Abstract
The structures of the catalytic domain of matrix metalloproteinase 12 in the presence of acetohydroxamic acid and N-isobutyl-N-[4-methoxyphenylsulfonyl]glycyl hydroxamic acid have been solved by x-ray diffraction in the crystalline state at 1.0 and 1.3-Å resolution, respectively, and compared with the previously published x-ray structure at 1.2-Å resolution of the adduct with batimastat. The structure of the N-isobutyl-N-[4-methoxyphenylsulfonyl]glycyl hydroxamic acid adduct has been solved by NMR in solution. The three x-ray structures and the solution structure are similar but not identical to one another, the differences being sizably higher in the loops. We propose that many of the loops show a dynamical behavior in solution on a variety of time scales. Different conformations of some flexible regions of the protein can be observed as “frozen” in different crystalline environments. The mobility in solution studied by NMR reveals conformational equilibria in accessible time scales, i.e., from 10–5 s to ms and more. Averaging of some residual dipolar couplings is consistent with further motions down to 10–9 s. Finally, local thermal motions of each frozen conformation in the crystalline state at 100 K correlate well with local motions on the picosecond time scale. Flexibility/conformational heterogeneity in crucial parts of the catalytic domain is a rule rather than an exception in matrix metalloproteinases, and its extent may be underestimated by inspection of one x-ray structure. Backbone flexibility may play a role in the difficulties encountered in the design of selective inhibitors, whereas it may be a requisite for substrate binding and broad substrate specificity.
Keywords: macrophage metalloelastase, protein mobility, solution structure, x-ray structure
Matrix metalloproteinases (MMPs) are enzymes involved in extracellular matrix degradation, which is a fundamental step in many physiological processes like tissue remodeling and repair (1, 2). Most MMPs are constituted by a prodomain that is removed upon activation, a catalytic domain responsible for hydrolytic activity, and a hemopexin-like domain that probably plays a role in substrate recognition. In several pathologies overexpression of MMPs, or the misregulation of their activity, is related to disease progression (3–5). MMPs thus are validated pharmaceutical targets. Many efforts have been devoted to develop inhibitors against these metalloenzymes (6) through rational drug design approaches, which require a detailed knowledge of the structural features of the active sites (7, 8). Up to now, drug candidates have failed the clinical trials, often because of side effects probably caused by their low selectivity (9, 10).
Over the years, it has become evident that, besides their intrinsic similarity, MMPs exhibit some capability of adapting the binding pocket to the inhibitor shape (11), thus suggesting another rationale for the lack of inhibitor selectivity. A more detailed picture of mobility of MMPs, its origin, and its implications for substrate and inhibitor binding would be extremely helpful at this point. With this in mind, we have used an integrated approach conjugating high-resolution x-ray structures and NMR structures to analyze the mobility of a macrophage metalloelastase (MMP12) and performed a systematic comparison with several other MMPs. The major finding is that there are certain loop regions that are subject to mobility and/or conformational heterogeneity in several MMPs. Some of these regions are known to be implicated in substrate and inhibitor binding, substrate recognition, and selectivity toward physiological and nonphysiological inhibitors. It is also apparent that a number of structural models are necessary to characterize a single MMP.
Materials and Methods
Protein Cloning and Expression. A form of the catalytic domain of MMP12, corresponding to the stretch Gly-106–Gly-263 with one additional methionine at position 105 and a Phe-171–Asp mutation, has been cloned and expressed as reported (12). After the refolding procedure, the catalytic domain was inhibited with either the strong inhibitor NNGH (N-isobutyl-N-[4-methoxyphenylsulphonyl]glycyl hydroxamic acid; Kd = 10 nM) or the weak inhibitor AHA (acetohydroxamic acid; Kd = 8 mM) by a multistep dialysis procedure. The details for the measurements of Kd are reported in Supporting Text, which is published as supporting information on the PNAS web site.
Crystallization, Data Collection, and Resolution of the Crystal Structures. For MMP12–AHA adduct an aliquot of 2 μl of protein solution (10 mM Tris/5 mM CaCl2/0.1 mM ZnCl2/300 mM NaCl/200 mM AHA, pH 8) was mixed with 2 μl of reservoir buffer (0.1 M Tris·HCl/25% PEG 6000/200 mM AHA, pH 8). The final protein concentration was 8 mg/ml. Crystallization was carried out with the hanging drop vapor diffusion method at 20°C. For MMP12–NNGH adduct, 3 mM of NNGH was added to protein solution before mixing with a reservoir buffer containing 0.1 M Tris·HCl, 20% PEG 6000, 200 mM AHA, and 1.0 M LiCl2 at pH 8.0.
