis, and transferred to an Immobilon P (Millipore) membrane, and membranes were probed with anti-phospho-p44/42 MAP kinase antibody (Cell Signaling Technology, Beverly, MA). Total HA-tagged proteins were detected in 50 μg (C. albicans) or 12 μg (S. cerevisiae) of extract with anti-HA mAb (Covance, Princeton, NJ). Actin, used as a loading control, was detected in 120 μg of extract with anti-actin mAb (Novus Biologicals, Littleton, CO). Blots were probed with secondary anti-rabbit-horseradish peroxidase (HRP) or anti-mouse-HRP (Cell Signaling Technology) and developed by using Ecl Plus chemiluminescent detection (Amersham Pharmacia) as described by the manufacturer.
Agar Invasion Assay. Cells were plated on YPS uridine–1% agar, yielding 30–50 colonies. Plates were incubated at 25°C, and after 4 days cells were washed off the surface and the agar was examined microscopically. Colonies in which the entire outline of the colony contained invading cells were scored as positive in contrast to colonies with only a few invading cells, which were scored as negative. Colonies were photographed either from the top or after cutting the agar and laying it on its side to observe the invading cells from the side with the 10× objective of a Nikon Eclipse E400 dissecting microscope and a SPOT Insight (Diagnostic Instruments, Sterling Heights, MI) color camera.
Biofilm Formation. The system of Ramage et al. (21) was used with minor modifications. Briefly, a cell suspension at 1 × 106 cells per ml in RPMI medium 1640/20 mM MOPS (pH 7.0) was incubated in polystyrene wells or Petri dishes. For cell harvesting, cells were plated at 3 × 106 cells per ml. Photography was performed with a Nikon Eclipse E400 dissecting microscope and a SPOT Insight color camera with the 10× objective.
Antifungal Resistance. Nikkomycin Z susceptibility was determined by using the standard Clinical and Laboratory Standards Institute [CLSI, formerly the National Committee for Clinical Laboratory Standards (NCCLS)] microdilution protocol M27-A (22) except that incubation was performed in yeast nitrogen base containing sucrose at 37°C. Susceptibility to fluconazole was analyzed by using the standard CLSI microdilution protocol M27-A (22). To determine the fluconazole susceptibility of sessile cells after 16 h of growth in Petri dishes at 37°C, sessile cells were harvested, vortexed, diluted, and incubated in round-bottom wells containing increasing concentrations of fluconazole for 3 days at 35°C. For each determination, the sample containing no fluconazole was repeated in triplicate, and the entire determination was done in duplicate for each strain. To quantitate adherent cells that had grown in the presence of drug, [2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide] (XTT) reduction activity was determined as described below.
XTT Reduction Assay. Relative numbers of sessile cells were measured by assaying XTT reduction, a reaction catalyzed by mitochondrial dehydrogenases (21). Briefly, adherent cells were washed once with PBS and then incubated with 0.5 mg/ml XTT and 1 μM menadione in PBS at 37°C for 2–4 h. A490 was determined by using a microtiter plate reader, and the minimum inhibitory concentration was defined as the minimal amount of fluconazole that reduced metabolic activity by 80%. Values for different strains were compared by using Student's t test with a two-tailed distribution.
Results
Activation of Mkc1p During Growth on a Semisolid Surface. When C. albicans cells grow on the surface of semisolid agar medium, some of the cells grow invasively, converting into filamentous hyphae and pseudohyphae and growing into the underlying medium (see Fig. 2). To determine whether activation of the C. albicans cell integrity MAP kinase Mkc1p occurred during growth under these conditions, as hypothesized, the phosphorylation state of Mkc1p was analyzed as described in Materials and Methods with strains deleted for Mkc1p or encoding either WT Mkc1p or HA-tagged Mkc1p (Mkc1p-HA). The MKC1-HA allele was less active than WT MKC1 and exhibited weak complementation of the hypersensitivity of the mkc1Δ-null mutant to Nikkomycin Z (14), a chitin synthase inhibitor that causes cell wall stress (Table 1).
