Abstract
Several of the polychlorinated biphenyls (PCBs), i.e. the dioxin-like PCBs, are known to induce the P450 enzymes CYP1A1, CYP1A2 and CYP1B1 by activating the aryl hydrocarbon receptor (Ah)-receptor. We evaluated if circulating levels of PCBs in a population sample were related to genetic variation in the genes encoding these CYPs. In the population-based Prospective Investigation of the Vasculature in Uppsala Seniors (PIVUS) study (1016 subjects all aged 70), 21 SNPs in the CYP1A1, CYP1A2 and CYP1B1 genes were genotyped. Sixteen PCB congeners were analysed by high-resolution chromatography coupled to high-resolution mass spectrometry (HRGC/HRMS). Of the investigated relationships between SNPs in the CYP1A1, CYP1A2 and CYP1B1 and six PCBs (congeners 118, 126, 156, 169, 170 and 206) that captures >80% of the variation of all PCBs measured, only the relationship between CYP1A1 rs2470893 was significantly related to PCB118 levels following strict adjustment for multiple testing (p=0.00011). However, there were several additional SNPs in the CYP1A2 and CYP1B1 that showed nominally significant associations with PCB118 levels (p-values in the 0.003–0.05 range). Further, several SNPs in the CYP1B1 gene were related to both PCB156 and PCB206 with p-values in the 0.005–0.05 range. Very few associations with p<0.05 were seen for PCB126, PCB169 or PCB170. Genetic variation in CYP1A1 was related to circulating PCB118 levels in the general elderly population. Genetic variation in CYP1A2 and CYP1B1 might also be associated with other PCBs.
Keywords: Gene, SNP, CYP, PCB, Metabolism, Epidemiology
1. Introduction
The cytochrome P450 superfamily (CYPs) includes a number of enzymes involved in the metabolism of xenobiotics and endogenous steroid hormones. One important feature of these enzymes is that the activity could be enhanced by high levels of the substrates, initiating a more rapid metabolism of the substrates in order to inactivate the biological action of the substrate. This is an important defence system against any deleterious actions of xenobiotics.
The polychlorinated biphenyls (PCBs) are a number of compounds heavily used in the middle of the last century. Due to their toxic properties regarding reproductive problems observed in animals, they were banned in the 70's and 80's in most high-income countries. Since many of the PCBs are highly lipophilic and accumulate in adipose tissue their estimated intrinsic human elimination half-lives are2–15 years (Ritter etal.,2011). Thus, although the major routes of exposure no longer are present, the commonly used PCBs are still measurable in the circulation in individuals, although they are gradually declining (Knobeloch et al., 2009; Schade and Heinzow, 1998).
Activation of the aryl hydrocarbon receptor(Ah)-receptor (AHR) is a potent way to induce the P450 enzymes CYP1A1, CYP1A2 and CYP1B1 (Poland etal.,1976,1989). The most potent activator of AHR is 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), but some co-planar, non-ortho PCBs also act as agonists of this receptor (Hennig et al., 2002; Safe et al., 1985; Shimada et al., 2002). Thus, it is likely that some PCBs could induce the enzyme activity of the CYP1A1, CYP1A2 and CYP1B1 genes and thereby enhance their own metabolism. In humans, CYP1A1 is mainly expressed in extra-hepatic cells, such as the lung and lymphocytes, while the CYP1A2 enzyme mainly is found in the liver (Schweikl et al., 1993; Shimada et al.,1992). The CYP1B1 enzyme was identified more recently and CYP1B1 is expressed in a large number of tissues, including the liver (Tang et al., 1996).
The associations between PCBs and P450 enzyme activities have only been studied in the experimental setting, and several single nucleotide polymorphisms (SNPs) in the P450 system have been discovered to influence the activity of different enzymes (McGraw and Waller, 2012). Therefore, we used data from the Prospective Investigation of the Vasculature in Uppsala Seniors (PIVUS) study (Lind et al., 2005) to study if SNPs in genes encoding the P450enzymes CYP1A1, CYP1A2 and CYP1B were related to circulating levels of PCBs in men and women.
2. Materials and methods
2.1. Subjects
Eligible subjects were all men and women aged 70 and lived in the community of Uppsala, Sweden at enrolment between April 2001 and June 2004. The subjects were randomly chosen from the national register of residence. A total of 2025 individuals were invited and 1016 subjects participated (participation rate of 50.1%) of which 50.2% were women. Details of the PIVUS study have been presented (Lind et al., 2005). The study was approved by the Ethics Committee of the University of Uppsala and all the participants gave their informed consent prior to the study.
The study subjects were investigated in the morning between 8 and 10am after an overnight fast. No medication or smoking was allowed after midnight. Blood samples were taken through an arterial cannula inserted in the brachial artery. The blood samples for PCB analyses were stored at -70°C until analysis while lipid variables were measured the same day as blood was drawn. The lipid variables HDL-cholesterol and triglycerides were measured on an Architect Ci8200 analyser (Abbott Laboratories, Abbott Park, Ill., USA). The participants were asked to answer a questionnaire about their medical history, education level, exercise habits, smoking habits and regular medication.
Approximately 10% of the cohort reported a history of coronary heart disease, 4% reported stroke and 9% reported diabetes mellitus. Almost half the cohort reported some sort of cardiovascular medication (45%), with antihypertensive medication being the most prevalent (32%). Fifteen per cent reported use of statins, while insulin and oral antiglycemic drugs were reported in 2 and 6%, respectively. Eleven per cent of the study subjects were current smokers. Further details on regular drug intake, basic characteristics and cardiovascular risk factors are presented elsewhere (Lind et al., 2005).
