Abstract
The genetic information is constantly challenged by genotoxic attacks. DNA repair mechanisms evolved early in evolution and recognize and remove the various lesions. A complex network of DNA damage responses (DDR) orchestrates a variety of physiological adaptations to the presence of genome instability. Erroneous repair or malfunctioning of the DDR causes cancer development and the accumulation of DNA lesions drives the aging process. For understanding the complex DNA repair and DDR mechanisms it is pivotal to employ simple metazoan as model systems. The nematode Caenorhabditis elegans has become a well-established and popular experimental organism that allows dissecting genome stability mechanisms in dynamic and differentiated tissues and under physiological conditions. We provide an overview of the distinct advantages of the nematode system for studying DDR and provide a range of currently applied methodologies.
Introduction
Organismal genome stability is constantly threatened by extrinsic or cell-intrinsic genotoxins that can cause various types of DNA damage, including helix-distorting “bulky” lesions, DNA double-strand (DSBs) and single-strand breaks (SSBs), interstrand-crosslinks (ICLs), as well as missing, mismatched and modified single bases. Failure in DNA repair can affect transcription and replication or directly interrupt or alter gene function, which can subsequently result in cell death, senescence or cancer (reviewed in (Helleday et al., 2014)). Organisms have evolved an array of lesion-specific DNA repair pathways to detect and counteract the damage: Nucleotide excision repair (NER) repairs UV-induced lesions, such as cyclobutane pyrimidine dimers (CPDs) and 6-4 photoproducts (6-4PPs). DSBs, which can be caused by ionizing radiation (IR), are commonly repaired by either of the two competing pathways non-homologous end-joining (NHEJ) or homologous recombination (HR). Base excision repair (BER) is the prevalent mechanism for removal and correction of single damaged or modified bases, which can be induced by reactive oxygen species (ROS) or nitrogen oxygen species (NOS). In addition to the highly specialized DNA repair mechanisms, cells and organisms are equipped with an intricate network of DNA damage signaling pathways that are referred to as the DNA damage response (DDR). The DDR mediates the DNA damage checkpoint mechanisms that can arrest cell cycle progression or trigger controlled cellular suicide via apoptosis (reviewed in (Ciccia and Elledge, 2010)). In humans, genetically impaired repair and DDR deficiency leads to a number of devastating diseases that are characterized by developmental growth failure, premature ageing and predisposition to cancer development (reviewed in (Torgovnick and Schumacher, 2015)). The cellular mechanisms of DNA repair and DDR are highly conserved across species and have been extensively studied in model systems ranging from bacteria and yeast, to mammals and human cell lines. Recently, increasing focus has been devoted to tissue-specific and systemic DDR mechanisms in the context of development and ageing, which requires the in vivo application of model organisms.
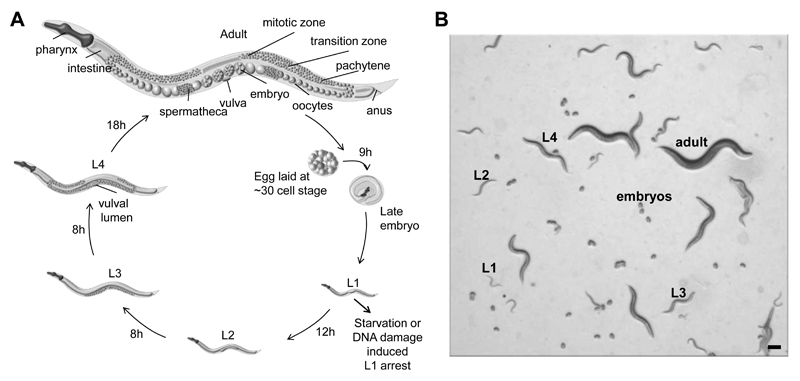
The transparent nematode C. elegans is a powerful experimental system that has been instrumental in deciphering the molecular basis of apoptotic cell death, RNA interference, development and ageing, as well as various systemic, non-cell-autonomous stress response mechanisms (Antebi, 2007; Blum et al., 2008; Grishok, 2005; Taylor et al., 2014). Animals are easy to maintain, the genome is fully sequenced and annotated, and there is an array of methods for genetic manipulation, including RNAi, transgenesis, genome editing, and random mutagenesis, which allow for a comparably simple design of forward and reverse genetic screens. Importantly, nematodes are susceptible to a variety of DNA damaging agents and most DNA repair mechanism are highly conserved (summarized in (Rieckher et al., 2016)). Developmental timing of C. elegans is highly reproducible: a full life cycle requires 2.5 days at 20°C during which animals develop from the fertilized oocyte to embryos, followed by the four larval stages (L1-L4) and finalized as adult hermaphrodite that produces 300 eggs by self-fertilization (Figure 1A). The C. elegans germline is formidable to study distinct DNA repair and signaling pathways occurring in meiotically and mitotically dividing germ cells, while somatic cells of adult animals are entirely postmitotic and more resistant to most DNA damaging agents (Figure 5; (Gartner et al., 2000). The different developmental stages can easily be distinguished under a stereomicroscope, allowing for detailed quantification of the consequences of DNA-damaging agents in somatic mitosis, cell growth, and developmental timing (Figure 1B and Figure 3; compare http://www.wormatlas.org).
Figure 1. C. elegans life cycle and developmental stages.
(A) Scheme of the life cycle from fertilized embryo, through the four larval stages L1-L4 and the fertile adult. One life-cycle is completed in 2.5 days when animals are grown at 20°C on OP50 bacteria-seeded NGM agar plates. Adults lay 300 eggs that hatch within 9 hours ex utero development. (B) Stereoscopic bright field image showing mixed, distinguishable developmental stages on an agar plate. Scale bar is 100 µm.
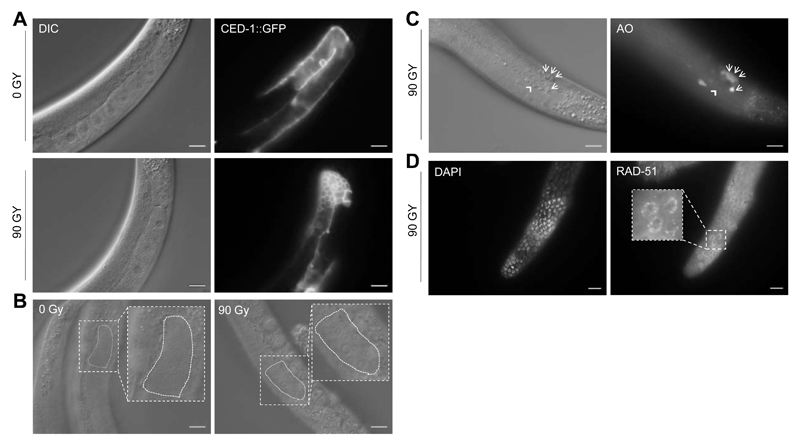
Figure 5. The C. elegans germline as model for cell cycle arrest and apoptosis upon IR-induced DSB-formation (see Methods).
