Abstract
Worldwide marine invaders, such as the brown alga Undaria pinnatifida, offer challenging models for unraveling the apparent paradox of sustainable settlement of exotic species over a large spectrum of environments. Two intergenic noncoding mitochondrial loci were found to be highly informative at the within-species level. Twenty-five haplotypes were found over the whole dataset (333 base pairs, 524 individuals, and 24 populations). The native range showed striking population genetic structure stemming from low diversity within and high differentiation among populations, a pattern not observed in the introduced range of this seaweed. Contrary to classical expectations of founding effects associated with accidental introduction of exotic species, most of the introduced populations showed high genetic diversity. At the regional scale, genetic diversity and sequence divergence showed contrasting patterns in the two main areas of introduction (Europe and Australasia), suggesting different processes of introduction in the two regions. Gene genealogy analyses point to aquaculture as a major vector of introduction and spread in Europe but implicate maritime traffic in promoting recurrent migration events from the native range to Australasia. The multiplicity of processes and genetic signatures associated with the successful invasion confirms that multiple facets of global change, e.g., aquaculture practices, alteration of habitats, and increased traffic, act in synergy at the worldwide level, facilitating successful pandemic introductions.
Keywords: coastal habitats, gene genealogy, introduced species, kelp, mitochondrial DNA
Biological invasions are closely linked to global change (1–3). The spread and success of exotic species may be facilitated by (i) climate change (4), (ii) increase in disturbed and/or anthropogenic habitats (5, 6), and (iii) synergistic effects resulting from increased biotic exchanges (7, 8). In turn, biological invasions may feed back into global change (9) by modifying interactions among species and even whole ecosystems (10, 11), inducing extinction in the native fauna/flora (12, 13) and homogenizing global diversity (14, 15). Whether consequence or cause of global change, the range expansion of exotic species depends on defying the barriers of natural dispersal (16, 17) and on adaptation to a novel local abiotic and biotic environment in which the organism did not evolve (6).
In the marine environment, where the rate of introduction of alien species has dramatically increased over the past century (e.g., refs. 18 and 19), ship-ballasting procedures, increased volume of transoceanic shipping and intracoastal boating, and modern aquaculture practices all contribute to the establishment of veritable invasion corridors (20, 21). Likewise, the propagule pressure hypothesis (sensu Williamson, ref. 17, p. 45; and ref. 6) postulates that, even if preceded by many failed attempts, the recurrence of introductions amplifies the very low initial probability of successful establishment. Furthermore, successive migration events may also increase overall genetic diversity, thereby improving the likelihood of encountering adaptive genetic variants and the rate of adaptive evolution (6, 22–24). Successful settlement of many marine invaders may indeed be explained by recurrent introductions (16). Alternatively, successful introductions of exotic species may be linked to changes in the recipient region (2, 25). For example, in the marine environment, many successful invaders have settled in anthropogenic habitats, such as marinas, commercial shipping ports, shellfish farms, or heavily disturbed/polluted natural areas that impose novel ecological conditions even for the native species community (6). In this context, the widely introduced brown alga Undaria pinnatifida provides an excellent opportunity for exploring the mechanisms and processes behind the apparent paradox (6) of rapid adaptation of exotic species to a wide spectrum of environments, competing with native species that have naturally evolved there.
U. pinnatifida (Laminariales, Alariaceae), otherwise known as the edible seaweed wakame, is an Asian species native to Japanese, Korean, and Chinese waters, where it is extensively cultivated. The first observation of this species outside its native Asian range was recent (1971, in the Mediterranean Sea). Since then, this kelp has been introduced to both Northern and Southern hemispheres, spreading to the Atlantic, Mediterranean, Adriatic, and North Sea coasts of Europe (see ref. 26); New Zealand (27); Australia (28); Argentina (29); and, most recently, to the American Pacific coast (California, ref. 26; Mexico, ref. 30). The patterns and intensity of invasion of U. pinnatifida vary around the world, ranging from recent rapid long-distance spread in Californian harbors (31); slow longdistance spread in European marinas, concomitant with the presence of wakame farms in Brittany, France (32); and heavy proliferation and naturalization in both harbors and natural subtidal zones in Australasia (28, 33, 34) and Argentina (35). The rapid spread of the species within regions of introduction is thought to be linked to its biological characteristics (i.e., annual life history, high growth rates, and high fecundity), typical of pioneer species (36). Because this kelp can be both accidentally transported by various vectors such as hull fouling, ballast water, and shellfish-farming transplants and also deliberately introduced for aquaculture purposes, the source, number, and diversity of introductions may contribute to the various degrees of successful settlement around the world (11, 25).