The data were measured for both data sets at the ELETTRA XRD-1 beamline (Trieste, Italy) at 100 K, and the crystals used for data collection were cryo-cooled without any cryo-protectant treatment. The MMP12–NNGH crystal had a mosaicity of ≈0.6° and diffracted to a maximum resolution of 1.3 Å, whereas the MMP12–AHA crystal, with mosaicity 0.3°, diffracted to 1.0-Å resolution. The crystal space group for the NNGH adduct is P21212 with a = 37.30, b = 62.63, c = 69.24 Å, α = β = γ = 90° with one molecule in the asymmetric unit. The AHA complex crystallizes in the C2 space group, with a = 50.92, b = 59.59, c = 53.49 Å, α = γ = 90°, and β = 115.14° with one molecule in the asymmetric unit. Details concerning data collection, structure solution, and refinement are given in Supporting Text and Tables 1 and 2, which are published as supporting information on the PNAS web site.
NMR Sample Preparation, Measurements, and Solution Structure Calculations. 13C/15N-enriched (for structure calculations) and 15N-enriched (for mobility measurements) samples of MMP12–NNGH were 0.9 and 0.6 mM, respectively. The pH was adjusted to 7.2.
All NMR experiments for protein assignment, structure calculation, and mobility measurements were acquired on Bruker AVANCE 900, AVANCE 800, AVANCE 700, and DRX 500 spectrometers. All instruments were equipped with a triple resonance (TXI) 5-mm probe with a z axis pulse field gradient, and the 500-MHz spectrometer was equipped with a triple resonance CRYO-probe. All NMR experiments were performed at 298 K. A detailed description of NMR experiments is reported in Supporting Text and Tables 3–10, which are published as supporting information on the PNAS web site.
The program dyana (13) was used to calculate a family of 200 structures starting from randomly generated conformers in 10,000 annealing steps. The quality of the structures calculated by dyana can be assessed by a properly defined energy function (target function) proportional to the squared deviations of the calculated constraints from the experimental ones, plus the standard covalent and nonbonded energy terms. The programs molmol (14) and procheck (15, 16) were subsequently used for secondary structure analysis and structural quality. The final family of 20 conformers was averaged and subjected to restrained energy minimization with amber 6.0 (17).
Results and Discussion
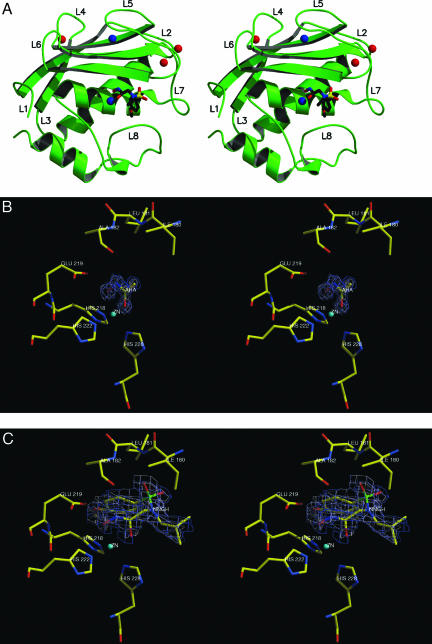
The crystal structures of the AHA and NNGH adducts of MMP12 have been determined at 100 K and 1.0- and 1.3-Å resolution, respectively. The structure of the latter adduct has been also determined by NMR in solution at room temperature [target function 1.07 ± 0.10 Å2, backbone (BB) RMS deviation (RMSD) = 0.74 ± 0.10 Å for residues 113–259, 0.41 ± 0.06 Å by excluding loop regions, overall G factor =–0.31]. The overall fold is very similar (but not identical with total BB RMSD = 1.37 Å; α-helices and β-sheets BB RMSD = 0.65 Å; the similarity being sizably lower in the loops with BB RMSD = 1.64 Å). The x-ray structure of the NNGH adduct is shown in Fig. 1A. Both MMP12 complexes display the classical structure of metalloproteinases (18, 19), which consist of three α-helices, a twisted five-stranded β-sheet, and eight intervening loops (L1–L8) of which L5 and L8 are very long. The resulting topology is L1-β1-L2-α1-L3-β2-L4-β3-L5-β4-L6-β5-L7-α2-L8-α3, where the five-stranded twisted β-sheet is formed by β2-β1-β3-β5-β4. In both structures three calcium-binding sites lie approximately in the plane of the β3, β5, and β4 strands of the β-sheet and are bound to residues from the β-turns of the sheet (Fig. 1 A). The catalytic zinc ion (Zn1) is bound to the active-site cleft between the β-sheet and loop L8. The zinc is five-coordinate, the ligands being the His-218 Nε2, His-222 Nε2, His-228 Nε2 atoms, and the two oxygen atoms of the chelating hydroxamate moiety of the AHA inhibitor (Fig. 1B) or the NNGH inhibitor (Fig. 1C). The second zinc ion (Zn2) is bound in a tetrahedral geometry by His-168 Nε2, Asp-170 Oδ1, His-183 Nε2, and His-196 Nδ1.