(A) CED-1::GFP strain labeling somatic sheath cells during the engulfment of germ cells death. Day 1 adult worms were treated with IR at 90 Gy and 6 hours post-IR treatment pictures are acquired. Arrow shows a single corpse in 0 Gy and the square displays accumulation of corpses upon 90 Gy treatment in the gonad loop. Scale bars are 20 µm. (B) Representative images of the most distal (mitotic) zone to determine apoptosis. Scale bars are 20 µm. (C) Acridine Orange (AO) staining 6 h post-γ-irradiation (90 Gy). AO-stained apoptotic corpses (arrows) become visible under fluorescent microscope. Note that corpse clusters often result in diffuse staining signals thus exacerbating corpse scoring. Importantly, air bubbles (arrowhead) do not take up the dye. Scale bars: 20 µm. (C) RAD-51 foci indicating persistent DNA damage 16 h post-UV irradiation (120 mJ/cm2) in the mitotic zone of an adult C. elegans germline. Scale bars: 10 µm.
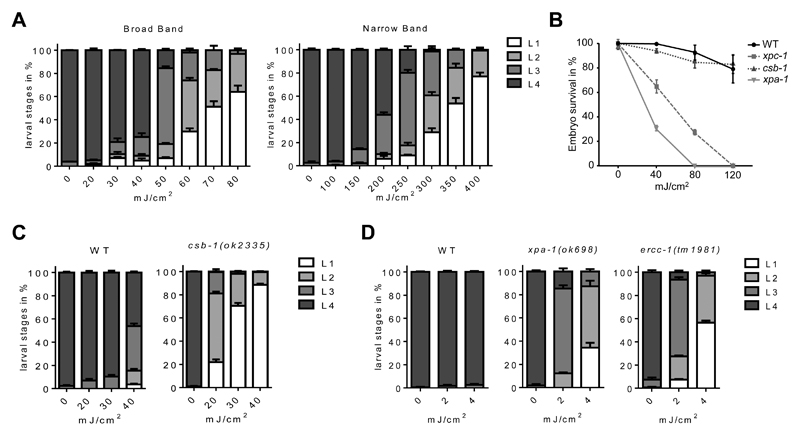
Figure 3. Determination of developmental delay and embryonic survival upon UVB-exposure (see Methods).
(A) Wild type irradiated at various doses with two different types of UVB irradiation sources. (B) Embryonic survival assay in the wild type, the GG-NER mutant for XPC-1, the TC-NER mutant for CSB-1 and the DNA unwinding factor XPA-1. (C) Developmental assay of the TC-NER mutant CSB-1 compared to wild type. (D) Developmental delay determined in the highly UV-sensitive mutants for XPA-1 or ERCC-1, acting in the core NER machinery. Errors bars indicate standard deviations.
A recent study underlines the strength of C. elegans as an emerging experimental system for exploring systemic DDR mechanisms: DNA lesions in germ cells activate the ERK1/2 MAP kinase MPK-1, which triggers an innate immune response through a transcriptional program that largely matches the profile upon pathogen infection. The innate immune response is comprised of numerous putative secreted peptides that emanate from damaged germ cells leading to the activation of the ubiquitin-proteasome system (UPS) in somatic tissues, which enhances protein homeostasis, thereby elevating somatic resistance to environmental stress factors, including heat and oxidative compounds (Figure 4D; Methods; (Ermolaeva et al., 2013). The elevated somatic endurance could provide extra time for arrested, genomically compromised germ cells to repair the DNA damage before offspring generation resumes. This “germline DNA damage-induced systemic stress resistance” (GDISR) is conceptually highly consistent with the disposable soma theory, which states that the resources of an organism need to be allocated between somatic maintenance and offspring generation in order to maximize its fitness (discussed in (Castells-Roca et al., 2015; Kirkwood, 2005)).
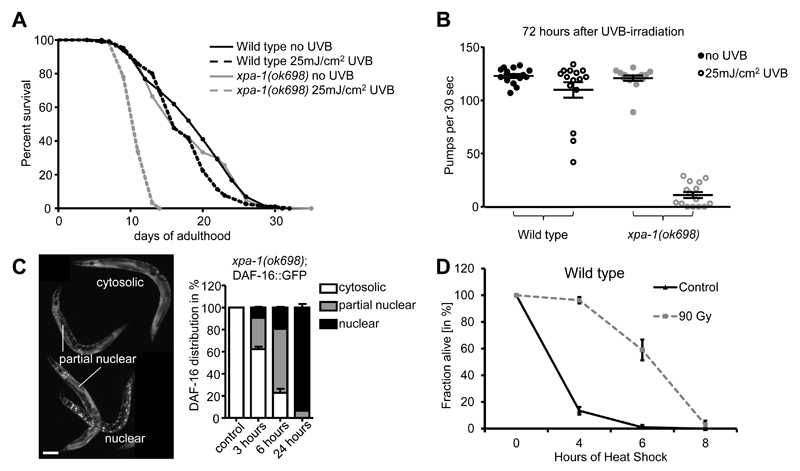
Figure 4. Commonly applied read-outs for somatic decline upon DNA damage induction (see Methods).
(A) Life span assay of wild type and xpa-1 mutants irradiated at day 1 of adulthood. (B) Pumping assay performed in adult-irradiated wild type and xpa-1 mutants 72 hours post-exposure. Error bars show SEM. (C) DAF-16::GFP nuclearization upon UVB-irradiation in xpa-1 mutant animals synchronized at L3 stage. The images are representative for the categories cytosolic, partially nuclear and fully nuclear. Size bars correspond to 100 µm. (D) Germline DNA damage induced stress resistance (GDISR) upon heat stress. Worms were treated either with 0 Gy (Control) or 90 Gy of IR at L4 larval stage before being shifted to 35°C 48 hours later. Experiment was done in biological triplicates. Error bars indicate standard error of the mean.
In this article we briefly review prominent DDR pathways and present various experimental procedures and read-outs that are commonly used to quantify the impact of the DNA damaging agents UVB and ionizing radiation (IR) in C. elegans. Furthermore, we provide representative data obtained from applying those methods in our laboratory.