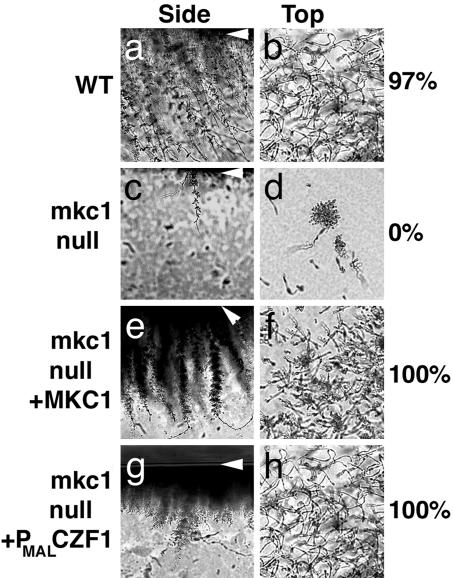
Fig. 2.
MKC1 is required for invasion. After 4 days at 25°C, surface cells were washed off the agar medium, and the remaining cells were photographed from the side and top. White arrowheads indicate the top of the agar. The percentage of colonies exhibiting extensive invasive growth (defined as invasive cells covering the entire outline of the colony) is shown. (a and b) CKY101 (MKC1+). (c and d) CKY357 (mkc1-null). (e and f) CKY358 (mkc1/mkc1/MKC1). (g and h) CKY355 (mkc1/mkc1/PMAL-CZF1).
Table 1. C. albicans strains used and minimum inhibitory concentration (MIC) for Nikkomycin Z.
| Strain | Source | Genotype | MIC |
|---|---|---|---|
| CAI-4 | Ref. 15 | SC5314 Δura3::λimm434/Δura3::λimm434 | ND |
| CKY136 | Ref. 10 | CAI-4, efg1::hisG/efg1::hisG ade2::pDBI52 (URA3) | ND |
| CKY101 | Ref. 2 | CAI-4, ade2::pDBI52 (URA3) | 6.20 |
| AJC6 | This study | CAI-4, mkc1Δ::hisG-URA3-hisG/MKC1 | 6.20 |
| CKY357 | This study | CAI-4, mkc1Δ::hisG/mkc1Δ::hisG mkc1::pCK70 (URA3) | 0.78 |
| CKY356 | This study | CAI-4, mkc1Δ::hisG/mkc1Δ::hisG ade2::pDBI52 (URA3) | 0.78 |
| CKY358 | This study | CAI-4, mkc1Δ::hisG/mkc1Δ::hisG mkc1::pCK71 (MKC1, URA3) | 3.10 |
| CKY359 | This study | CAI-4, mkc1Δ::hisG/mkc1Δ::hisG mkc1::pCK72 (MKC1-HA, URA3) | 1.60 |
| CKY355 | This study | CAI-4, mkc1Δ::hisG/mkc1Δ::hisGade2::pDB212 (PMAL-CZF1, URA3) | 0.78 |
Minimum inhibitory concentration (μg/ml) was determined by using the CLSI microdilution method. ND, not determined.
In other yeasts, the cell integrity MAP kinase is activated by phosphorylation in response to cell wall or membrane stress (11, 23); therefore, phosphorylation of C. albicans Mkc1p was first assessed under cell wall stress conditions. Analysis of extracts of MKC1+ cells treated with Nikkomycin Z revealed a phosphorylated MAP kinase protein (Fig. 1, lane 4) that was not present in untreated cells (Fig. 1, lane 3) or mkc1-null mutant cells (Fig. 1, lanes 1 and 2). When a strain expressing Mkc1p-HA as the sole form of Mkc1p was treated with Nikkomycin Z, the protein exhibited slower mobility (Fig. 1, lane 8) and was also detected with anti-HA antibody (Fig. 1, lanes 7 and 8). Anti-HA antibody failed to detect a band when untagged extracts were analyzed (Fig. 1, lanes 3 and 4). Probing with anti-actin demonstrated that proteins were present in all lanes. These results demonstrated that the detected band corresponds to phospho-Mkc1p. Phospho-Mkc1p was also detected after treatment with chlorpromazine, an agent that causes membrane curvature (11) (Fig. 1, lanes 5, 6, 9, and 10). Therefore, in C. albicans, activated Mkc1p was detected after growth in cell wall stress or membrane-perturbing conditions, as expected for a component of the cell integrity pathway.