2.2. PCB analyses
PCBs were measured in stored serum samples collected at enrolment. Analyses of POPs were performed using a Micromass Autospec Ultima (Waters, Milford, MA, USA) high-resolution gas chromatography coupled to a high resolution mass spectrometry (HRGC/HRMS) system based on the method by Sandau and co-workers with some modifications (Sandau et al., 2003). A total of 16 polychlorinated biphenyls (PCBs) congeners were evaluated. The evaluated PCBs were those where we found detectable levels in >90% of the population. An established summation formula based on total serum cholesterol and serum triglyceride concentrations was used to calculate the total amount of lipids in each plasma sample (Rylander et al., 2006). Thereafter the wet-weight concentrations of the POPs were divided by this estimation of lipids to obtain lipid-normalized values (i.e. POPs values given in ng/g lipid) which was used in the analyses. Median values and interquartile ranges (IQR) for the POPs are given in Table1. A more detailed description of the POP analysis in this sample has previously been presented (Salihovic et al., 2012).
Table 1.
Distribution of the polychlorinated biphenyls (PCBs).
| Variable | N | Mean (SD) | Median (25th and 75th percentile) |
|---|---|---|---|
| PCB126 | 982 | 8.4 (7.5) | 6.2 (3.3, 10.8) |
| PCB169 | 982 | 27.7 (11.3) | 26 (20.1, 34.2) |
| PCB118 | 987 | 34.1 (20.1) | 30 (21.6, 41.7) |
| PCB156 | 988 | 24.9 (9.7) | 23.6 (18.2, 29.9) |
| PCB170 | 988 | 80 (31.3) | 74.8 (59.6, 96.3) |
| PCB206 | 988 | 4.4 (1.9) | 4.2 (3.1, 5.4) |
Mean and SD as well as median and 25th and 75th percentile are given for the investigated PCBs (ng/g lipid), since the persistent organic pollutants are skewed towards high levels.
We have previously found that levels of some low-chlorinated PCBs are highly correlated forming a cluster, and a set of high-chlorinated PCBs are highly correlated forming another cluster, while the PCBs 126 and PCB169 were not related to any of those clusters (Lampa et al., 2012). Therefore, we performed the statistical analysis of only 6PCBs that acted as markers for the exposure, out of the 16 measured PCBs (the dioxin-like congeners 118, 126, 156, 169, and congeners 170 and 206) to reduce the risk of false positive findings due to multiple testing. PCB118 represented the low-chlorinated cluster whereas PCBs 156, 170 and 206 represented the high-chlorinated cluster. The variation in the six selected PCBs represents >80% of the variation in the total set of the 16 measured PCBs (Lampa et al., 2012).
2.3. Genotyping
A total of 21 SNP in the CYP1A1, CYP1A2 and CYP1B genes were genotyped using the Illumina Omni Express and a custom Illumina Golden Gate array at the SNP&SEQ Technology Platform at Uppsala University (www.genotyping.se). Sample exclusion criteria included: (1) genotype call rate <95%; (2) heterozygosity >3 standard deviations (SD); (3) gender discordance; (4) duplicated samples; and (5) no identity-by-descent match. SNP exclusion criteria included: (1) monomorphic SNPs; (2) Hardy–Weinberg equilibrium (HWE) p-value <1x10-6; (3) genotype call rate <0.99 for SNPs with minor allele frequency ((MAF) <5%) or <0.95 for SNPs with MAF ≥5%.
The average call-rate per SNP was 99.06% (♯ of no calls: 47; ♯ of calls: 4950). No SNPs showed a HWE chi-squared >3.84, presenting no deviation from the HWE assumption, i.e. that allele and genotype frequencies remain constant from generation to generation, and no indication of genotyping errors. A 100% reproducibility was seen (0 duplicate conflicts in 119 duplicate tests), and a 100% consistent inheritance was found (0 inheritance conflicts in 30 inheritance tests).
Using the European 1000 Genomes genotypes as reference (population codes: CEU, FIN, GBR, IBS, TSI), the gene regions for CYP1A1 (of length 21.1kb), CYP1A2 (22.3 kb) and CYP1B1 (23.7kb) contained 14, 31 and 61 SNPs with MAF >5% respectively. The SNPs genotyped in this study could tag 7 of the 14 SNPs in CYP1A1, 15 of the 31 in CYP1A2 and 47 of the 61 SNPs in CYP1B1 at r2>0.8. All SNPs genotyped in the study were present in the 1000 Genomes data.
2.4. Statistics
Non-normally distributed variables, i.e. the lipid-normalized PCBs, were transformed using the natural logarithm before analysis. Relationships between the CYP SNPs and the 6 PCBs were evaluated by linear regression analysis with a PCB congener as dependent variable adjusting for gender. To evaluate any effect modification by gender, each model with significant associations between SNP and a PCB were rerun including an interaction term between gender and the SNP. Also, for significant associations between SNP and a PCB, the models were rerun including (i) smoking as a confounder (ii) an interaction term between smoking and SNP to evaluate whether results were robust regarding confounding and effect modification. Each SNP was coded as 0, 1 or 2 copies of the minor allele assuming additive allele effect. All combinations of SNP and PCB congeners were evaluated in separate multi-variable models. Since 6x21 tests were performed, we adjusted the critical p-value accordingly using the Bonferroni correction (0.05/126=0.000397). The use of this strict threshold, not taking into account the high correlations of some of the SNPs within the same LD blocks which would warrant a more liberal threshold, was motivated by the lack of suitable replication samples (see further in the discussion). STATA (version 11; StataCorp, College Station, TX, USA) was used for calculations.
3. Results
Medians and IQR for the six investigated PCBs are given in Table 1. The PCB values are lipid-normalized.