C. elegans as model organism to study the removal of UV-induced DNA lesions by NER
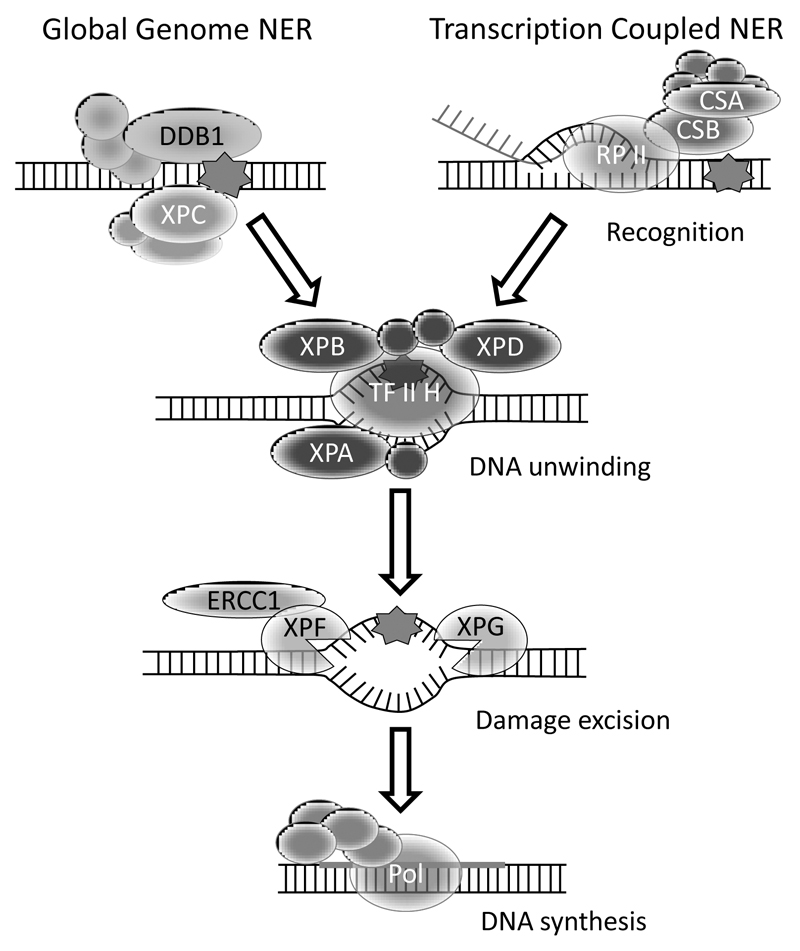
UVB (320-290nm) and UVC (290-100nm) irradiation are potent inducers of bulky DNA lesions 6-4PPs, CPDs and their Dewar valence isomers, which are commonly removed by NER (Figure 2) (Rastogi et al., 2010). Moderate UV irradiation doses result in a transient arrest of cell division and DNA replication in human cell lines, and delay the development of a stage-synchronized C. elegans wild type population (Figure 3A; Methods). Studies primarily conducted in S. cerevisiae and mammalian systems revealed that NER proceeds in four consecutive steps: DNA damage detection followed by DNA unwinding for sterically accessing the damaged site, excision of 25-30 nucleotides around the lesion and subsequent gap-filling through DNA synthesis and ligation. DNA damage detection is achieved by two distinct mechanisms: Transcription-Coupled NER (TC-NER) or Global-Genome NER (GG-NER). TC-NER is activated when RNA polymerase II stalls upon encountering a bulky lesion during transcription. This leads to the recruitment of the chromatin remodeling proteins Cockayne syndrome protein B (CSB) and Cockayne syndrome protein A (CSA); GG-NER is initiated upon lesion detection by the UV-damaged DNA-binding protein (UV-DDB) complex and Xeroderma pigmentosum group C (XPC), which constantly scan the genome for lesions. Both detection mechanisms subsequently activate the same downstream core machinery to repair the damage (Kamileri et al., 2012).
Figure 2. Simplified summary of the NER pathway in C. elegans.
The scheme shows the NER factors that have been studied most extensively in the nematode. The damage is recognized either via GG-NER factors DDB-1 or XPC-1, or via TC-NER, in which CSB-1 and CSA-1 interact with the stalled RNA polymerase II. Both pathways employ a common core mechanism that is distinguished in DNA unwinding, damage excision and DNA synthesis to fill the gap.
In humans, inherited mutations in the NER genes lead to distinct devastating congenital disorders: TC-NER deficiency leads to growth and mental retardation and symptoms of premature aging in Cockayne Syndrome (CS) patients, while defects primarily affecting GG-NER result in pigmentation abnormalities, atrophic skin and an increased skin cancer susceptibility in Xeroderma Pigmentosum (XP) patients. XP patients carrying mutations in NER genes that are operating in the core mechanism of the repair reaction additionally display neurodegenerative pathologies (Edifizi and Schumacher, 2016). Despite this generalized picture, the complexity of these congenital disorders and underlying pathomechanisms should not be underappreciated as the genotype-phenotype correlation between NER gene mutations and associated disease symptoms remains rather poorly understood.
The expediency of C. elegans as a model to study genome instability upon UV-exposure was early-on revealed by the isolation of a number of UV radiation-sensitive mutants (rad) (Hartman and Herman, 1982). In the last decades, a large number of NER homologs and orthologues were identified and functionally verified in the nematode (Figure 2; summarized in (Lans and Vermeulen, 2011). Importantly, it was shown that the two NER subpathways act in different tissues to repair UV-induced lesions: GG-NER and its damage sensor XPC-1 resolve damage specifically in proliferating cells in the germline and during early embryogenesis, while TC-NER majorly displays activity in post-mitotic somatic tissues. Consequently, UVB-exposure of xpc-1 deficient animals results in a dramatic reduction of embryonic survival, while csb-1 deficiency does not result in germline-specific defects but impaired larval developmental growth and survival (Figure 3B; Methods). The simultaneous abrogation of the lesion-recognition factors CSB-1 and XPC-1, or factors acting in the NER core machinery, including the DNA unwinding factor XPA-1 or the endonucleases ERCC-1, XPG-1 or XPF-1 that perform lesion excision, results in reduced embryonic survival and decline of somatic function upon exposure to UVB (Figure 3; Methods; (Lans et al., 2010; Mueller et al., 2014).
The robust UVB susceptibility of NER deficient C. elegans has been successfully exploited to identify novel regulators of DDR: Transcriptome analysis of UVB-treated xpa-1 mutants revealed that the insulin/insulin-like growth factor signaling (IIS) is attenuated and the major transcription factor of the IIS pathway DAF-16, member of the FOXO transcription factor family, becomes active to mount a DDR that alleviates developmental arrest and elevates tissue-functionality. DAF-16 activity in response to UVB in C. elegans is co-regulated by the GATA-transcription factor EGL-27, homolog of the metastasis tumour antigen 1 (MTA1) (Mueller et al., 2014). Those results are congruent with previous findings in Ercc1 and Xpa/Csbm/m-deficient mice, which show elevated anti-oxidant defenses and a shift to anabolism that are consistent with observed changes in the insulin-like signaling (IIS) pathway (Niedernhofer et al., 2006; van der Pluijm et al., 2007). Indeed, mammalian cells downregulate the central IIS modules, the GH and IGF-1 receptors, in response to persistent UV-induced DNA lesion (Garinis et al., 2009). Mueller et al. employed several read-outs to explore the role of the stress and ageing regulator DAF-16 in DDR: functional tissue-decline of NER mutants upon UVB is determined by developmental timing studies, lifespans, pharyngeal pumping, and the activation of DAF-16 is visualized in transgenic animals carrying a full length DAF-16::GFP fusion (Figure 3D and 4A-C; Methods; (Lin et al., 2001)). Repair kinetics of CPDs can be directly quantified in a slot blot using a CDP-specific antibody (Methods; (Babu and Schumacher, 2016; Mueller et al., 2014; Wolters et al., 2014)) and immunofluorescence staining (Lans et al. 2010).