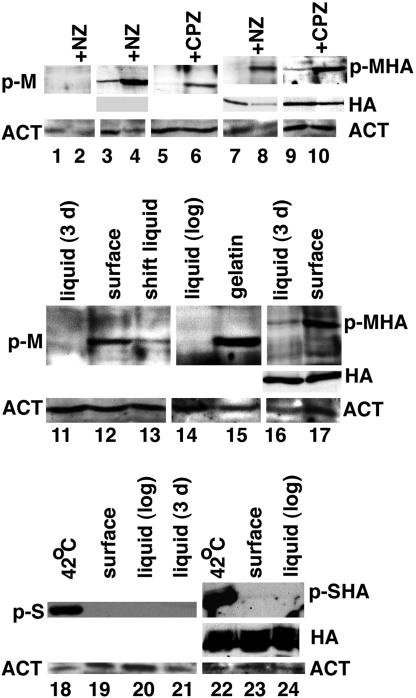
Fig. 1.
Surface-grown cells accumulate phospho-Mkc1p. Cells of strains CKY357 (mkc1-null) (lanes 1 and 2), CKY358 (MKC1) (lanes 3–6 and 11–15), CKY359 (MKC1-HA) (lanes 7–10, 16, and 17), S. cerevisiae strain L5487 (lanes 18–21), or S. cerevisiae strain CKY338 (pMPK1-HA) (lanes 22–24) were extracted as described in Materials and Methods, after growth under conditions described below. Samples were normalized by using total protein, determined by the Pierce Micro BCA assay, and analyzed by Western blot analysis on a full-size gel with either anti-phospho-p44/42 MAP kinase antibody (Cell Signaling Technology) (Top for each lane), anti-actin antibody (Novus Biologicals) (Bottom for each lane), or anti-HA monoclonal antibody (Covance) (Middle for lanes 3, 4, 7–10, 16–17, and 22–24). HRP-conjugated secondary antibody and ECL Plus chemiluminescent detection were used. Actin was used as a loading control. Lanes 1, 3, and 7, SD liquid at 37°C; lanes 2, 4, and 8, SD liquid plus Nikkomycin Z (NZ; 3.1 μg/ml) at 37°C for 4 h; lanes 5 and 9, CM liquid at 25°C; lanes 6 and 10, CM liquid plus 0.5 mM chlorpromazine at 25°C for 1 h; lane 11, YPS liquid at 25°C for 3 days; 12, YPS-agarose surface at 25°C for 3 days; lane 13, YPS-agarose surface, then liquid YPS for 1 h; lane 14, YPS liquid (log phase) at 25°C; lane 15, YPS-gelatin surface at 25°C for 3 days; lane 16, YPS liquid at 25°C for 3 days; lane 17, YPS-agarose surface at 25°C for 3 days; lane 18, YPD liquid at 42°C for 1 h; lane 19, YPD agar surface at 25°C for 3 days; lane 20, YPD liquid at 25°C (log phase); lane 21, YPD liquid at 25°C for 3 days; lane 22, CM-leu liquid at 42°C for 1 h; lane 23, CM-leu agar surface at 25°C for 3 days; and lane 24, CM-leu liquid at 25°C (log phase). p-M, phospho-Mkc1p; p-MHA, phospho-Mkc1p-HA; HA, total Mkc1p-HA; ACT, actin; p-S, S. cerevisiae phospho-Mpk1p; p-SHA, S. cerevisiae phospho-Mpk1p-HA; CPZ, chlorpromazine.