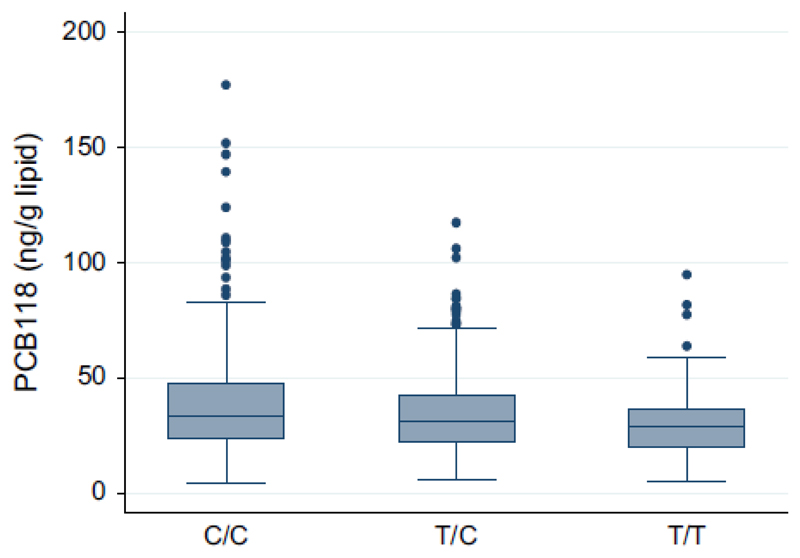
Of the investigated relationships between SNPs in the CYP1A1, CYP1A2 and CYP1B1 genes and the six PCBs, only the relationship between rs2470893 in the CYP1A1 and circulating levels of PCB118 was significant following strict adjustment for multiple testing (p=0.00011, Table 2). Details on the association between PCB118 and rs2470893 in the CYP1A1 gene are shown in Figure 1. The allele distribution for rs2470893 was very similar between genders with the proportion of the number of copies of the minor allele being 44, 46 and 9% for women and 45, 44 and 11% for men. Inclusion of an interaction term between gender and rs2470893 did not change any point estimates and the term was highly non-significant (p=0.96). Evaluation of smoking as a confounder or effect modifier did not show any influence of smoking on the association between PCB118 and rs2470893 as results were stable with identical point estimates of both rs2470893 and gender up until the third decimal when smoking was included as a confounder and a non-significant interaction term, respectively (data not shown). In addition, many associations showed p-values in the 0.05–0.003 range and it could be noted that also SNPs in the CYP1A2 and CYP1B1 genes showed such associations versus PCB118 (Table 2).
Table 2.
Relationships between SNPs and polychlorinated biphenyls (PCBs) 118, 126 and 156.
| SNP | MAF | PCB118 | PCB126 | PCB156 | |||
|---|---|---|---|---|---|---|---|
| Beta (95% CI) | P-value | Beta (95% CI) | P-value | Beta (95% CI) | P-value | ||
| CYP1A1 | |||||||
| rs6495121 | 0.14 | 0.0039 (−0.066, 0.074) | 0.91 | 0.037 (−0.083, 0.16) | 0.55 | −0.034 (−0.086, 0.018) | 0.20 |
| rs1048943 | 0.03 | 0.011 (−0.11, 0.14) | 0.86 | 0.17 (−0.048, 0.38) | 0.13 | −0.037 (−0.13, 0.057) | 0.44 |
| rs1799814 | 0.04 | −0.056 (−0.17, 0.063) | 0.36 | −0.10 (−0.30, 0.10) | 0.33 | −0.071 (−0.16, 0.017) | 0.11 |
| rs4646421 | 0.10 | 0.012 (−0.069, 0.094) | 0.77 | 0.11 (−0.028, 0.25) | 0.12 | −0.023 (−0.083, 0.038) | 0.47 |
| rs2470893 | 0.33 | −0.10 (−0.15, −0.051) | 0.00011 | −0.056 (−0.14, 0.032) | 0.21 | −0.017 (−0.056, 0.021) | 0.38 |
| CYP1A2 | |||||||
| rs2472299 | 0.27 | 0.056 (0.00042, 0.11) | 0.049 | 0.069 (−0.025, 0.16) | 0.15 | 0.0068 (−0.034, 0.048) | 0.74 |
| rs2472304 | 0.34 | 0.048 (−0.0043, 0.10) | 0.072 | 0.084 (−0.0049, 0.17) | 0.064 | −0.012 (−0.051, 0.026) | 0.53 |
| rs11854147 | 0.33 | 0.047 (−0.0021, 0.095) | 0.061 | 0.066 (−0.017, 0.15) | 0.12 | 0.0022 (−0.035, 0.039) | 0.91 |
| rs3743484 | 0.05 | 0.014 (−0.09, 0.12) | 0.79 | 0.067 (−0.11, 0.25) | 0.47 | −0.032 (−0.11, 0.048) | 0.44 |
| rs16972208 | 0.01 | −0.022 (−0.22, 0.18) | 0.83 | 0.048 (−0.29, 0.39) | 0.78 | −0.018 (−0.17, 0.13) | 0.82 |
| CYP1B1 | |||||||
| rs1157185 | 0.42 | −0.0091 (−0.057, 0.039) | 0.71 | 0.045 (−0.037, 0.13) | 0.28 | 0.045 (0.0094, 0.081) | 0.014 |
| rs163078 | 0.35 | −0.045 (−0.094, 0.0044) | 0.075 | −0.043 (−0.13, 0.041) | 0.31 | −0.043 (−0.080, −0.0055) | 0.025 |
| rs9341266 | 0.03 | 0.055 (−0.069, 0.18) | 0.38 | 0.098 (−0.11, 0.31) | 0.36 | 0.046 (−0.046, 0.13) | 0.33 |
| rs162562 | 0.24 | −0.030 (−0.086, 0.025) | 0.28 | −0.064 (−0.16, 0.03) | 0.18 | −0.038 (−0.079, 0.0028) | 0.068 |
| rs1800440 | 0.18 | −0.036 (−0.10, 0.027) | 0.27 | −0.056 (−0.16, 0.051) | 0.31 | −0.0058 (−0.052, 0.041) | 0.81 |
| rs1056836 | 0.42 | 0.020 (−0.029, 0.069) | 0.42 | −0.016 (−0.099, 0.067) | 0.71 | −0.031 (−0.067, 0.0055) | 0.097 |
| rs162561 | 0.20 | −0.027 (−0.086, 0.032) | 0.38 | −0.026 (−0.13, 0.076) | 0.62 | −0.017 (−0.060, 0.027) | 0.46 |
| rs4646430 | 0.31 | 0.0082 (−0.043, 0.059) | 0.75 | 0.066 (−0.023, 0.15) | 0.15 | 0.054 (0.016, 0.092) | 0.0052 |
| rs162556 | 0.48 | −0.068 (−0.12, −0.021) | 0.0050 | −0.080 (−0.16, 0.0022) | 0.057 | −0.039 (−0.075, −0.0036) | 0.031 |
| rs162555 | 0.17 | 0.096 (0.033, 0.16) | 0.0028 | 0.021 (−0.087, 0.13) | 0.70 | −0.0026 (−0.050, 0.044) | 0.91 |
| rs10175368 | 0.31 | 0.0068 (−0.045, 0.058) | 0.79 | 0.067 (−0.021, 0.15) | 0.14 | 0.051 (0.013, 0.089) | 0.0085 |