The C. elegans germline as model to study DSB-induced DDR
DSBs in the DNA are considered a highly toxic form of DNA damage, as a single DSB can lead to cell cycle arrest (Bennett et al., 1993). The C. elegans germline has been employed as a model to delineate pathways regulating DSB-induced checkpoint mechanisms regulating cell cycle arrest and apoptosis. The transparency feature of C. elegans allows for in vivo observation via differential interference contrast (DIC) microscopy. The germline is easily discerned from somatic tissues and germ cells can be observed as they are passing through the mitotic and meiotic stages in distinct areas of the germline: the distal mitotic zone, in which germ stem cells proliferate, the transition zone that contains germ cells entering meiosis prophase I, the pachytene zone, in which meiotic recombination is completed. From here, germ cells further progress through the diplotene stage, in which chromosomes remain condensed and held together by the chiasmata, followed by the diakinesis stage, into oocytes, which are fertilized in the spermatheca (Figure 1A; summarized in (Lemmens and Tijsterman, 2011)). Commonly, DSB-formation and chromosome rearrangements are experimentally induced by IR (inflicted by X-rays or γ irradiation) or following replication fork breakdown for example at unrepaired UV-induced lesions, resulting in cell cycle arrest in mitotic cells and apoptosis in meiotic pachytene cells (Gartner et al., 2000). A basic level of physiological germ cell death occurs in the pachytene zone, which gets significantly elevated upon irradiation, can be visualized by DIC imaging (Gumienny et al., 1999). Apoptotic corpses can be stained via simple protocols applying the membrane-permeable dye acridine orange (AO) that robustly fluoresces in engulfed apoptotic cells (Figure 5C; Methods; (Craig et al., 2012)). To obtain more precise quantification of apoptosis kinetics, the transgenic expression of the engulfment protein CED-1::GFP can be visualized, which appears as a halo around cells during apoptosis (Figure 5A; Methods; (Zhou et al., 2001)). Cell cycle arrest can be observed and quantified in the mitotic region, since size and number of mitotic germ cells are significantly altered upon DNA damage (Figure 5B; Methods; (Craig et al., 2012)).
DSB repair is mediated by at least three repair pathways: homologous recombination (HR), non-homologous end-joining (NHEJ) as well as through alternative end-joining that includes polymerase Theta (POLQ)-mediated end-joining (TMEJ) (Clejan et al., 2006; Johnson et al., 2013; Roerink et al., 2014). During NHEJ, the loose break ends of the DNA double helix are enzymatically protected from resection, processed and directly religated. Errors during the processing and ligation steps can lead to nucleotide loss or misincorporation, thus potentially producing genetic mutations. In consequence, NHEJ is often considered a fast, yet error-prone repair mechanism. Error-free HR, in contrast, requires an identical, homologous template to drive DSB repair. HR involves enzymatic resection of the DSBs to generate 3’ single-stranded DNA overhangs that are bound by RAD-51 to form a nucleoprotein filament that can invade into homologous double-stranded DNA, which can subsequently serve as repair template (summarized in (Kakarougkas and Jeggo, 2014; Kass and Jasin, 2010)). Since the germline is highly accessible for immunostaining, RAD-51 antibody staining has proven to be a reliable tool to monitor and quantify DSB-induction (Figure 5D; Methods; (Alpi et al., 2003)). Whether DSBs arising in cells are repaired via NHEJ or HR primarily depends on the current phase of the cell cycle. HR is mainly restricted to S and G2 phase when replication has taken place and homologous sister chromatids are available for recombination. NHEJ acts throughout the cell cycle but primarily during G1, thus compensating for the absence of HR (summarized in (Kakarougkas and Jeggo, 2014)).
HR and NHEJ, are consecutively active during embryonic and larval development (Clejan et al., 2006). Early embryonic cells during the first six hours of development divide rapidly and directly switch between S and M-phases, omitting both G1 and G2. DSBs induced during that period are almost exclusively repaired via HR (summarized in (Kipreos, 2005)). In consequence, IR irradiation of C. elegans embryos harboring mutations in essential HR genes such as brc-1 fail to complete embryonic development and die before hatching (Methods). NHEJ is of major importance during late development as well as in arrested L1 larvae when cells persist predominantly in G1 phase. For instance, IR treatment of NHEJ-depleted worms lacking the CKU-80 dsDNA-binding factor results in developmental arrest at early larval stages (Johnson et al., 2013). Therefore, monitoring C. elegans development after DSB induction provides a valuable tool to decipher the involvement of novel factors in either of the two repair pathways.
Methods
C. elegans strains and genetics
C. elegans is maintained following standard protocols (Brenner, 1974). The strains used for this manuscript are N2 (Bristol; wildtype), csb-1(ok2335) X., xpa-1(ok698) I., ercc-1(tm1981) I., xpc-1(tm3886) IV., zIs356 (pDAF-16::DAF-16-GFP;rol-6) and were received from the Caenorhabditis Genetics Center (CGC; https://cbs.umn.edu/cgc/home), University of Minnesota, MN, or Shared Information of Genetic Resources (SHIGEN; http://shigen.nig.ac.jp/shigen/index.jsp), Center of Genetic Resource Information, National Institute of Genetics, Mishima, Japan. Recipes for common C. elegans maintenance and experiments can be found in (He, 2011).
Bleach synchronization
Adult worms are collected in a 15 mL tube with 4 mL of M9 buffer. 1 mL of alkaline hypochlorite solution is added (5% Sodium hypochlorite solution, 2.5 M KOH) and the tubes are incubated at room temperature (RT) for 4-5 minutes while vortexing. The released embryos are pelleted by centrifuging 1 minute at 800 g, hypochlorite solution is removed and the pellet is washed three times with 5 mL of M9 buffer. 10 mL of M9 is added to the tube and the embryos are incubated overnight at 20 °C in a rotating mixer at medium speed (CAT Roller RM10W-80V). Subsequently, obtained L1 staged worms are filtered using an 11 µm hydrophilic filter (Millipore, NY1104700). Concentration of worms is calculated by averaging the number of worms in three 10 µL drops per strain under a stereomicroscope.
Developmental arrest upon UVB irradiation
C. elegans undergoes a life cycle consisting of an embryonic stage, followed by four larval stages that lead to adulthood (Figure 1A). DNA damage present in the somatic tissues during larval development can delay the developmental progression in a damage-dependent manner, and therefore can be used to evaluate the proficiency of different strains in the response to and the repair of the damage (Mueller et al., 2014). For this assay, animals are bleach-synchronized at L1 larval stage. 50 worms per strain are plated in three unseeded 60 mm NGM plates per condition and treated with UVB light. L1 worms can be irradiated with UVB 310nm light using broadband PL-L 36W/UVB UV6 bulbs (Waldmann, 451436623-00005077), or narrowband PL-L 36W/UVB TL01 bulbs (Waldmann, 451436600-00003109). Upon preheating of the lamps, the intensity can be measured using a UVX-31 sensor with a UVX radiometer (UVP, Part Number 97-0015-02 and 97-0016-04). To obtain a moderate to high developmental arrest in wild type animals, UVB doses ranging from 40 mJ/cm2 to 60 mJ/cm2 are recommended with broadband bulbs, and 300 mJ/cm2 to 400 mJ/cm2 when using the narrowband bulbs (Figure 3A). During the irradiation of the worms, the lids of the plates must be removed. Also, the plates should not be moist since the liquid shields against UV irradiation. We recommend drying the plates one hour prior to usage with open lid on a sterile bench. Upon irradiation, 50 µL OP50 E. coli is added to the plates and worms are allowed to develop at 20°C. Worm stages are assessed 48 h and 72 h later using a stereomicroscope (Figure 1B). NER deficient strains should be used as UVB sensitive controls or to assess possible genetic interactions between the repair pathway and the genes of interest. Here, we exposed NER mutants for csb-1, xpa-1 or ercc-1 genes to various doses of UVB, which severely delays developmental timing and causes larval arrest (Figure 3C and 3D).