To determine the effects of contact with a semisolid surface, cells were grown on the surface of agarose plates or in liquid medium at low temperature and tested for the presence of phospho-Mkc1p. As shown in Fig. 1, lanes 11, 14, and 16, cells grown in YPS liquid medium (stationary or log phase) contained low levels of phospho-Mkc1p (both tagged and untagged). In contrast, cells grown on the surface of YPS agarose medium contained higher levels of activated phospho-Mkc1p (tagged and untagged) (Fig. 1, lanes 12 and 17) with minimal change in the total level of tagged Mkc1p (Fig. 1, lane 17). When cells growing on the surface of agarose medium were scraped from the plate and returned to growth in liquid medium for 1 h, the level of phospho-Mkc1p decreased (Fig. 1, lane 13). Activation of Mkc1p was also demonstrated after growth on gelatin medium (Fig. 1, lane 15), showing that the chemical nature of agarose was not critical. In summary, growth on the surface of semisolid medium resulted in activation of the cell integrity pathway, and the effects of contact were rapidly reversed when the cells lost contact with the surface.
In contrast to the response of the pathogen C. albicans, phosphorylation of the S. cerevisiae cell integrity MAP kinase, Mpk1p/Slt2p, did not increase when cells were grown on agar medium (Fig. 1, lanes 18–24). Therefore, under these conditions, S. cerevisiae lacks the ability to respond to contact by activation of the cell integrity pathway.
Signaling for Invasive Filamentous Growth Requires Mkc1p. To determine whether Mkc1p conveys signals that lead to invasive filamentous growth, C. albicans colonies were grown on the surface of YPS agar medium, cells were washed off the surface, and the agar below the colonies was examined. Cross sections of the agar below WT colonies showed filaments extending from the surface into the agar (Fig. 2a). When viewed from the top, invading cells covered the entire outline of the colony (Fig. 2b), and 97% of the WT colonies exhibited this extensive invasion. In contrast, the mkc1-null mutant colonies produced few invading cells (Fig. 2 c and d). In addition, the mkc1/mkc1-null mutant was defective in producing filamentous colonies when cells were embedded within agar medium (data not shown). Introduction of WT MKC1 into the null mutant restored extensive invasive filamentation (Fig. 2 e and f), demonstrating complementation of the defect.
Measurement of growth rates demonstrated that the invasive filamentation defect was not caused by slow growth of the mutant (data not shown). In addition, the mkc1-null mutant filaments in liquid medium (Fig. 3d and ref. 14), demonstrating that the defect in invasive filamentation did not reflect an inability to form a filament per se. Thus, the mkc1-null mutant is defective in invasive filamentation triggered by contact.
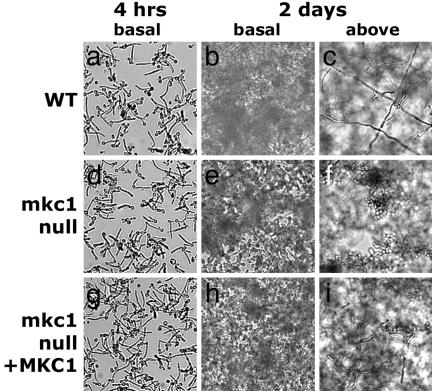
Fig. 3.
Defects in biofilm morphology in the mkc1-null mutant. CKY101 (MKC1+) (a–c), CKY357 (mkc1-null) (d–f), and CKY358 (mkc1/mkc1/MKC1) (g–i) were grown for 4 h (a, d, and g) or 2 days (b, c, e, f, h, and i) and photographed with the 10× objective. (b, e, and h) The basal layer of cells adhering to the surface. In mature biofilms, individual cells are difficult to discern because of the 3D nature of the biofilm and the presence of matrix. (c, f, and i) Cells growing away from the surface.