Fig. 1.
Stereoview of the overall fold of the MMP12–NNGH adduct (A) and enlargements of the active site-bound inhibitors AHA (B) and NNGH (C). In B and C the ligand electron densities are also shown.
The AHA adduct is a close analog to a noninhibited MMP, as the small AHA molecule only interacts with the catalytic zinc ion and its immediate neighbors. The two oxygens of AHA bind the catalytic Zn at a distance of ≈2.1 Å (Fig. 1B). The AHA CO oxygen is 2.9 Å from His-228 Nε2, 3.1 Å from His-222 Nε2, and 3.0 and 3.1 Å from two water molecules; the AHA NOH oxygen is 2.9 Å from His-218 Nε2, 2.5 Å from Glu-219 Oδ1, 3.2 Å from His-222 Nε2, and 3.0 Å from a water molecule. The AHA nitrogen is 2.9 Å from Ala-182 O.
The binding mode of the NNGH inhibitor is similar to that recently reported for MMP10 (19). The interaction of the inhibitor with the active site is limited to the Zn1 ion and the S1′ subsite (the latter formed by loop L8 and the beginning of strand β4). In addition to the metal chelation, NNGH is held in the active site by H bonds and hydrophobic interactions (Fig. 1C). The hydroxamic acid head chelates the zinc ion and is further involved in two H bonds. The protonated oxygen atom (O4), in addition to the zinc coordination, donates a strong H bond to the carboxylate Oε2 of Glu-219 (2.4 Å) (Fig. 1C). In other hydroxamate-inhibited MMPs this distance ranges from 2.7 to 3.1 Å. In contrast, the carbonyl hydroxamate oxygen O5 of NNGH is not involved in any H bond, whereas the hydroxamate NH has significant electrostatic interaction with the Ala-182 carbonyl oxygen. The NNGH sulfonyl oxygen O2 makes an important contribution to inhibitor binding energy by establishing H bonds with Leu-181 N. Finally, the phenolic methylether group of NNGH is inserted in the S1′ pocket. A water molecule is buried in the pocket and makes a weak H bond with the methylether oxygen of the inhibitor.
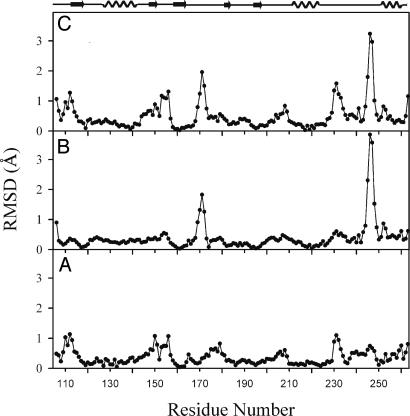
Conformational Heterogeneity and Mobility of MMP12. The present x-ray data at 1.0- and 1.3-Å resolution, together with the structure at 1.2-Å resolution of the batimastat adduct of MMP12 (20), constitute a set of extremely high-resolution structures that are unique among all MMPs. Moreover, the structure of the AHA adduct is particularly interesting, as it shows the protein without any exogenous ligand in the S1′ site. A comparison of these three structures should thus allow a direct assessment of the flexibility of the hydrophobic ligand binding pocket, and possibly other parts of the molecule, as a function of the presence and type of the exogenous ligand. The least-squares superposition between MMP12–AHA and MMP12–NNGH yields a global BB RMSD of 0.45 Å, and no regions where the RMSD goes beyond 1.2 Å (Fig. 2A). The least-squares superposition between MMP12–AHA and MMP12–batimastat (PDB ID code 1JK3) yields a global BB RMSD of 0.46 Å; however, there are two narrow regions in which the deviation is significantly high: region 170–173 where it goes up to 2 Å, and region 245–248 where it goes up to ≈4 Å (Fig. 2B). Finally, the least-squares superposition between MMP12–NNGH and MMP12–batimastat gives a somewhat higher BB RMSD of 0.62 Å. The RMSD goes up to ≈1.3 Å in region 153–157, ≈2.0 Å in region 168–172, between 1 and 1.5 Å in region 230–234, and up to 3 Å in region 245–248 (Fig. 2C). Because of the extremely high resolution of the three structures (RMSD on atomic positions between 0.03 and 0.07 Å), any deviation among atomic coordinates of different molecules >0.5 Å (a conservative estimate) is meaningful and can be analyzed in terms of molecular flexibility.