Measurement of Embryonic Lethality
Animals are bleach-synchronized and grown for 72 hr at 20°C into day 1 adults, and then treated with UVB at 0, 40, 80 and 120 mJ/cm2 of irradiation. Animals are allowed to recover for 24 hr on OP50-seeded plates. Subsequently, 5 animals of each treatment are transferred in triplicates to fresh OP50 seeded 60 mm plates and allowed to lay eggs for 2 hr. After removal of the adults, the number of eggs is counted, and 48 hr later the number of offspring is determined to quantify hatching efficiency. GG-NER-deficient animals, such as the xpc-1 and xpa-1 mutants, show high embryonic lethality in comparison to TC-NER mutant csb-1 or the repair-proficient wild type (Figure 3B).
Lifespan assay
Lifespan changes in C. elegans upon genotoxic insults, like UVB light, is a meaningful way of assessing the involvement in the DNA damage response of the different mutants or conditions of interest (Babu and Schumacher, 2016; Mueller et al., 2014). For this assay, bleach-synchronized worms grown to day 1 adult worms are exposed to UVB light to analyze the effects of the DNA damage on C. elegans lifespan. The lifespan of wild type animals can be significantly shortened upon irradiation with 100 mJ/cm2 and higher doses, while NER-deficient mutants display a robust lifespan reduction at 25 mJ/cm2 with the broadband machine (Figure 4A). After UVB irradiation, 10 times 20 worms per condition and strain are picked to fresh 60 mm OP50-seeded NGM plates. To avoid progeny overgrowth at the beginning of the experiment, worms must be transferred to new seeded NGM plates every second day when incubated at 20°C, or daily when grown at 25°C respectively. It is noteworthy that the induction of DNA damage can lead to an increased amount of worms with internal hatching, protruding and/or ruptured vulva, events that are censored from the experiment. NER deficient mutants, such as xpa-1(ok698), show an approximately 50% decrease in lifespan upon UVB exposure at 25 mJ/cm2, while the wild type stays largely unaffected (Figure 4A).
Pharyngeal pumping assay
The worm’s capacity of clearing and responding to the DNA lesions can also be assessed by measuring “healthspan” indicators upon induction of the damage. Particularly, the pharyngeal pumping rate has been established as an indicator of general muscular function and health of C. elegans that can be assessed with ease by focusing on the movement of the grinder in the terminal bulb of the pharynx (Collins et al., 2008; Huang et al., 2004). To perform the pharyngeal pumping assay upon DNA damage induction, bleach-synchronized day 1 adults are treated with UVB at 25 mJ/cm2 with the broadband bulb. Pumping ratio is assessed 24, 48 and 72 h after the DNA damage induction, using 15 worms per strain, condition and time point. Counting of the pumps is done at a stereomicroscope for 30 or 60 seconds periods per worm, after which the worm will be excluded from the experiment to avoid repetition. As the experiment will be prolonged up to 72 h, it might be necessary to transfer the worms to new plates to avoid starvation due to progeny overgrowth. It is of relevance to perform the assay in tightly controlled temperatures, as shuttle changes on it can lead to appreciable differences in the pumping rate. Therefore, a temperature-controlled room is recommended. The chosen dosage of 25 mJ/cm2 does not significantly affect the pumping rate of wild type but severely reduces xpa-1 mutant animals (Figure 4B).
DNA damage-induced DAF-16 nuclear localization
DAF-16, the main transcription factor acting in C. elegans insulin/insulin-like growth factor signaling, has been described to translocate into the nucleus upon different stresses, including heat, mechanical stress and DNA damage (Lin et al., 2001; Mueller et al., 2014). The analysis of DAF-16::GFP (strain: TJ365) cellular localization over time can be used as an indicator of the stress response to the induced damage. In this assay, bleach-synchronized L1 are plated onto 60mm NGM plates seeded with OP50. A minimum of 15 transgenic animals expressing DAF-16::GFP are plated in triplicates for every condition. After incubating the worms at 20°C for 24 hours, L3 stage animals are irradiated with 150 and 200 mJ/cm2 of UVB (broadband), and thereafter scored for DAF-16::GFP nuclearization directly on the plate after 3, 5, 7 and 24 hours post UVB-irradiation using a fluorescent stereomicroscope (Zeiss Axio Zoom.V16). For the analysis, animals are distinguished in three groups depending on the severity of DAF-16::GFP localization: no nuclearization (only cytoplasmic), partial nuclearization and full nuclearization. Non-irradiated transgenics almost exclusively show cytoplasmic DAF-16::GFP, while a partial nuclear localization might indicate stress-induction via starvation or other intrinsic or extrinsic stress factors (Figure 3C). We show that in an xpa-1(ok698) mutant background, DAF-16::GFP gradually nuclearizes over time and remains in the nucleus after 24 hours, indicating its role in response to persistent UVB-induced DNA damage (Figure 4C, (Mueller et al., 2014)).
Slot blot to measure DNA repair capacity
To measure DNA damage repair capacity, the genetic material of a bleach-synchronized L1 population is extracted right after damage induction and at a later time point, and immunolabelled for the accumulation of helix-distorting lesions (Mueller et al., 2014). For this experiment, large amounts of synchronized L1 animals (≥ 30.000 per plate) are plated on unseeded 60 mm NGM plates. After UVB treatment at 60 mJ/cm2 (broadband), worms are collected in M9 buffer and split into two samples: Sample 1 correspond to the time point 0, in which induced damage remains unrepaired, and Sample 2 is collected 24 hours after irradiation, allowing ample time for DNA repair. Animals of Sample 1 are immediately processed by pelleting in a centrifuge (2 minutes 800 g), the supernatant M9 is removed, subsequently quick-frozen on dry ice or liquid nitrogen and stored at -80°C. Animals of Sample 2 are plated on 60 mm NGM plates seeded with OP50 and incubated at 20°C for 24 hours. After completion of the incubation time, worms are collected in M9, washed 5 times (centrifuge at 200 g for 2 minutes, remove supernatant M9 and add 5 mL of fresh M9) and left 2 hours at 20°C in a rotating mixer to permit the removal of the intestinal bacteria. Thereafter, washing is repeated another 5 times before the sample is pelleted, quick-frozen and stored at -80°C.