Normal Biofilm Formation Requires Mkc1p. When C. albicans cells are grown on a solid surface, they produce biofilms, attached 3D communities of cells that contain exopolymeric matrix, and exhibit high resistance to antifungal drugs (3). To test the hypothesis that Mkc1p-dependent sensing of contact with a solid surface contributes to the development of biofilms, cells were grown in polystyrene wells in RPMI medium 1640 as described (21). After 4 h of incubation, there were no obvious differences in adherence between the WT and the mkc1-null mutant (Fig. 3 a, d, and g). After 48 h of incubation, WT cells formed a mature biofilm with filamentous cells projecting away from the surface (Fig. 3c), creating a 3D structure. In contrast, the mkc1-null mutant produced an abnormal biofilm with reduced filamentation (Fig. 3f). Biofilm formation was improved when MKC1 was reintroduced into the mkc1-null mutant (Fig. 3 h and i).
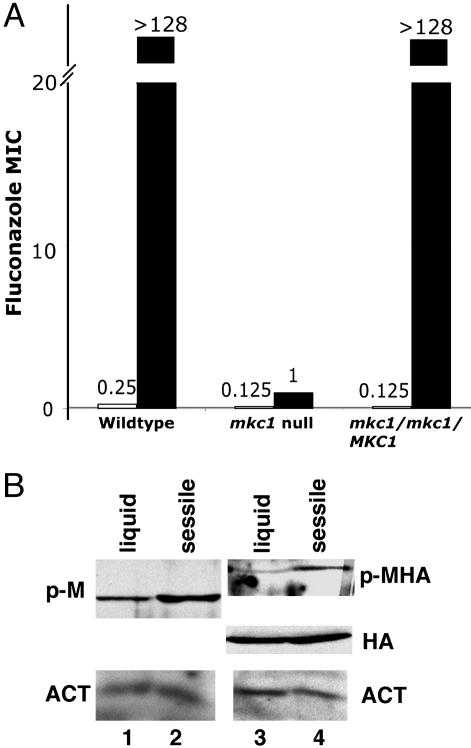
Sessile, WT cells were extremely resistant to fluconazole (Fig. 4A). In contrast, sessile mkc1-null mutant cells were fluconazole-sensitive (Fig. 4A). When measured under standard CLSI conditions (22), WT and mkc1-null mutant cells exhibited very similar sensitivity to fluconazole (Fig. 4A and ref. 14), demonstrating that mkc1-mutant cells were not inherently more susceptible to fluconazole. In combination, these results demonstrate that, although mkc1-null mutant cells adhered to plastic, they did not develop the normal biochemical or morphological phenotype of sessile cells.
Fig. 4.
Fluconazole resistance and activation of Mkc1p during sessile growth. (A) Fluconazole minimum inhibitory concentration (MIC) for strains CKY101 (MKC1+), CKY357 (mkc1-null), and CKY358 (mkc1/mkc1/MKC1) was determined by the standard CLSI protocol (open bars). After 16 h of incubation at 37°C, fluconazole MIC of sessile cells was determined by XTT reduction after growth in drug as described in Materials and Methods (filled bars). (B) Cells of strains CKY358 (MKC1) (lanes 1 and 2) or CKY359 (MKC1-HA) (lanes 3 and 4) were extracted and analyzed as in Fig. 1. Lanes 1 and 3, RPMI medium 1640 liquid at 37°C for 4 h; and lanes 2 and 4, RPMI medium 1640 sessile growth at 37°C for 4 h. p-M, phospho-Mkc1p; p-MHA, phospho-Mkc1p-HA; HA, total Mkc1p-HA; ACT, actin.
These findings predict that activation of Mkc1p would occur during sessile growth. As shown in Fig. 4B, after 4 h of incubation at 37°C, sessile cells exhibited higher levels of phospho-Mkc1p than liquid-grown cells. In this experiment, the shift from low to higher temperature resulted in some production of phospho-Mkc1p in the liquid-grown cells. These results are consistent with the model that adherence to a plastic surface activates the cell integrity pathway and that this activation contributes to the development of some features of the biofilm.