Fig. 2.
Global BB RMSD between the x-ray structures of MMP12–NNGH and MMP12–AHA (A), MMP12–AHA and MMP12–batimastat (Protein Data Bank ID code 1JK3) (B), and MMP12–NNGH and MMP12–Batimastat (Protein Data Bank ID code 1JK3) (C).
We should first of all comment on the discrepancy between the present two structures and the batimastat adduct in region 168–174 (part of loop L5), which is not catalytically relevant. The present MMP12 domain contains a Phe-171–Asp mutation aimed at increasing its solubility (12). Although this mutation prevents us from attaching any functional significance to the variability in this particular region, it serves the purpose of illustrating how a single mutation on a surface residue may translate into a relatively marked BB movement involving seven residues of a loop region.
On the contrary, the differences observed in the other parts of the molecule are certainly meaningful. The most striking is that between the batimastat and the other adducts in the L8 loop region (which constitutes a wall of the S1′ site) centered around residues 245–248 (C-terminal part of the loop). It is possible that this difference is caused by a different mode of interaction of the hydrophobic part of the ligand (an isopropyl for batimastat vs. a methoxyphenyl for NNGH, and no hydrophobic moiety at all for AHA). However, residues 245–248 are located very far from the entrance of the S1′ site and certainly out of reach for the relatively short hydrophobic parts of either batimastat or NNGH.
Other differences are observed between NNGH and the other two structures in loop regions 110–116 (L1+β1), 146–157 (L3+β2+L4), 176–180 (C-terminal part of loop L5), 206–209 (part of loop L7), and 230–236 (central part of loop L8) (Fig. 1 A). In all cases, these differences are unrelated to the different inhibitor bound to the protein. We can conclude that the conformational heterogeneity observed among these three high-resolution MMP12 structures is largely independent of the presence and type of inhibitor, with the possible exception of the 245–248 region. Other x-ray structures of MMP12 where the S1′ pocket is occupied by the side chains of methionines bound in a product-analog mode (18) (Protein Data Bank ID codes 1OS2 and 1OS9) show that even with the same moiety bound to the S1′ site, the 245–248 (C-terminal part of loop L8) region still shows conformational heterogeneity.
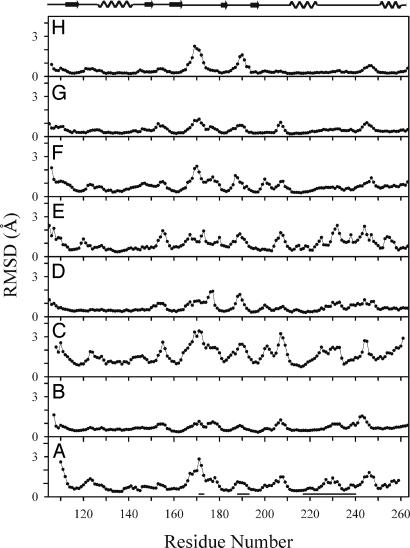
Fig. 3A shows the RMSD per-residue of the family of the NMR structures of MMP12–NNGH. It is apparent that there are several disordered regions in the structure of the above adduct in solution. Disorder in NMR-derived structures arises from the lack of medium- to long-range nuclear Overhauser effects (NOEs), which in turn may be caused by low data quality and/or severe overlap of NOESY cross peaks or intrinsic lack of NOEs caused by mobility. Careful inspection of the 2D and 3D NOESY spectra clearly shows that in all disordered regions medium- to long-range NOEs are indeed present, but the intensity of those arising from side chains is far too small compared with the expectations from the present x-ray and solution structures. Also the NOEs arising from BB NHs are reduced, mostly because the NH signals are reduced in intensity due to partial exchange with water protons, whereas the short-range NOEs arising from BB Hα are of approximately normal intensity. Small NOEs typically arise from equilibrium among different conformations, which rapidly quenches the Overhauser effect of the involved protons. We conclude that disorder in the solution structure of MMP12–NNGH is indeed largely caused by protein mobility.
Fig. 3.