For DNA extraction from the samples we can use the Gentra® Puregene® Tissue Kit (Qiagen, 158667) and the protocol for DNA Purification from Tissue. The protocol should be followed as if processing 5-10 mg of tissue (Gentra® Puregene® Handbook, page 39-40). The Cell Lysis Solution is directly added to the thawed sample and the additional step with proteinase K is performed to obtain a maximum yield. The DNA concentration is measured by using the Qubit® dsDNA HS Assay Kit (Invitrogen, Q32851). An amount of 2 and 4 µg of DNA can be used to label CPDs or 6-4PPs, respectively. To aid the visualization of the damage, different amounts of DNA are prepared by diluting the DNA serially from six to eight times (1:1). These serial dilutions of DNA are denatured at 95°C for 5 minutes, put directly on ice or at 4°C, and transferred onto a Hybond nylon membrane (Amersham, RPN119B) by using a Convertible Filtration Manifold System (Life Technologies, 11055). To crosslink the DNA, the membrane is incubated at 80°C for 2 hours and then blocked for 30 minutes in 3% milk/PBS-T (0.1%) at RT. To label the lesions, the membrane can be incubated overnight at 4°C with antibodies anti-CPDs (Clone TDM-2, 1:10.000, Cosmo Bio, CAC-NM-DND-001) or anti-6-4PPs (Clone 64M-2, 1:3.000, Cosmo Bio, CAC-NM-DND-002). Posteriorly, the membrane is washed three times with PBS-T (5 minutes, RT) and blocked for 30 minutes with 3% milk/PBS-T. Secondary antibody incubation is performed using a peroxidase-conjugated AffiniPure secondary antibody (1:10.000, Jackson Immuno Research, 115-035-174), followed by 3 washes in PBS-T and incubation of the membrane with ECL Prime (Amersham, RPN2232). Finally, the DNA lesions can be visualized in a Hyperfilm ECL (Amersham, 28906836).
Assessing mitotic germ cell cycle arrest
To assess the cell cycle arrest response to DNA damage, worms are treated with DNA damage sources such as IR-inducing cesium-137 or X-ray with 0, 30, 60, 90 and 120 Gy at the late L4 larval stage. 20 worms per condition are mounted 12 to 16 hours post-IR on 2% agarose pad for DIC microscopy (for more details on mounting, see: http://www.wormatlas.org/agarpad.htm; (Gartner et al., 2000)). Worms need to be immobilized during the mounting using the anesthetic levamisole 5 mM (Sigma-Aldrich, Germany) or with a nanoparticle-solution (Kim et al., 2013). A defined region of the distal germ line can be scored for the presence of enlarged germ cell nuclei (Figure 5B). This defined field corresponds to an area of 3.125 µm x 6.25 µm in the most distal region of the germ line. For a precise and convenient counting of mitotic germ cells within a specific scale, a netmicrometer can be applied to the microscope ocular (d=26 mm, 12,5 x 12,5/5;10, Zeiss, Germany), considering that the germ cell nuclei in all focal planes should be counted (Gartner et al., 2004; Gartner et al., 2000; Schumacher et al., 2005).
Quantification of apoptotic corpses in the meiotic germline
To quantify the number of apoptotic corpses in the meiotic germline, synchronized late L4s are exposed to IR at 0, 60 and 90 Gy (Gartner et al., 2000). 20 worms per condition should be mounted on 2% agarose pad at each time point following the treatment. Mounting is achieved via nano-particles on an agarose pad (Kim et al., 2013). The number of apoptotic corpses can be assessed under DIC microscopy based on refractive morphological changes in the pachytene region in the gonad loop of the germline (Figure 5A and 5C; DIC). In case of application of CED-1::GFP reporter for evaluating germline apoptosis, fluorescence microscopy can be used to verify induction of apoptosis by the surrounding GFP signal (Figure 5A; GFP signal).
Acridine Orange staining for visualization of meiotic germ cell corpses
To stain for apoptotic corpses with acridine orange (AO) after IR treatment, 5 µL of 2% AO stock solution (Sigma-Aldrich, Cat.-No. A9231-10ML) are mixed with 1mL M9 buffer (Gumienny et al., 1999). 300-500 µL of the final staining solution are added to each 60 mm OP50-seeded NGM plate containing adult worms. The solution is evenly distributed on the plates by rotating. Thereafter, plates are incubated in the dark for 1 hour at RT. Animals are rinsed off the plates using 5 mL M9 buffer and are washed at least 2x with fresh M9 to remove excess AO staining solution. Subsequently, animals are transferred to fresh OP50-seeded NGM plates and are incubated in the dark for at least 45 minutes to remove residual AO from the intestines. Finally, corpses can be assessed within in the late pachytene region close to the germline loop using fluorescence microscopy (Figure 5C). Apoptotic cells stained with AO emit light at about 520 nm and thus display green fluorescence.
RAD-51 immunofluorescence in dissected C. elegans germlines
To prepare worms for germline dissection, L4 larvae are picked onto fresh NGM plates seeded with OP50 and incubated for 24 hours at 20°C. The next day, young adult worms are irradiated with UV (60 - 80 mJ/cm2) or IR (30 Gy) and are allowed to recover at 20°C for 2 or 16 hours, respectively. Germline dissection is performed with syringe needles after each chosen time point in dissection buffer containing 1.1x egg salts buffer (10x egg salts buffer: 250 mM HEPES pH 7.4, 1.18 M NaCl, 480 mM KCl, 20 mM CaCl2, 20 mM MgCl2), 0.2% Tween 20 ,20 mM NaN23 and H2O. For the dissection, 20 worms are picked into a 15 µL drop of dissection buffer. Worms need to be carefully decapitated with the syringe needle so that one germline arm and parts of the intestine are released. Make sure to not injure the germline during the cutting. The use of a second needle to keep the worm in a stable position during dissection might be helpful to optimize both speed and precision. Dissected germlines are fixed on poly lysine-coated microscope slides (Menzel-Gläser Polysine® Slides, Thermo Scientific, Art.-No. J2800AMNZ) using a para-formaldehyde solution containing 1.1x egg salts buffer, 0.2% Tween 20 and 3.7% of freshly prepared para-formaldehyde. Slides are incubated 5 minutes at RT before being shock-frozen for approximately 10 minutes in liquid nitrogen or on dry ice. After freeze cracking, slides need to be incubated for 1 minute in methanol at -20°C. Subsequently, slides are washed twice for 10 minutes in PBS-T. To prevent unspecific binding of the antibody, samples are blocked for 30 minutes with PBS-T containing 10% donkey serum. RAD-51 can be detected with a rabbit anti-RAD-51 antibody (Novus Biologicals, Cat.-No. 29480002, dilution 1:350 in PBS-T containing 10% donkey serum). Slides are incubated with the antibody at 4°C over night. The next day, slides are washed 3x with PBS-T before being incubated for 2 hours with Alexa-Fluor@594 donkey anti-rabbit secondary antibody (Thermo Fisher Scientific, Cat.-No. A21207, dilution 1:350 in PBS-T containing 10% donkey serum) at RT in the dark. Slides are washed again 3x with PBS-T and are subsequently mounted using DAPI Flouromount-G (SouthernBiotech,Cat. No. 0100-20). Slides are assessed with a Zeiss Axio Imager M1 at 63x magnification, using the 43 HE DsRed channel. We recommend RAD-51 staining 1-2 hours post irradiation to monitor immediate RAD-51 recruitment to sites of DNA damage, as well as 16-24 hours post-irradiation to detect persistent, unrepaired DNA lesions. For quantification, count cell nuclei displaying RAD-51 fluorescence. In addition, total amounts of RAD-51 foci per nucleus can be scored if required (Figure 5D).