Distinct Regulation of Biofilm Formation and Invasive Hyphal Growth. The data presented here demonstrate that biofilm formation and invasive hyphal growth involve signaling by Mkc1p. However, there are differences in the regulation of the two processes. The transcription factor Efg1p performs a positive function needed for development of a stable, multilayered biofilm (Fig. 5 and ref. 24). In contrast, Efg1p has a repressive effect on invasive filamentation (10). One model to explain these results is that the common step, Mkc1p-dependent contact sensing, occurs before biofilm- or invasion-specific steps regulated by Efg1p.
Fig. 5.
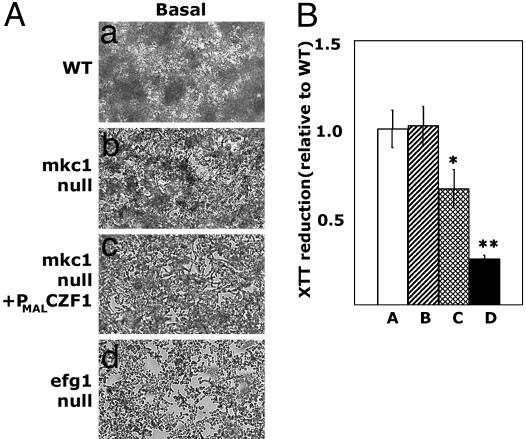
Ectopic expression of CZF1 antagonizes biofilm development. (A) Biofilms formed by CKY101 (MKC1+), CKY357 (mkc1-null), CKY355 (mkc1/mkc1/PMAL-CZF1), and CKY136 (efg1/efg1) cells were washed and photographed with the 10× objective. (a and b) Individual cells are indistinct because of the 3D nature of biofilms. (c and d) Individual cells adherent to the plastic are easily seen because a well developed biofilm is not present. (B) XTT reduction activity of adherent cells grown as in A was determined as described in Materials and Methods. The average value for each strain relative to WT and the standard deviation is shown. Statistically significant differences are indicated by asterisks. *, P < 0.02 by using Student's t test with a two-tailed distribution; **, P < 0.0003 by using Student's t test with a two-tailed distribution. Bar A, CKY101 (MKC1+); bar B, CKY357 (mkc1-null); bar C, CKY355 (mkc1/mkc1/PMAL-CZF1); and bar D, CKY136 (efg1/efg1).
During growth on semisolid medium, the DNA-binding protein Czf1p relieves repression mediated by Efg1p and promotes invasive filamentation (10). Ectopic expression of Czf1p with a construct encoding CZF1 controlled by the promoter of the C. albicans maltase gene (PMAL-CZF1) restored filamentation in the absence of Mkc1p (Fig. 2). However, expression of CZF1 did not bypass all defects of the mkc1-null mutant: Nikkomycin Z sensitivity was unchanged by the introduction of PMAL-CZF1 (Table 1), demonstrating that CZF1 was not substituting for MKC1. These results suggest that, during growth on a semisolid surface, Mkc1p acts before Czf1p or in a parallel pathway, consistent with the above model.
To determine whether ectopic expression of CZF1 would similarly bypass defects in biofilm development caused by the absence of Mkc1p, CKY355 (mkc1/mkc1/PMAL-CZF1) cells were incubated in polystyrene wells. After 2 days of growth, the biofilm formed by this strain was less extensive than a WT biofilm (Fig. 5A) and showed decreased XTT reduction activity (P < 0.02) (Fig. 5B), indicating lower numbers of cells (21). Because XTT reduction activity reflects numbers of metabolically active cells rather than the total amount of biomaterial in the biofilm, a difference between WT and mkc1-null biofilms was not observed (Fig. 5B). Therefore, in the mkc1-null mutant, ectopically expressed Czf1p antagonized biofilm formation. Thus, although biofilm formation and invasive filamentation are both contact-dependent responses signaled by Mkc1p, Czf1p functions in distinct ways in the two processes.