Relationships between SNPs in the CYP1A1, CYP1A2, and CYP1B1 genes and circulating levels of PCB118, PCB126 and PCB156 given as the regression coefficient (beta) in linear regression models. PCB levels were transformed using the natural logarithm and then lipid-normalized before analysis. MAF= Minor allele frequency.
Figure 1.
Levels of polychlorinated biphenyl (PCB) 118 in the SNP CYP1A1 rs2470893 genotypes. Medians and interquartile range for PCB118 levels in the different genotypes for rs2470893 in the CYP1A1 gene (n=424 for C/C, n=426 for T/C and n=97 for T/T, p=0.00011 for relationship between the genotype and PCB118 levels).
It could also be noted that several SNPs in the CYP1B1 gene were related to both PCB156 and PCB 206 with p-values in the 0.05–0.005 range. Very few associations with p<0.05 were seen for PCB126, PCB169 or PCB170 (Tables 2 and 3).
Table 3.
Relationships between SNPs and polychlorinated biphenyls (PCBs) 169, 170 and 206.
| SNP | MAF | PCB169 | PCB170 | PCB206 | |||
|---|---|---|---|---|---|---|---|
| Beta (95%CI) | P-value | Beta(95%CI) | P-value | Beta(95%CI) | P-value | ||
| CYP1A1 | |||||||
| rs6495121 | 0.14 | −0.024 (−0.084, 0.035) | 0.43 | −0.026 (−0.075, 0.023) | 0.29 | −0.024 (−0.083, 0.034) | 0.41 |
| rs1048943 | 0.03 | 0.02 (−0.086, 0.13) | 0.71 | 0.012 (−0.075, 0.10) | 0.78 | −0.012(−0.12, 0.092) | 0.82 |
| rs1799814 | 0.04 | 0.011 (−0.089, 0.11) | 0.83 | −0.018 (−0.10, 0.064) | 0.67 | −0.022 (−0.12, 0.076) | 0.66 |
| rs4646421 | 0.1 | −0.037 (−0.11, 0.032) | 0.30 | −0.028 (−0.084, 0.029) | 0.33 | −0.025 (−0.09, 0.043) | 0.47 |
| rs2470893 | 0.33 | −0.0022 (−0.046, 0.041) | 0.92 | −0.0083 (−0.044, 0.028) | 0.65 | −0.022 (−0.065, 0.021) | 0.31 |
| CYP1B1 | |||||||
| rs2472299 | 0.27 | −0.0032 (−0.050, 0.044) | 0.89 | 0.0074 (−0.031, 0.046) | 0.71 | 0.039 (−0.0071, 0.085) | 0.097 |
| rs2472304 | 0.34 | −0.022 (−0.066, 0.022) | 0.33 | −0.012 (−0.048, 0.025) | 0.53 | 0.014 (−0.029, 0.058) | 0.51 |
| rs11854147 | 0.33 | 0.0052 (−0.036, 0.046) | 0.81 | 0.0064 (−0.028, 0.041) | 0.72 | 0.030 (−0.011, 0.070) | 0.16 |
| rs3743484 | 0.05 | −0.0038 (−0.093, 0.086) | 0.93 | 0.0020 (−0.073, 0.077) | 0.96 | 0.0017 (−0.087, 0.091) | 0.97 |
| rs16972208 | 0.01 | −0.0064 (−0.18, 0.16) | 0.94 | −0.0025 (−0.15, 0.14) | 0.97 | −0.054 (−0.22, 0.12) | 0.54 |
| CYP1B1 | |||||||
| rs1157185 | 0.42 | 0.017 (−0.023, 0.058) | 0.40 | 0.031 (−0.0027, 0.065) | 0.072 | 0.046 (0.0059, 0.087) | 0.025 |
| rs163078 | 0.35 | −0.021 (−0.06, 0.021) | 0.33 | −0.035 (−0.070, 0.00001) | 0.050 | −0.055 (−0.097, −0.014) | 0.0089 |
| rs9341266 | 0.03 | 0.0086 (−0.096, 0.11) | 0.87 | 0.022 (−0.064, 0.11) | 0.62 | −0.0030 (−0.11, 0.10) | 0.95 |
| rs162562 | 0.24 | −0.017 (−0.064, 0.030) | 0.48 | −0.032 (−0.07, 0.0067) | 0.11 | −0.043 (−0.089, 0.0028) | 0.066 |
| rs1800440 | 0.18 | −0.0047 (−0.058, 0.048) | 0.86 | −0.0087 (−0.052, 0.035) | 0.70 | −0.018 (−0.070, 0.034) | 0.49 |
| rs1056836 | 0.42 | −0.0056 (−0.047, 0.036) | 0.79 | −0.023 (−0.056, 0.011) | 0.19 | −0.033 (−0.074, 0.0071) | 0.11 |
| rs162561 | 0.2 | −0.0062 (−0.056, 0.044) | 0.81 | −0.015 (−0.056, 0.026) | 0.46 | −0.040 (−0.089, 0.0092) | 0.11 |
| rs4646430 | 0.31 | 0.021 (−0.022, 0.064) | 0.34 | 0.036 (0.00051, 0.071) | 0.047 | 0.058 (0.02, 0.10) | 0.0077 |
| rs162556 | 0.48 | −0.034 (−0.075, 0.0064) | 0.10 | −0.036 (−0.070, −0.0034) | 0.031 | −0.055 (−0.094, −0.015) | 0.0069 |
| rs162555 | 0.17 | −0.0035 (−0.057, 0.050) | 0.90 | 0.0062 (−0.038, 0.050) | 0.78 | 0.019 (−0.033, 0.072) | 0.48 |
| rs10175368 | 0.31 | 0.026 (−0.018, 0.069) | 0.24 | 0.033 (−0.0021, 0.069) | 0.066 | 0.059 (0.017, 0.10) | 0.0065 |
Relationships between SNPs in the CYP1A1, CYP1A2, and CYP1B1 genes and circulating levels of PCB169, PCB170 and PCB206 given as the regression coefficient (beta) in linear regression models. MAF= Minor allele frequency.
4. Discussion
4.1. Main findings