Homologous recombination repair assay
To assess HR repair capacity in C. elegans, worms are synchronized by picking L4 larvae and are maintained at 20°C (Clejan et al., 2006; Johnson et al., 2013). The next day, for each condition and genotype used in the experiment, 5 adult hermaphrodites are transferred onto a single 35 mm NGM plate seeded with a small drop of OP50 and are subsequently allowed to lay eggs for 1-1.5 hours. Afterwards, hermaphrodites have to be removed from the plates as fast as possible. Freshly laid eggs need to be irradiated immediately with 0, 20 and 40 Gy of IR. 24 hours later, count hatched vs. non-hatched eggs under a dissecting stereoscope and calculate the percentage of embryo survival. We recommend to include a brc-1(tm1145) HR-deficient mutant strain as positive control (Boulton et al., 2004; Wolters et al., 2014). The experiment has to be performed in biological triplicates.
Assay to determine GDISR
To analyze the GDISR in C. elegans, 35 worms per condition are synchronized at L4 larval stage on 35 mm NGM-plates seeded with OP50 and are subsequently treated with 0 or 90 Gy of IR (Ermolaeva et al., 2013). We allow worms to recover for 48 hours at 20°C before exposing them to heat stress at 35°C in a Sanyo MIR-154 incubator. Plate dehydration can be prevented by keeping the plates in closed carton boxes during the heat shock treatment. Dead vs. alive worms are scored after 4, 6 and 8 hours of heat shock. If required, more time points can be added, especially while working with stress-resistant or long-lived mutants. As in all survival experiments, worms displaying a protrusion of the vulva should be excluded from the analysis. The experiment should be performed in triplicates for each condition.
Concluding Remarks
DDR and the specialized DNA repair mechanisms are essential to maintain genome stability of all organisms. In humans, DDR-deficiency results in a number of devastating genetic diseases, including high cancer-susceptibility and progeroid symptoms. Most DNA repair mechanisms are conserved across taxa and thus present in C. elegans. The nematode has been exemplary in providing insight into many biological processes, including apoptosis, development, structure and function of the neuronal system, systemic RNAi and ageing. The ease of maintenance, the availability of genetic methodologies and clear-cut phenotypic read-outs make C. elegans an impeccable model to study DDR in vivo and in the context of a multicellular organism. DNA damage accumulation in the germline and its consequences, namely mitotic arrest or meiotic apoptosis, can easily be detected. The consequences of DDR in the germline affect the animal in a systemic fashion though GDISR. Somatic DNA damage leads to developmental delay or permanent arrest and physiological decline of tissue-functionality during ageing. The nematode has successfully served as a platform to derive mechanistic data on prominent DNA repair pathways including HR, NHEJ, and NER among others. C. elegans presents multiple ways to address complex questions, such as cell- or tissue-specific DNA repair or system-wide DDR mechanisms, in a simple way, and will contribute to produce novel and unanticipated insights into cellular and organismal mechanisms maintaining genome stability.
Acknowledgements
B.S. acknowledges funding from the Deutsche Forschungsgemeinschaft (CECAD, SFB 829, SFB 670, and KFO 286), the European Commission (ERC Starting Grant 260383, FP7-PEOPLE-2012-ITN CodeAge 316354, aDDRess 316390, MARRIAGE 316964, Flag-Era-JTC2015 GRAPHENE), and the Bundesministerium für Bildung und Forschung (Sybacol FKZ0315893A-B).
References
- Alpi A, Pasierbek P, Gartner A, Loidl J. Genetic and cytological characterization of the recombination protein RAD-51 in Caenorhabditis elegans. Chromosoma. 2003;112:6–16. doi: 10.1007/s00412-003-0237-5. [DOI] [PubMed] [Google Scholar]
- Antebi A. Ageing: when less is more. Nature. 2007;447:536–537. doi: 10.1038/447536a. [DOI] [PubMed] [Google Scholar]
- Babu V, Schumacher B. A C. elegans homolog for the UV-hypersensitivity syndrome disease gene UVSSA. DNA Repair (Amst) 2016;41:8–15. doi: 10.1016/j.dnarep.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CB, Lewis AL, Baldwin KK, Resnick MA. Lethality induced by a single site-specific double-strand break in a dispensable yeast plasmid. Proc Natl Acad Sci U S A. 1993;90:5613–5617. doi: 10.1073/pnas.90.12.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum ES, Driscoll M, Shaham S. Noncanonical cell death programs in the nematode Caenorhabditis elegans. Cell Death Differ. 2008;15:1124–1131. doi: 10.1038/cdd.2008.56. [DOI] [PubMed] [Google Scholar]
- Boulton SJ, Martin JS, Polanowska J, Hill DE, Gartner A, Vidal M. BRCA1/BARD1 orthologs required for DNA repair in Caenorhabditis elegans. Curr Biol. 2004;14:33–39. doi: 10.1016/j.cub.2003.11.029. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castells-Roca L, Mueller MM, Schumacher B. Longevity through DNA damage tolerance. Cell Cycle. 2015;14:467–468. doi: 10.1080/15384101.2015.1006543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clejan I, Boerckel J, Ahmed S. Developmental modulation of nonhomologous end joining in Caenorhabditis elegans. Genetics. 2006;173:1301–1317. doi: 10.1534/genetics.106.058628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins JJ, Huang C, Hughes S, Kornfeld K. The measurement and analysis of age-related changes in Caenorhabditis elegans. WormBook. 2008:1–21. doi: 10.1895/wormbook.1.137.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig AL, Moser SC, Bailly AP, Gartner A. Methods for studying the DNA damage response in the Caenorhabdatis elegans germ line. Methods Cell Biol. 2012;107:321–352. doi: 10.1016/B978-0-12-394620-1.00011-4. [DOI] [PubMed] [Google Scholar]
- Ermolaeva MA, Segref A, Dakhovnik A, Ou HL, Schneider JI, Utermohlen O, Hoppe T, Schumacher B. DNA damage in germ cells induces an innate immune response that triggers systemic stress resistance. Nature. 2013;501:416–420. doi: 10.1038/nature12452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garinis GA, Uittenboogaard LM, Stachelscheid H, Fousteri M, van Ijcken W, Breit TM, van Steeg H, Mullenders LH, van der Horst GT, Bruning JC, et al. Persistent transcription-blocking DNA lesions trigger somatic growth attenuation associated with longevity. Nat Cell Biol. 2009;11:604–615. doi: 10.1038/ncb1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner A, MacQueen AJ, Villeneuve AM. Methods for analyzing checkpoint responses in Caenorhabditis elegans. Methods Mol Biol. 2004;280:257–274. doi: 10.1385/1-59259-788-2:257. [DOI] [PubMed] [Google Scholar]