Mean RMSD values per residue for the NMR structure families of MMP-12 (NNGH adduct, this work) (A), MMP-1 (Protein Data Bank ID code 1AYK) (B), MMP-2 (Protein Data Bank ID code 1HOV) (C), MMP-3 (Protein Data Bank ID code 2SRT) (D), MMP-3 (Protein Data Bank ID code 1UMS) (E), MMP-3 (Protein Data Bank ID code 1BM6) (F), MMP-13 (Protein Data Bank ID code 1EUB) (G), and MMP-13 (Protein Data Bank ID code 1FM1) (H). In A the regions showing signal splitting (local conformational heterogeneity) are indicated by horizontal bars.
A comparison of Figs. 2 and 3A shows that there is a remarkable correspondence between protein loop regions that show conformational heterogeneity among different structures in the solid state and loop regions that are mobile in solution. This correspondence allows us to draw further conclusions on the motions that characterize these loops. From the NMR point of view, the fact that mostly side-chain NOEs are strongly reduced in intensity indicates that the side chains are very mobile. In turn, this feature means that there are little or no contacts between these side chains and other parts of the protein. Likewise, partial exchange of NH protons is also indicative of the absence of hydrogen bonds involving BB NHs, whereas the few short-range NOEs involving Hα protons are of normal intensity. Collectively, these findings mean that there are little energetic constraints that can define the conformational preferences of these loops.
Residual dipolar couplings (RDCs) are potentially sensitive indicators of the presence of mobility on any submillisecond time scale (21), giving rise to reduction in RDC absolute values upon conformational averaging, provided that the spanned conformations differ sensibly in their RDCs. Accordingly, several measured RDCs are smaller in absolute values than the calculated ones, particularly at the hinges between loops and elements of secondary structure (i.e., between β1 and L2 and L2 and α1, before and after β2, and in loops L5 and L7). Finally, seven of nine residues in the C-terminal part of loop L8 also exhibit quenched RDC values.
All of these observations suggest, although they do not prove, the presence of ample motions of each loop as a whole. From the x-ray point of view, the high resolution of all of the loop regions shows that both static and dynamic disorder is small within each structure in the solid state, and the comparison of the different structures fully enlightens the conformational heterogeneity. It is likely that what we observe in solution are motions that sample within each molecule a conformational space that has an amplitude similar to that sampled by different molecules in different crystalline environments.
Time Scales of Mobility. Protein motions occur on a variety of different time scales, from nanosecond to picosecond ranges typical of local motions involving few atoms, to microseconds for conformational changes involving protein segments, and to milliseconds or longer for very large interdomain motions or, for example, cis-trans proline isomerizations. To better characterize the time scales of the present MMP dynamics we have performed heteronuclear R1, R2, and NOE measurements (Fig. 4, which is published as supporting information on the PNAS web site) and Carr–Purcell–Meiboom–Gill and off-resonance R1ρ experiments. We have also detected and analyzed a number of signal splittings that witness the presence of conformations that interconvert on a time scale of tens of milliseconds or longer.
The analysis of 1H-15N relaxation data (Fig. 4) shows values in the normal range for a protein of the size of MMP12 at room temperature. The R2/R1 ratio indicates the protein to be monomeric; only two R2 values (His-172 and Ile-191) exceed the average value significantly, already indicating that there are no extensive regions of the protein undergoing motions on the millisecond time scale. Carr–Purcell–Meiboom–Gill measurements provide a semiquantitative estimate of the time scale of the motions experienced by residue Ile-191 (252 ± 41 μs), whereas from off-resonance R1ρ experiments we could evaluate a τex for His-172 of 117 ± 22 μs. 15N NOEs for most residues belonging to secondary structure elements are also within the expected range for a well folded, rigid protein. On the other hand, 15N NOEs for residues in the loop regions are systematically lower, but still normal for NH groups of loops when not involved in hydrogen bonds. The lower NOE values for loop NHs are ascribed (22–25) to local motions occurring on the nanosecond time scale or shorter. As such, the decrease in the NOEs should correspond to an increase of the group average temperature factors obtained from the x-ray structure (26, 27). Indeed, the correspondence between these two parameters is very good (see Fig. 5, which is published as supporting information on the PNAS web site).