The present study showed that a SNP in the CYP1A1 gene was significantly related to the circulating concentration of PCB118. In addition, there were several additional SNPs in the CYP1A2 and CYP1B1 genes that showed nominally significant associations with PCB118 levels (p-values in the 0.003–0.05 range). Furthermore, several SNPs in the CYP1B1 gene were related to both PCB156 and PCB 206 levels with p-values in the 0.05–0.005 range, being non-significant following multiple comparisons. Very few associations with p<0.05 were seen for PCB126, PCB169 or PCB170 concentrations.
4.2. Comparisons with the literature
The cytochrome P450 CYP1A1, CYP1A2 and CYP1B1 are xenobiotic metabolising enzymes playing an important role in e.g. the detoxification of xenobiotics such as PCBs (Nebert and Dalton, 2006). CYP1A1 has been suggested as a leading genetic candidate that may influence susceptibility to PCBs (Laden et al., 2002). Different alleles may give rise to different proteins which may affect the function of the produced enzyme and alter the carriers phenotype (Kumar et al., 2009). To the best of our knowledge, this is one of the first studies in which circulating levels of PCBs have been related to variation in the CYP1A1, CYP1A2 and CYP1B1 genes in humans. Previous experimental studies have suggested that PCBs could induce enzyme activities in these genes (Roy et al., 2011; Yamazaki et al., 2011), possibly by activation of the AHR (Hennig et al.,2002; Safe etal., 1985; Shimada et al., 2002). However, other studies have failed to see an association between the allele frequency for SNPs in CYP1A1 and CYP1B1 and any effects on expression or activation of the corresponding enzymes although this might be due to small sample sizes (van Duursen et al., 2005). Significant difference in serum PCB153 levels according to CYP1A1 polymorphism in rs1048943 and in CYP1B1 rs1056836 has been reported in Inuits (Ghisari et al., 2013) which was not repeated for those SNPs in this study. CYP1A1 is known to be polymorphic and several SNPs has been identified of which two common SNPs have been found to be functionally significant; rs4646903 and the missense rs1048943 (Fang et al., 2014). However, these two SNPs are in low linkage disequilibrium (LD) with the top SNP of this study, rs2470893 (r2=0.05 and r2=0.02, respectively). In addition, the CYP1A1 gene has been associated with diastolic and systolic blood pressure with rs1378942 as the top SNP (International Consortium for Blood Pressure Genome-Wide Association et al., 2011; Newton-Cheh et al., 2009). This SNP which has low LD with our top SNP(r2=0.16), was also investigated in this study with no detected association with the PCB levels. There is scarce information available on the SNP rs2470893. However, it is a synonymous (silent) variant (i.e. believed to not affect protein sequence) located between CYP1A1 and CYP1A2 and it has been associated with coffee consumption in two recent prior genome-wide association studies (Amin et al., 2012; Cornelis et al., 2011). This SNP has been found to be in strong LD with another SNP, rs2472297, in the same region (Amin et al., 2012), although the magnitude of the LD may vary between populations (Josse et al., 2012). Further, this SNP is in LD with several other SNPs (rs35107470, rs12910558 and rs12910841, r2=0.44–0.53) of which rs12919841 is located in an intron. Traditionally, non-synonymous single nucleotide polymorphism occurring in a coding gene has been considered the explanation to the production of a different protein and altering of the phenotype of the host organism (Kumar et al., 2009). However, there is growing evidence that synonymous single nucleotide variants affect protein splicing, expression and function (Edwards et al., 2012; Sauna and Kimchi-Sarfaty, 2011), and some of these variants contribute to disease, for example cancer, diabetes and liver disease (Sauna and Kimchi-Sarfaty, 2011). Therefore, additional studies are needed to replicate the findings of this study and to investigate the role of synonymous variants in general and rs2470893 in particular, on genetic function and physiological/biological consequences.