Despite increased public awareness and, in some cases, legislative measures, until now the few empirical studies of U. pinnatifida have concentrated on spread and local ecological dynamics (26, 31, 33). However, phylogeographic analyses lend insight into the mechanisms of introduction, colonization, and spread of invaders (23). Although the mitochondrial genome of a kelp has been entirely sequenced (37), no gene genealogy studies have been attempted in brown algae at the population level. Here, we use two polymorphic mitochondrial markers (i.e., intergenic spacer regions) to compare the diversity, sequence divergence (gene genealogies), and patterns of invasion among regions of introduction of native, cultivated, and spontaneous introduced populations of U. pinnatifida from around the world.
Materials and Methods
Samples representing the present-day distribution of U. pinnatifida were collected from five natural populations located in the native range (Japan, Korea, and China), 16 natural populations in all documented (before 2004) regions of introduction (Europe, mainland Australia, Tasmania, New Zealand, Argentina, and California; Fig. 1; details in Table 2, which is published as supporting information on the PNAS web site), and cultivated populations from Japan and Korea (commercial specimens) and in France (sampled directly from the farm). In each population, 23–24 individuals were collected, but commercial specimens comprised fewer individuals. Excised thallus tissue (≈5cm2) was dried in silica gel before DNA analysis. Total DNA was extracted from ≈5 mg of dried tissue by using the Nucleospin Multi96 Plant extraction kit (Macherey & Nagel) according to the manufacturer's instructions. Using primers designed from the complete sequence of the mitochondrial genome of Laminaria digitata (Laminariales, Laminariaceae) (37), two mitochondrial intergenic spacer regions were PCR-amplified: (i) between the atp8 and trnS genes and (ii) between the trnW and trnI genes, hereafter called atp8-S and W-I, respectively. Primers were designed in highly conserved coding regions flanking each spacer. For atp8-S, 239 base pairs were amplified by using atp8-trnS-F (3' end of atp8, 5'-TGTACGTTTCATATTACCTTCTTTAGC-3') and atp8-trnS-R (5' end of trnS, 5'-TAGCAAACCAAGGCTTTCAAC-3') primers and for W-I, 292 base pairs were amplified by using trnW-trnI-F (3' end of trnW, 5'-GGGGTTCAAATCCCTCTCTT-3') and trnW-trnI-R (5' end of trnI, 5'-CCTACATTGTTAGCTTCATGAGAA-3') primers.
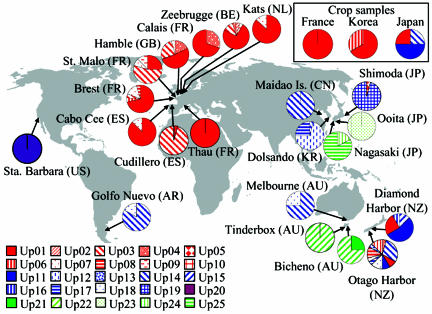
Fig. 1.
Distribution of the 25 mtDNA haplotypes detected in the 24 wild and cultivated U. pinnatifida populations sampled around the world. Color-motif pie charts display relative frequencies of each haplotype in each population. Foreground colors of the pie charts refer to the clusters defined as in Fig. 2: red, cluster A; red and blue, cluster B1; and green, cluster B2. The international code of the country of origin of each population is indicated in parentheses.
For both loci, PCRs were carried out in 20 μl containing 5 μl of sample DNA diluted 1:100/0.1 mg/ml BSA/2.5 mM MgCl2/75 mM Tris·HCl/20 mM (NH4)2SO4/200 μM dNTPs/250 nM each primer/0.5 unit Thermoprime Plus DNA polymerase (Abgene, Epsom, U.K.) and run on an MJ Research (Cambridge, MA) PTC200 thermocycler. After an initial denaturation step (95°C, 5 min), touchdown PCR was carried out for five cycles of 30 s at 95°C, 30 s at 60° reduced by 1°C each subsequent cycle, and 45 s at 72°C, followed by 30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 45 s.