By analyzing in detail the splitting of 1H-15N heteronuclear single-quantum coherence (HSQC) signals in the light of the atomic resolution x-ray structures it appears that in most cases signal splitting is paralleled by local conformational heterogeneity of protein side chains and not of the BB, because the RDC values of both components (where measurable) are never strikingly different. Resonance splitting for BB amides is clearly observed in 1H-15N HSQC and 1H-15N HSQC–NOESY spectra in the case of residues 171–173 (on loop L5), 188–193 (on loop L6), and 217–240 (N-terminal part of loop L8). In the x-ray structure, side-chain conformational heterogeneity is found for two of these three protein fragments. For example, in the NNGH adduct, double conformation is observed for BB N of residue 170 and BB carbonyl of Glu-219 and Ile-220 that belong to helix α2, as well as for the side chain of Tyr-239 in loop L8; other side chains in the same loop (residues 229, 233, 241, 245–247, and 249) appear very disordered. The x-ray structure of the AHA adduct shows very similar behavior. It should be noted that loop L5 contains the mutation Phe-171–Asp. As discussed, residues 171–173 have double conformation. Notably, the preceding residues 167–170 have BB NHs that could not be detected in HSQC spectra. Exchange phenomena on the time scale of tens to hundreds of μs are detected for residues Ala-164, His-172, Phe-174, Asp-175, Gly-178, and Ile-180, as it results from Carr–Purcell–Meiboom–Gill and R1ρ measurements. This finding is consistent with differential mobility along this loop: on the submillisecond scale for the hinge with β3, intermediate exchange, which causes line broadening beyond detection, for residues 167–170, >10 ms for residues 171 and 173, and again on the submillisecond time scale for the C terminal. Notably, the second part of loop L8, which does not show double conformation, is characterized by sizably low values of the experimental RDC with respect to the expectation, suggesting mobility on the submillisecond time scale. For the C-terminal part of loop L8, seven of nine RDCs are smaller in absolute value than the calculated ones (Fig. 6, which is published as supporting information on the PNAS web site). The motions experienced by these residues are neither in the very slow subsecond range (with the exception of residue 241 that shows evidence of conformational exchange from R1ρ), nor in the subnanosecond range (again, no dramatic decrease of 15N NOEs).
By recalling that (i) the proton–proton NOEs in the loop regions are very small in the side chains because of large mobility and scarcity of long-range contacts, (ii) the NOEs involving NH groups are only somewhat smaller than expected because of partial exchange of NH protons, (iii) the NOEs involving Hα protons are generally of normal intensity, and (iv) the measured RDCs for signals that do not show double conformation on the NMR time scale are sizably smaller than the expected ones, the picture that emerges is that at least the C-terminal part of loop L8 may undergo relatively wide but collective BB motions. We are left with a still relatively wide time window extending between ≈10–5 and 10–9 s for the motions that cause reduction of the RDC values in the C-terminal part of loop L8. The higher end of this window is perfectly consistent with motions that are ample enough to encompass the 1–4 Å of both the RMSD between loops in different x-ray structures (Fig. 2) and the RMSD in the loop regions of the structure family in solution.
Other MMPs. It is worth attempting a comparative analysis of the published x-ray and NMR structures of other human MMPs. So far, NMR data exist for MMP1 (28), MMP2 (29), MMP3 (30–32), MMP12 (this work), and MMP13 (33, 34). In some cases the issue of mobility has been specifically addressed, in some others it has not. In any case, the families of structures deposited in the Protein Data Bank for all of the above molecules turn out to be instructive. Fig. 3 shows a collection of BB RMSD per residue for each NMR structure family. All structures have been realigned by using structure-based rather than sequence-based algorithms. It is apparent, beyond uncertainty, that all profiles are remarkably similar, with similar increases in disorder in the same loop regions for all of these MMPs. The qualities of the various structures differ, but even the best ones show the same trend, confirming that disorder is indeed caused by an intrinsic high mobility of the loop regions of MMPs. Many different Protein Data Bank entries for x-ray structures of the same MMP are available for MMP1, MMP3, and MMP13. For MMP1 and MMP3 again relatively high RMSD between the different structures is obtained, corroborating the present conclusions. The RMSDs are instead small for the different MMP13 structures, and there are at least three loop regions that are disordered in solution (Fig. 3 G and H). It is possible that for MMP13 one conformation is more energetically favored that the others, and the crystallization process always freezes the same seemingly ordered structure.
Summary Considerations. In this article, the solution structure of an adduct of MMP12 has been analyzed in terms of conformational exchange of loops in time scales not directly available through NMR experiments (microseconds to nanoseconds). However, the exchange is supported by the different “frozen” conformations displayed by three x-ray structures, two of them being solved during this research, and also by the reduction of several RDC values in solution. The different frozen conformations provide an estimate of the amplitude of the conformational motions. It appears that this property is characteristic of many, if not all, MMPs. This research shows the power of coupling x-ray structures (possibly more than one) and the solution structure, the latter with an analysis of NOE intensities, RDC values, and relaxation properties, to obtain a structural model that is beyond a single structural datum. The adaptability of loops (and cavities whose walls are constituted by loops) to ligands and other external perturbations are a further challenge to drug discovery bioinformatic procedures, as these slow-exchanging conformations should be sampled before the interaction with the ligand is studied.