Tobacco smoking is a known inducer of CYP1A1 enzyme (Hakkola et al., 1997) and has been reported as the major effect modifier when evaluating the relationship between the assessed CYP1A1 activity in the placenta and PCB153 (Pereg et al., 2002). However, we did not see any influence of tobacco smoking on the relationship between the main SNP and PCB118 when adding smoking status as a confounder or including an interaction term in the model (data not shown). This discrepancy may be explained by the different tissues being examined in the different studies and the different age groups of study subjects.
4.3. Strengths and limitations of the study design
The major strength of the study is the large number of subjects with measurements of PCBs at the same age, which increases the likelihood to disclose true relationships.
In the ideal study on the topic of the role of CYP induction on the metabolism of a xenobiotic, such as PCB118, a fixed dose of the compound should be given at a certain time point and the circulating concentrations of the compounds should be evaluated thereafter in a standardized fashion. This is obviously not the case in the present study. Therefore, the associations of the concentrations of the different PCBs and the different SNPs are highly dependent on the amount and timing of the exposure to the PCBs in relation to the measurement of the compound. The consequence of this is that an epidemiological study like this, with limited information on the amount and timing of the exposure, could never rule out an association of a compound and a SNP in a P450 enzyme. Only the identified associations could be taken as evidence that a certain P450 enzyme is of importance for the metabolism of that particular compound.
There was an indication of an association between several SNPs in the CYP1B1 gene and PCB156 and PCB 206 with p-values in the 0.05–0.005 range, however, the p-values were below the Bonferroni-adjusted level of statistical significance (p<0.000397). Thus, it cannot be concluded from the present study if these relationships represent valid relationships. Since we are not aware of any other population-based studies with PCB measurements and DNA, we cannot replicate the present findings in an independent cohort. Hence, we acknowledge the lack of replication as an important limitation, but believe that the study still is worthwhile due to the high a priori probability for involvement of CYPs in the metabolism of POPs.
4.4. AHR binding affinity and CYP induction
There is strong evidence that the induction of CYP1A1 gene is mediated through the AHR receptor (Nebert and Jones, 1989). According to previous experimental studies (Hennig et al., 2002; Safe et al.,1985; Shimada et al., 2002), it would be suspected that PCBs with a high affinity to the AHR (i.e. the dioxin-like PCBs), like PCB126 and PCB169, would show the closest relationship versus any functional SNP in the CYP1A1, CYP1A2 and CYP1B1 genes. This was however not the case, and PCB118 is only a rather weak agonist to the AHR (Hestermann et al., 2000). PCB118 is one of the most significant congeners in humans in terms of prevalence and magnitude of concentration (Kang et al.,2008; Rudge et al., 2012) and as such may act as a marker for all mono-ortho coplanar PCBs (i.e. PCBs 105, 118, 156, 157 and 189). However, for this study, two congeners were selected to act as markers for this group (PCB118 and PCB156) based on the known covariation of the different PCBs in this study population (Lampa et al., 2012). It has also been suggested that PCB118 may be used as marker for PCBs 105and 126, and PCB153 as marker for PCBs 156, 157, 167 and 169 (van den Berg et al., 1995). However, this recommendation would not be appropriate in the present study with a different covariation pattern (Lampa et al., 2012). In this study the PCB118 levels were the second highest and considerably higher than the PCB126 levels. Low concentration of a PCB may reduce the possibility to observe any relationship due to smaller variation in the PCB concentrations, unless sample sizes are very large. The reasons described above regarding the timing and amount of exposure might explain the lack of relationships for the more potent AHR activators PCBs 126 and169.
5. Conclusions
Genetic variation in the CYP1A1 gene was related to circulating PCB118 levels in the general elderly population. Genetic variation in CYP1A2 and CYP1B1 might be associated with other PCBs.
Acknowledgements
Funding
The study was funded by the Swedish Research Council FORMAS.
This work was supported by the Swedish Research Council (Vetenskapsrådet) and the Swedish Research Council Formas.
Footnotes
Ethics statement
The study was approved by the Ethics Committee of the University of Uppsala (diary number 2007/302) and all the participants gave their informed consent prior to the study.
All authors have read and approved the content of the manuscript and approve its submission. The authors have no conflict of interests.