Discussion
The conclusion from these studies is that C. albicans uses the cell integrity pathway to mediate multiple contact-dependent responses. During growth on a semisolid surface, activation of the cell integrity pathway leads to invasion of the underlying medium. This behavior is repressed by Efg1p and promoted by Czf1p. On a solid surface, activation of the cell integrity pathway contributes to biofilm structure and expression of the drug-resistant phenotype. Biofilm formation is promoted by Efg1p and antagonized by Czf1p. Because invasive filamentation and biofilm formation are distinct processes occurring on different types of surfaces, C. albicans cells must sense more than one feature of the surface to determine its nature, so that the correct response is produced. By integrating information received from multiple signaling pathways, the response of the organism to its environment can be finely tuned.
The demonstration that ectopic expression of Czf1p restores invasive filamentation in an mkc1-null mutant strain suggests that Mkc1p acts upstream of Czf1p. Because Czf1p relieves repression of filamentation by Efg1p (10), Mkc1p may regulate invasive filamentation by phosphorylating either Czf1p or Mkc1p, contributing to relief of repression. Mkc1p may also affect derepression of filamentation indirectly by phosphorylating a target protein that affects the activity of Efg1p or Czf1p.
An important task for a pathogenic microorganism is to determine its location within the host. In the case of C. albicans, the organism may be found on mucous membranes or skin or within the blood stream or visceral organs. Results presented here demonstrate that, without Mkc1p, the response of cells to the presence of a surface is aberrant, and thus the cell integrity pathway is a source of important information that allows monitoring of the immediate environment. A null mutant lacking Mkc1p exhibits reduced virulence (25), suggesting that virulence of the organism is associated with the ability to initiate contact-dependent responses.
The ability to sense and respond to surface contact is wide-spread in biology. Contact-dependent regulatory mechanisms, such as anoikis, ensure that only the correct cells divide and help bring about the controlled proliferation that is essential for metazoan development. Loss of contact-dependent regulation is associated with cell transformation, which leads to uncontrolled proliferation and invasive growth of transformed cells into tissues. As shown here, the pathogen C. albicans shares with its host the ability to use sensing of contact to activate signaling pathways that control cellular responses.
Acknowledgments
I thank Federico Navarro-Garcia for gifts of strains and helpful discussion; Paula Watnick, Deborah Zucker, Peter Kraus, David Levin, Perry Riggle, Marcelo Vinces, Xi Chen, and Igor Bruzual for helpful discussions; Irene Kaplow, Sarah Jane White, and Alana Benjamin for technical assistance; and Dean Dawson, Joan Mecsas, Cathy Squires, Andrew Wright, and Ralph Isberg for comments on the manuscript. This work was supported by National Institutes of Health Grant AI038591 (to C.A.K.).
Author contributions: C.K. designed research, performed research, analyzed data, and wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: MAP, mitogen-activated protein; HA, hemagglutinin; YPD, yeast extract/peptone/dextrose; YPS, yeast extract/peptone/sucrose; SD medium, synthetic defined medium; CM, complete medium; log phase, logarithmic phase; CLSI, Clinical and Laboratory Standards Institute; XTT, [2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide].