- Gartner A, Milstein S, Ahmed S, Hodgkin J, Hengartner MO. A conserved checkpoint pathway mediates DNA damage--induced apoptosis and cell cycle arrest in C. elegans. Mol Cell. 2000;5:435–443. doi: 10.1016/s1097-2765(00)80438-4. [DOI] [PubMed] [Google Scholar]
- Grishok A. RNAi mechanisms in Caenorhabditis elegans. FEBS Lett. 2005;579:5932–5939. doi: 10.1016/j.febslet.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Gumienny TL, Lambie E, Hartwieg E, Horvitz HR, Hengartner MO. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development. 1999;126:1011–1022. doi: 10.1242/dev.126.5.1011. [DOI] [PubMed] [Google Scholar]
- Hartman PS, Herman RK. Radiation-sensitive mutants of Caenorhabditis elegans. Genetics. 1982;102:159–178. doi: 10.1093/genetics/102.2.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F. Common Worm Media and Buffers. Bio-protocl. 2011;Bio101:e55. [Google Scholar]
- Helleday T, Eshtad S, Nik-Zainal S. Mechanisms underlying mutational signatures in human cancers. Nat Rev Genet. 2014;15:585–598. doi: 10.1038/nrg3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Xiong C, Kornfeld K. Measurements of age-related changes of physiological processes that predict lifespan of Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2004;101:8084–8089. doi: 10.1073/pnas.0400848101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson NM, Lemmens BB, Tijsterman M. A role for the malignant brain tumour (MBT) domain protein LIN-61 in DNA double-strand break repair by homologous recombination. PLoS Genet. 2013;9:e1003339. doi: 10.1371/journal.pgen.1003339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakarougkas A, Jeggo PA. DNA DSB repair pathway choice: an orchestrated handover mechanism. Br J Radiol. 2014;87:20130685. doi: 10.1259/bjr.20130685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamileri I, Karakasilioti I, Garinis GA. Nucleotide excision repair: new tricks with old bricks. Trends Genet. 2012;28:566–573. doi: 10.1016/j.tig.2012.06.004. [DOI] [PubMed] [Google Scholar]
- Kass EM, Jasin M. Collaboration and competition between DNA double-strand break repair pathways. FEBS Lett. 2010;584:3703–3708. doi: 10.1016/j.febslet.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Sun L, Gabel CV, Fang-Yen C. Long-term imaging of Caenorhabditis elegans using nanoparticle-mediated immobilization. PLoS One. 2013;8:e53419. doi: 10.1371/journal.pone.0053419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipreos ET. C. elegans cell cycles: invariance and stem cell divisions. Nat Rev Mol Cell Biol. 2005;6:766–776. doi: 10.1038/nrm1738. [DOI] [PubMed] [Google Scholar]
- Kirkwood TB. Understanding the odd science of aging. Cell. 2005;120:437–447. doi: 10.1016/j.cell.2005.01.027. [DOI] [PubMed] [Google Scholar]
- Lans H, Marteijn JA, Schumacher B, Hoeijmakers JH, Jansen G, Vermeulen W. Involvement of global genome repair, transcription coupled repair, and chromatin remodeling in UV DNA damage response changes during development. PLoS Genet. 2010;6:e1000941. doi: 10.1371/journal.pgen.1000941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lans H, Vermeulen W. Nucleotide Excision Repair in Caenorhabditis elegans. Mol Biol Int. 2011;2011:542795. doi: 10.4061/2011/542795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmens BB, Tijsterman M. DNA double-strand break repair in Caenorhabditis elegans. Chromosoma. 2011;120:1–21. doi: 10.1007/s00412-010-0296-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K, Hsin H, Libina N, Kenyon C. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet. 2001;28:139–145. doi: 10.1038/88850. [DOI] [PubMed] [Google Scholar]
- Mueller MM, Castells-Roca L, Babu V, Ermolaeva MA, Muller RU, Frommolt P, Williams AB, Greiss S, Schneider JI, Benzing T, et al. DAF-16/FOXO and EGL-27/GATA promote developmental growth in response to persistent somatic DNA damage. Nat Cell Biol. 2014;16:1168–1179. doi: 10.1038/ncb3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer LJ, Garinis GA, Raams A, Lalai AS, Robinson AR, Appeldoorn E, Odijk H, Oostendorp R, Ahmad A, van Leeuwen W, et al. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 2006;444:1038–1043. doi: 10.1038/nature05456. [DOI] [PubMed] [Google Scholar]
- Rastogi RP, Richa, Kumar A, Tyagi MB, Sinha RP. Molecular mechanisms of ultraviolet radiation-induced DNA damage and repair. J Nucleic Acids. 2010;2010:592980. doi: 10.4061/2010/592980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieckher M, Lopes AFC, Schumacher B. Genome Stability in C. elegans. Elsevier Book. In: Kovalchuk I, Kovalchuk O, editors. Genome Stability - From Virus to Human Application. 2016. pp. 163–186. [Google Scholar]
- Roerink SF, van Schendel R, Tijsterman M. Polymerase theta-mediated end joining of replication-associated DNA breaks in C. elegans. Genome Res. 2014;24:954–962. doi: 10.1101/gr.170431.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher B, Hanazawa M, Lee MH, Nayak S, Volkmann K, Hofmann ER, Hengartner M, Schedl T, Gartner A. Translational repression of C. elegans p53 by GLD-1 regulates DNA damage-induced apoptosis. Cell. 2005;120:357–368. doi: 10.1016/j.cell.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Taylor RC, Berendzen KM, Dillin A. Systemic stress signalling: understanding the cell non-autonomous control of proteostasis. Nat Rev Mol Cell Biol. 2014;15:211–217. doi: 10.1038/nrm3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torgovnick A, Schumacher B. DNA repair mechanisms in cancer development and therapy. Front Genet. 2015;6:157. doi: 10.3389/fgene.2015.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Pluijm I, Garinis GA, Brandt RM, Gorgels TG, Wijnhoven SW, Diderich KE, de Wit J, Mitchell JR, van Oostrom C, Beems R, et al. Impaired genome maintenance suppresses the growth hormone--insulin-like growth factor 1 axis in mice with Cockayne syndrome. PLoS Biol. 2007;5:e2. doi: 10.1371/journal.pbio.0050002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolters S, Ermolaeva MA, Bickel JS, Fingerhut JM, Khanikar J, Chan RC, Schumacher B. Loss of Caenorhabditis elegans BRCA1 promotes genome stability during replication in smc-5 mutants. Genetics. 2014;196:985–999. doi: 10.1534/genetics.113.158295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Hartwieg E, Horvitz HR. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell. 2001;104:43–56. doi: 10.1016/s0092-8674(01)00190-8. [DOI] [PubMed] [Google Scholar]