References
- Amin N, Byrne E, Johnson J, Chenevix-Trench G, Walter S, Nolte IM, kConFab I, Vink JM, Rawal R, Mangino M, Teumer A, et al. Genome-wide association analysis of coffee drinking suggests association with CYP1A1/CYP1A2 and NRCAM. Mol Psychiatry. 2012;17:1116–1129. doi: 10.1038/mp.2011.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis MC, Monda KL, Yu K, Paynter N, Azzato EM, Bennett SN, Berndt SI, Boerwinkle E, Chanock S, Chatterjee N, Couper D, et al. Genome-wide meta-analysis identifies regions on7p21 (AHR) and 15q24 (CYP1A2) as determinants of habitual caffeine consumption. PLoS Genet. 2011;7:e1002033. doi: 10.1371/journal.pgen.1002033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards NC, Hing ZA, Perry A, Blaisdell A, Kopelman DB, Fathke R, Plum W, Newell J, Allen CE, Geetha S, Shapiro A, et al. Characterization of coding synonymous and non-synonymous variants in ADAMTS13 using ex vivo and in silico approaches. PLoS One. 2012;7:e38864. doi: 10.1371/journal.pone.0038864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Wang S, Wang H, Zhang S, Su S, Song Z, Deng Y, Qian J, Gu J, Liu B, Cao J, et al. The Cytochrome P4501A1 gene polymorphisms and idiopathic male infertility risk: a meta-analysis. Gene. 2014;535:93–96. doi: 10.1016/j.gene.2013.11.011. [DOI] [PubMed] [Google Scholar]
- Ghisari M, Long M, Bonefeld-Jorgensen EC. Genetic polymorphisms in CYP1A1, CYP1B1 and COMT genes in Greenlandic Inuit and Europeans. Int J Circumpolar Health. 2013;72:21113. doi: 10.3402/ijch.v72i0.21113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakkola J, Pasanen M, Pelkonen O, Hukkanen J, Evisalmi S, Anttila S, Rane A, Mantyla M, Purkunen R, Saarikoski S, Tooming M, et al. Expression of CYP1B1 in human adult and fetal tissues and differential inducibility of CYP1B1 and CYP1A1 by Ah receptor ligands in human placenta and cultured cells. Carcinogenesis. 1997;18:391–397. doi: 10.1093/carcin/18.2.391. [DOI] [PubMed] [Google Scholar]
- Hennig B, Meerarani P, Slim R, Toborek M, Daugherty A, Silverstone AE, Robertson LW. Proinflammatory properties of coplanar PCBs: in vitro and in vivo evidence. Toxicol Appl Pharmacol. 2002;181:174–183. doi: 10.1006/taap.2002.9408. [DOI] [PubMed] [Google Scholar]
- Hestermann EV, Stegeman JJ, Hahn ME. Relative contributions of affinity and intrinsic efficacy to aryl hydrocarbon receptor ligand potency. Toxicol Appl Pharmacol. 2000;168:160–172. doi: 10.1006/taap.2000.9026. [DOI] [PubMed] [Google Scholar]
- International Consortium for Blood Pressure Genome-Wide Association S. Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, Smith AV, Tobin MD, Verwoert GC, Hwang SJ, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josse AR, DaCosta LA, Campos H, El-Sohemy A. Associations between polymorphisms in the AHR and CYP1A1–CYP1A2 gene regions and habitual caffeine consumption. Am J Clin Nutr. 2012;96:665–671. doi: 10.3945/ajcn.112.038794. [DOI] [PubMed] [Google Scholar]
- Kang JH, Park H, Chang YS, Choi JW. Distribution of organochlorine pesticides (OCPs) and polychlorinated biphenyls (PCBs) in human serum from urban areas in Korea. Chemosphere. 2008;73:1625–1631. doi: 10.1016/j.chemosphere.2008.07.087. [DOI] [PubMed] [Google Scholar]
- Knobeloch L, Turyk M, Imm P, Schrank C, Anderson H. Temporal changes in PCB and DDE levels among a cohort of frequent and infrequent consumers of Great Lakes sportfish. Environ Res. 2009;109:66–72. doi: 10.1016/j.envres.2008.08.010. [DOI] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Laden F, Ishibe N, Hankinson SE, Wolff MS, Gertig DM, Hunter DJ, Kelsey KT. Polychlorinated biphenyls, cytochrome P4501A1, and breast cancer risk in the Nurses’ Health Study. Cancer Epidemiol Biomarkers Prev. 2002;11:1560–1565. [PubMed] [Google Scholar]
- Lampa E, Lind L, Hermansson AB, Salihovic S, van Bavel B, Lind PM. An investigation of the co-variation in circulating levels of a large number of environmental contaminants. J Expo Sci Environ Epidemiol. 2012;22:476–482. doi: 10.1038/jes.2012.41. [DOI] [PubMed] [Google Scholar]
- Lind L, Fors N, Hall J, Marttala K, Stenborg A. A comparison of three different methods to evaluate endothelium-dependent vasodilation in the elderly: the Prospective Investigation of the Vasculature in Uppsala Seniors (PIVUS) study. Arterioscler Thromb Vasc Biol. 2005;25:2368–2375. doi: 10.1161/01.ATV.0000184769.22061.da. [DOI] [PubMed] [Google Scholar]
- McGraw J, Waller D. Cytochrome P450 variations in different ethnic populations. Expert Opin Drug Metab Toxicol. 2012;8:371–382. doi: 10.1517/17425255.2012.657626. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Dalton TP. The role of cytochrome P450 enzymes in endogenous signalling pathways and environmental carcinogenesis. Nat Rev Cancer. 2006;6:947–960. doi: 10.1038/nrc2015. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Jones JE. Regulation of the mammalian cytochrome P1-450 (CYP1A1) gene. Int J Biochem. 1989;21:243–252. doi: 10.1016/0020-711x(89)90182-1. [DOI] [PubMed] [Google Scholar]
- Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L, Najjar SS, Zhao JH, Heath SC, Eyheramendy S, Papadakis K, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereg D, Dewailly E, Poirier GG, Ayotte P. Environmental exposure to polychlorinated biphenyls and placental CYP1A1 activity in Inuit women from northern Quebec. Environ Health Perspect. 2002;110:607–612. doi: 10.1289/ehp.02110607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland A, Glover E, Kende AS. Stereospecific, high affinity binding of 2,3,7,8-tetrachlorodibenzo-p-dioxin by hepatic cytosol. Evidence that the binding species is receptor for induction of aryl hydrocarbon hydroxylase. J Biol Chem. 1976;251:4936–4946. [PubMed] [Google Scholar]