References
- 1.Ostrosky-Zeichner, L., Rex, J. H., Bennett, J. & Kullberg, B. J. (2002) Infect. Dis. Clin. North Am. 16, 821–835. [DOI] [PubMed] [Google Scholar]
- 2.Brown, D. H., Jr., Giusani, A. D., Chen, X. & Kumamoto, C. A. (1999) Mol. Microbiol. 34, 651–662. [DOI] [PubMed] [Google Scholar]
- 3.Douglas, L. J. (2003) Trends Microbiol. 11, 30–36. [DOI] [PubMed] [Google Scholar]
- 4.Mukherjee, P. K., Chandra, J., Kuhn, D. M. & Ghannoum, M. A. (2003) Infect. Immun. 71, 4333–4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies, J. M., Stacey, A. J. & Gilligan, C. A. (1999) FEMS Microbiol. Lett. 171, 245–249. [DOI] [PubMed] [Google Scholar]
- 6.Migliaccio, F. & Piconese, S. (2001) Trends Plant Sci. 6, 561–565. [DOI] [PubMed] [Google Scholar]
- 7.Hoch, H. C., Staples, R. C., Whitehead, B., Comeau, J. & Wolf, E. D. (1987) Science 235, 1659–1662. [DOI] [PubMed] [Google Scholar]
- 8.Liu, H. (2002) Int. J. Med. Microbiol. 292, 299–311. [DOI] [PubMed] [Google Scholar]
- 9.Riggle, P. J., Andrutis, K. A., Chen, X., Tzipori, S. R. & Kumamoto, C. A. (1999) Infect. Immun. 67, 3649–3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giusani, A. D., Vinces, M. & Kumamoto, C. A. (2002) Genetics 160, 1749–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamada, Y., Jung, U. S., Piotrowski, J. & Levin, D. E. (1995) Genes Dev. 9, 1559–1571. [DOI] [PubMed] [Google Scholar]
- 12.Heinisch, J. J., Lorberg, A., Schmitz, H. P. & Jacoby, J. J. (1999) Mol. Microbiol. 32, 671–680. [DOI] [PubMed] [Google Scholar]
- 13.Navarro-Garcia, F., Sanchez, M., Pla, J. & Nombela, C. (1995) Mol. Cell. Biol. 15, 2197–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Navarro-Garcia, F., Alonso-Monge, R., Rico, H., Pla, J., Sentandreu, R. & Nombela, C. (1998) Microbiology 144, 411–424. [DOI] [PubMed] [Google Scholar]
- 15.Fonzi, W. A. & Irwin, M. Y. (1993) Genetics 134, 717–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cannon, R. D., Jenkinson, H. F. & Shepherd, M. G. (1990) Mol. Gen. Genet. 221, 210–218. [DOI] [PubMed] [Google Scholar]
- 17.Longtine, M. S., McKenzie, A., III, Demarini, D. J., Shah, N. G., Wach, A., Brachat, A., Philippsen, P. & Pringle, J. R. (1998) Yeast 14, 953–961. [DOI] [PubMed] [Google Scholar]
- 18.Ausubel, F., Brent, R., Kingston, R., Moore, D., Seidman, J., Smith, J. & Struhl, K. (1989) Current Protocols in Molecular Biology (Wiley, New York).
- 19.Miller, J. H. (1972) Experiments in Molecular Genetics (Cold Spring Harbor Lab. Press, Plainview, NY).
- 20.Martin, H., Rodriguez-Pachon, J. M., Ruiz, C., Nombela, C. & Molina, M. (2000) J. Biol. Chem. 275, 1511–1519. [DOI] [PubMed] [Google Scholar]
- 21.Ramage, G., Vande Walle, K., Wickes, B. L. & Lopez-Ribot, J. L. (2001) Antimicrob. Agents Chemother. 45, 2475–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.National Committee for Clinical Laborator y Standards (1997) Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts: Approved Standard M27-A (Natl. Committee Clin. Lab. Standards, Wayne, PA).
- 23.Kraus, P. R., Fox, D. S., Cox, G. M. & Heitman, J. (2003) Mol. Microbiol. 48, 1377–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramage, G., VandeWalle, K., Lopez-Ribot, J. L. & Wickes, B. L. (2002) FEMS Microbiol. Lett. 214, 95–100. [DOI] [PubMed] [Google Scholar]
- 25.Diez-Orejas, R., Molero, G., Navarro-Garcia, F., Pla, J., Nombela, C. & Sanchez-Perez, M. (1997) Infect. Immun. 65, 833–837. [DOI] [PMC free article] [PubMed] [Google Scholar]