To detect single-site substitutions at the atp8-S locus, all PCR products were sequenced in both directions by using the amplification primers on an Applied Biosystems PRISM 3100 Automated DNA Sequencer (PE Applied Biosystems). For W-I, a more efficient screening strategy was developed by using single-stranded DNA conformation polymorphism (SSCP) analysis (38). PCRs were performed by using a modified forward primer (trnW-trnI-F2, 5'-ATGAGGGTAAGTTTTTCACAGT-3'), designed based on preliminary sequencing of seven individuals. To check the accuracy of SSCP typing, an average of nine individuals possessing each distinct W-I SSCP profile were sequenced. Each SSCP profile was found to have the same unique sequence, and some differed by as little as 1 base pair.
Sequences were aligned by using clustalw (39) in bioedit (40). No ambiguous base compositions were encountered, suggesting that heteroplasmy is absent in Undaria (see ref. 41). Due to the nonindependence of the two loci (D'= 0.77, P < 0.001), statistical analyses were carried out on concatenated intergenic sequences with a total length of 333 bp (see Table 3, which is published as supporting information on the PNAS web site). Estimates of haplotypic (He) and nucleotide (π) diversity were calculated for each population, within three main regional groups [native region (Far-East Asia, i.e., Japanese, Korean, and Chinese populations); Europe, Australasia (mainland Australia, Tasmania, and New Zealand)]; and for the entire dataset by using arlequin v. 2.0 (http://anthropologie.unige.ch/arlequin). This software was also used to carry out an analysis of molecular variance by computing Φ statistics, analogous to Wright's fixation indices, F statistics, and exact tests of differentiation to investigate population structure (42) (i) among populations within each region, (ii) among and within native and introduced regions, and (iii) among and within the two introduced regions. Gene genealogies were reconstructed with a reduced-median network (43) by using network 4.1 (www.fluxusengineering.com/sharenet.htm). According to the author's recommendations, superfluous links were eliminated by using the maximum parsimony calculation after down-weighting by half rapidly mutating sites (two single-site substitutions). Analyses were carried out treating the two multiple-site insertion deletions (indels; see Results) either as tandem independent sites (total sequence length, 333 bp) or as two single blocks (i.e., one block coding a 2-bp indel and one, a 16-bp indel, decreasing the total sequence length to 316 positions) by using several block-weighting schemes. Because we obtained the same results with either coding method, only values from the more conservative block-coding method are explicitly given and discussed.
Results
Genetic Diversity Across Populations and Regional Groups. Over the whole dataset (524 individuals), 10 polymorphic sites, nine substitutions, and one indel were identified at the atp8-S locus and 24 polymorphic sites, five substitutions, and three indels of 16, 1, and 2 bp at the W-I locus. At each locus, nine different sequences were identified. The combined dataset (concatenated sequence) was made up of 25 haplotypes, resulting in high overall haplotype (He = 0.86) and nucleotide (π = 0.0139) diversity. More haplotypes were detected than would be expected in the complete absence of homoplasy (17; i.e., the sum of the number of haplotypes at both loci minus one). In addition, although highly significant, the linkage disequilibrium value between the two loci was not close to unity, probably reflecting the occurrence of several evolutionary pathways among haplotypes rather than recombination. Statistical parsimony associated with network reconstruction is particularly recommended for managing homoplasy (43). In any case, the limited homoplasy detected (see loops in Fig. 2) was not uniquely due to the concatenation of the two loci (data not shown).
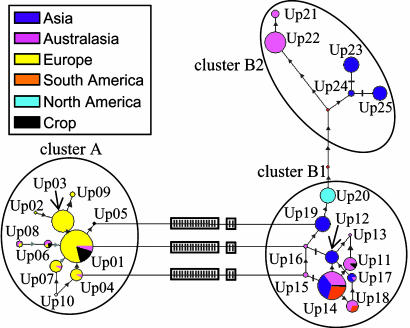
Fig. 2.
Haplotypic network and regional distribution of the 25 U. pinnatifida mtDNA haplotypes. Circle sizes are proportional to the number of individuals observed for each haplotype, and color pie charts indicate the distribution of the haplotype among wild populations in the five world-wide regions and crops. Connecting lines show mutational pathways among haplotypes; triangles represent single-site substitutions; perpendicular bars, indels; and boxed bars, blocks of 16 and 2 tandem indels. Unlabeled red nodes indicate inferred steps not found in the sampled populations. Clusters group closely related haplotypes.