- Poland A, Teitelbaum P, Glover E, Kende A. Stimulation of in vivo hepatic uptake and in vitro hepatic binding of [125I]2-lodo-3,7,8-trichlorodibenzo-p-dioxin by the administration of agonist for the Ah receptor. Mol Pharmacol. 1989;36:121–127. [PubMed] [Google Scholar]
- Ritter R, Scheringer M, MacLeod M, Moeckel C, Jones KC, Hungerbuhler K. Intrinsic human elimination half-lives of polychlorinated biphenyls derived from the temporal evolution of cross-sectional biomonitoring data from the United Kingdom. Environ Health Perspect. 2011;119:225–231. doi: 10.1289/ehp.1002211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy NK, Walker N, Chambers RC, Wirgin I. Characterization and expression of cytochrome P4501A in Atlantic sturgeon and short nose sturgeon experimentally exposed to coplanar PCB126 and TCDD. Aquat Toxicol. 2011;104:23–31. doi: 10.1016/j.aquatox.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudge CV, Sandanger T, Rollin HB, Calderon IM, Volpato G, Silva JL, Duarte G, Neto CM, Sass N, Nakamura MU, Odland JO, et al. Levels of selected persistent organic pollutants in blood from delivering women in seven selected areas of Sao Paulo State, Brazil. Environ Int. 2012;40:162–169. doi: 10.1016/j.envint.2011.07.006. [DOI] [PubMed] [Google Scholar]
- Rylander L, Nilsson-Ehle P, Hagmar L. A simplified precise method for adjusting serum levels of persistent organohalogen pollutants to total serum lipids. Chemosphere. 2006;62:333–336. doi: 10.1016/j.chemosphere.2005.04.107. [DOI] [PubMed] [Google Scholar]
- Safe S, Bandiera S, Sawyer T, Robertson L, Safe L, Parkinson A, Thomas PE, Ryan DE, Reik LM, Levin W, et al. PCBs: structure–function relationships and mechanism of action. Environ Health Perspect. 1985;60:47–56. doi: 10.1289/ehp.856047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salihovic S, Mattioli L, Lindstrom G, Lind L, Lind PM, van Bavel B. A rapid method for screening of the Stockholm Convention POPs in small amounts of human plasma using SPE and HRGC/HRMS. Chemosphere. 2012;86:747–753. doi: 10.1016/j.chemosphere.2011.11.006. [DOI] [PubMed] [Google Scholar]
- Sandau CD, Sjodin A, Davis MD, Barr JR, Maggio VL, Waterman AL, Preston KE, Preau JL, Jr, Barr DB, Needham LL, Patterson DG., Jr Comprehensive solid-phase extraction method for persistent organic pollutants. Validation and application to the analysis of persistent chlorinated pesticides. Anal Chem. 2003;75:71–77. doi: 10.1021/ac026121u. [DOI] [PubMed] [Google Scholar]
- Sauna ZE, Kimchi-Sarfaty C. Understanding the contribution of synonymous mutations to human disease. Nat Rev Genet. 2011;12:683–691. doi: 10.1038/nrg3051. [DOI] [PubMed] [Google Scholar]
- Schade G, Heinzow B. Organochlorine pesticides and polychlorinated biphenyls in human milk of mothers living in northern Germany: current extent of contamination, time trend from 1986 to 1997 and factors that influence the levels of contamination. Sci Total Environ. 1998;215:31–39. doi: 10.1016/s0048-9697(98)00008-4. [DOI] [PubMed] [Google Scholar]
- Schweikl H, Taylor JA, Kitareewan S, Linko P, Nagorney D, Goldstein JA. Expression of CYP1A1 and CYP1A2 genes in human liver. Pharmacogenetics. 1993;3:239–249. doi: 10.1097/00008571-199310000-00003. [DOI] [PubMed] [Google Scholar]
- Shimada T, Inoue K, Suzuki Y, Kawai T, Azuma E, Nakajima T, Shindo M, Kurose K, Sugie A, Yamagishi Y, Fujii-Kuriyama Y, et al. Aryl hydrocarbon receptor-dependent induction of liver and lung cytochromes P450 1A1, 1A2, and 1B1 by polycyclic aromatic hydrocarbons and polychlorinated biphenyls in genetically engineered C57BL/6J mice. Carcinogenesis. 2002;23:1199–1207. doi: 10.1093/carcin/23.7.1199. [DOI] [PubMed] [Google Scholar]
- Shimada T, Yun CH, Yamazaki H, Gautier JC, Beaune PH, Guengerich FP. Characterization of human lung microsomal cytochrome P-4501A1 and its role in the oxidation of chemical carcinogens. Mol Pharmacol. 1992;41:856–864. [PubMed] [Google Scholar]
- Tang YM, Wo YY, Stewart J, Hawkins AL, Griffin CA, Sutter TR, Greenlee WF. Isolation and characterization of the human cytochrome P450 CYP1B1gene. J Biol Chem. 1996;271:28324–28330. doi: 10.1074/jbc.271.45.28324. [DOI] [PubMed] [Google Scholar]
- van den Berg M, Sinnige TL, Tysklind M, Bosveld AT, Huisman M, Koopmans-Essenboom C, Koppe JG. Individual PCBs as predictors for concentrations of non and mono-ortho PCBs in human milk. Environ Sci Pollut Res Int. 1995;2:73–82. doi: 10.1007/BF02986720. [DOI] [PubMed] [Google Scholar]
- van Duursen MB, Sanderson JT, van den Berg M. Cytochrome P450 1A1 and 1B1 in human blood lymphocytes are not suitable as biomarkers of exposure to dioxin-like compounds: polymorphisms and interindividual variation in expression and inducibility. Toxicol Sci. 2005;85:703–712. doi: 10.1093/toxsci/kfi089. [DOI] [PubMed] [Google Scholar]
- Yamazaki K, Suzuki M, Itoh T, Yamamoto K, Kanemitsu M, Matsumura C, Nakano T, Sakaki T, Fukami Y, Imaishi H, Inui H. Structural basis of species differences between human and experimental animal CYP1A1s in metabolism of 3,30,4,40,5-pentachlorobiphenyl. J Biochem. 2011;149:487–494. doi: 10.1093/jb/mvr009. [DOI] [PubMed] [Google Scholar]