Supplementary Material
Acknowledgments
We thank Dr. Renato Barbieri for help with the RDC measurements. This research was financially supported by European Union Contract QLG2-CT-2002-00988 (Structural Proteomics in Europe), Ente Cassa di Risparmio di Firenze, Fondo per Gli Investimenti della Ricerca di Base(Ministero dell'Istruzione, dell'Università e della Ricerca Contract RBNE01TTJW), Fondo Integrativo Speciale Ricerca (Ministero dell'Istruzione, dell'Università e della Ricerca: “Modeling di Structure di Metalloproteine e delle Internazioni Proteina-Farmaco e Proteina-Proteina”), and Ministero dell'Istruzione, dell'Università e della Ricerca Ricerca Industriale Project 10537/P/01.
Author contributions: I.B. designed research; V.C., M.C., M.F., Y.-M.L., B.T., and P.T. performed research; V.C., M.F., Y.-M.L., C.L., S.M., and P.T. analyzed data; and I.B., C.L., and S.M. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: MMP, matrix metalloproteinase; NNGH, N-isobutyl-N-[4-methoxyphenylsulfonyl]glycyl hydroxamic acid; AHA, acetohydroxamic acid; BB, backbone; NOE, nuclear Overhauser effect; RDC, residual dipolar coupling; RMSD, RMS deviation.
Data deposition: The atomic coordinates and structure factors for the x-ray crystal structures of the AHA and NNGH adducts have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 1Y93 and 1RMZ, respectively). The solution structure of the NNGH and the list of upper experimental constraints used for structure calculations have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 1YCM and 1Z3J). The NMR chemical shift values have been deposited in the BioMagResBank, www.bmrb.wisc.edu (accession no. 6444).
References
- 1.Birkedal-Hansen, H. (1995) Curr. Opin. Cell Biol. 7, 728–735. [DOI] [PubMed] [Google Scholar]
- 2.Murphy, G. J. P., Murphy, G. & Reynolds, J. J. (1991) FEBS Lett. 289, 4–7. [DOI] [PubMed] [Google Scholar]
- 3.Stetler-Stevenson, W. G., Hewitt, R. & Corcoron, M. (1996) Semin. Cancer Biol. 7, 147–154. [DOI] [PubMed] [Google Scholar]
- 4.Borkakoti, N. (2000) J. Mol. Med. 78, 261–268. [DOI] [PubMed] [Google Scholar]
- 5.Steinman, L. (1996) Cell 85, 299–302. [DOI] [PubMed] [Google Scholar]
- 6.Whittaker, M., Floyd, C. D., Brown, P. & Gearing, A. J. H. (1999) Chem. Rev. 99, 2735–2776. [DOI] [PubMed] [Google Scholar]
- 7.Lukacova, V., Zhang, Y., Mackov, M., Baricic, P., Raha, S., Calvo, J. A. & Balaz, S. (2004) J. Biol. Chem. 279, 14194–14200. [DOI] [PubMed] [Google Scholar]
- 8.Borkakoti, N. (2004) Biochem. Soc. Trans. 32, 17–20. [DOI] [PubMed] [Google Scholar]
- 9.Coussens, L. M., Fingleton, B. & Matrisian, L. M. (2002) Science 295, 2387–2392. [DOI] [PubMed] [Google Scholar]
- 10.Pavlaki, M. & Zucker, S. (2003) Cancer Metast. Rev. 22, 177–203. [DOI] [PubMed] [Google Scholar]
- 11.Moy, F. J., Chanda, P. K., Chen, J., Cosmi, S., Edris, W., Levin, J. I., Rush, T. S., Wilhelm, J. & Powers, R. (2002) J. Am. Chem. Soc. 124, 12658–12659. [DOI] [PubMed] [Google Scholar]
- 12.Banci, L., Bertini, I., Ciulli, A., Fragai, M., Luchinat, C. & Terni, B. (2003) J. Mol. Cat. A Chem. 204–205, 401–408. [Google Scholar]
- 13.Güntert, P., Mumenthaler, C. & Wüthrich, K. (1997) J. Mol. Biol. 273, 283–298. [DOI] [PubMed] [Google Scholar]
- 14.Koradi, R., Billeter, M. & Wüthrich, K. (1996) J. Mol. Graphics 14, 51–55. [DOI] [PubMed] [Google Scholar]
- 15.Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993) J. Appl. Crystallogr. 26, 283–291. [Google Scholar]
- 16.Laskowski, R. A., Rullmann, J. A. C., MacArthur, M. W., Kaptein, R. & Thornton, J. M. (1996) J. Biomol. NMR 8, 477–486. [DOI] [PubMed] [Google Scholar]
- 17.Case, D. A., Pearlman, D. A., Caldwell, J. W., Cheatham, T. E., Ross, W. S., Simmerling, C. L., Darden, T. A., Merz, K. M., Stanton, R. V., Cheng, A. L., et al. (1999) amber (University of California, San Francisco), Version 6.0.
- 18.Bertini, I., Calderone, V., Fragai, M., Luchinat, C. Mangani, S. & Terni, B. (2003) Angew. Chem. Int. Ed. 42, 2673–2676. [DOI] [PubMed] [Google Scholar]
- 19.Bertini, I., Calderone, V., Fragai, M., Luchinat, C., Mangani, S. & Terni, B. (2004) J. Mol. Biol. 336, 707–716. [DOI] [PubMed] [Google Scholar]
- 20.Lang, R., Kocourek, A., Braun, M., Tschesche, H., Huber, R., Bode, W. & Maskos, K. (2001) J. Mol. Biol. 312, 731–742. [DOI] [PubMed] [Google Scholar]
- 21.Prestegard, J. H., Bougault C. M. & Kishore A. I. (2004) Chem. Rev. 104, 3519–3540. [DOI] [PubMed] [Google Scholar]
- 22.Kay, L. E., Torchia, D. A. & Bax, A. (1989) Biochemistry 28, 8972–8979. [DOI] [PubMed] [Google Scholar]
- 23.Palmer, A. G., III, Skelton, N. J., Chazin, W. J., Wright, P. E. & Rance, M. (1992) Mol. Phys. 75, 699–711. [Google Scholar]
- 24.Barbato, G., Ikura, M., Kay, L. E., Pastor, R. W. & Bax, A. (1992) Biochemistry 31, 5269–5278. [DOI] [PubMed] [Google Scholar]
- 25.Kay, L. E., Nicholson, L. K., Delaglio, F., Bax, A. & Torchia, D. A. (1992) J. Magn. Reson. 97, 359–375. [Google Scholar]
- 26.Longhi, S., Nicolas, A., Creveld, L., Egmond, M., Verrips, C. T., de Vlieg, J., Martinez, C. & Cambillau, C. (1996) Proteins Struct. Funct. Genet. 26, 442–458. [DOI] [PubMed] [Google Scholar]
- 27.Parri, L. & Mangani, S. (1998) Inorg. Chim. Acta 275–276, 349–358. [Google Scholar]
- 28.Moy, F. J., Chanda, P. K., Cosmi, S., Pisano, M. R., Urbano, C., Wilhelm, J. & Powers, R. (1998) Biochemistry, 37, 1495–1504. [DOI] [PubMed] [Google Scholar]
- 29.Feng, Y., Likos, J. J., Zhu, L., Woodward, H., Munie, G., McDonald, J. J., Stevens, A. M., Howard, C. P., De Crescenzo, G. A., Welsch, D., et al. (2002) Biochim. Biophys. Acta 1598, 10–23. [DOI] [PubMed] [Google Scholar]
- 30.Van Doren, S. R., Kurochkin, A. V., Hu, W., Ye, Q.-Z., Johnson, L. L., Hupe, D. J. & Zuiderweg, E. R. P. (1995) Protein Sci. 4, 2487–2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, Y.-C., Zhang, X., Melton, R., Ganu, V. & Gonnella, N. C. (1998) Biochemistry 37, 14048–14056. [DOI] [PubMed] [Google Scholar]
- 32.Gooley, P. R., O'Connell, J. F., Marcy, A. I., Cuca, G. C., Salowe, S. P., Bush, B. L., Hermes, J. D., Esser, C. K., Hagmann, W. K., Springer, J. P. & Johnson, B. A. (1994) Nat. Strut. Biol. 1, 111–118. [DOI] [PubMed] [Google Scholar]
- 33.Moy, F. J., Chanda, P. K., Chen, J. M., Cosmi, S., Edris, W., Levin, J. I. & Powers, R. (2000) J. Mol. Biol. 302, 671–689. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, X., Gonnella, N. C., Koehn, J., Pathak, N., Ganu, V., Melton, R., Parker, D., Hu, S.-I. & Nam, K.-Y. (2000) J. Mol. Biol. 301, 513–524. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.