The distribution of the 25 haplotypes is detailed in Fig. 1. At the regional scale, the native region (Asia) showed high genetic diversity with eight haplotypes and He = 0.84 (π = 0.0121; Table 1). Introduced regions displayed contrasting levels of genetic diversity. Australasia, with 14 different haplotypes, showed the highest genetic diversity (He = 0.81; π = 0.0166; Table 1). In Europe, where only nine different haplotypes were observed, haplotype diversity was only moderate (He = 0.55), and nucleotide diversity was reduced (π = 0.0021; Table 1). This European pattern was due to the high frequency of one haplotype (Up01) in all but two populations (St. Malo and Cudillero; Fig. 1).
Table 1. Molecular diversity indices for each geographic region and crops.
| Region | Nind | Npop | Ppop | Nhap | He | S | π × 102 (SD) |
|---|---|---|---|---|---|---|---|
| Asia | 118 | 5 | 3 | 8 | 0.84 | 11 | 1.21 (0.68) |
| Europe | 208 | 9 | 8 | 9 | 0.55 | 4 | 0.21 (0.18) |
| Australasia | 120 | 5 | 4 | 14 | 0.81 | 14 | 1.66 (0.90) |
| South America | 24 | 1 | 1 | 2 | 0.29 | 1 | 0.09 (0.11) |
| North America | 23 | 1 | 0 | 1 | 0.00 | 0 | 0.00 (0.00) |
| Crops | 31 | 3 | 2 | 4 | 0.24 | 6 | 0.29 (0.23) |
| Total | 524 | 24 | 18 | 25 | 0.86 | 17 | 1.39 (0.76) |
Nind, number of individuals; Npop, number of populations; Ppop, number of polymorphic populations; Nhap, number of haplotypes; He, haplotypic diversity; S, number of polymorphic sites; π, nucleotide diversity.
At the population level, He ranged from 0.0 to 0.88 (Table 2). Monomorphic populations were found in both native and introduced groups. Nevertheless, highly contrasting patterns were observed in Asia compared with Europe and Australasia. Native populations showed the lowest within-population genetic diversity [mean He, 0.17 ± 0.20, number of populations (Npop) = 5], whereas the two main introduced regions showed considerably higher values (Europe, mean He, 0.36 ± 0.23, Npop = 9; Australasia, mean He, 0.46 ± 0.33, Npop = 5), with the most genetically diverse population found in Australasia (i.e., Dunedin; Table 2). Over all cultivated populations, the genetic diversity was very low (He = 0.24, Table 1) due to both the monomorphism of the French crop (Fig. 1) and the consistent presence of the Up01 haplotype whatever the sample.
Among-Region Effects in the Population Genetic Structure. Genetic differentiation was significant among populations within each region, but differentiation was at least twice as great among populations in either Far-East Asia or Australasia as compared with Europe (native, among-populations fixation index ΦST = 0.960; Australasia, ΦST = 0.800; Europe, ΦST = 0.386; all P < 0.001). When comparing Australasia with native populations, significant genetic structure was found at the within-region level (among-populations within-regions fixation index, ΦSC = 0.868, P < 10–5) but not at the among-regions level (among-regions fixation index, ΦCT = –0.019, P = 0.533) due to very high variation among-populations within each region as well as shared haplotypes (e.g., Up14) among regions. Conversely, European populations were highly genetically differentiated from the populations located in the native range (ΦCT = 0.622, P < 10–5). Furthermore, European and Australasian populations were found to be significantly differentiated from each other (ΦCT = 0.553, P < 0.001).
Haplotypic Network. The reduced-median network revealed two major groups, essentially defined by insertion or deletion of a 16-bp block (Fig. 2, no intermediate haplotypes observed). Although branch lengths varied between groups according to indel-coding method and weighting scheme, the overall topology of the network always remained the same. The first group, cluster A, formed a group of closely related haplotypes with deletion of the 16-bp block. The second group, cluster B, constituted by haplotypes with insertion of the 16-bp block, was split into two subclusters. Cluster B1 groups closely related haplotypes, some of which differ from cluster A only by the presence of the 16-bp indel (i.e., Up15, Up16, and Up19). Cluster B2 included five haplotypes (Up21–Up25) that were separated from all of the other B haplotypes by a minimum of five mutational steps (Fig. 2). Finally, there were no alternative links among clusters A and B2 or between B1 and B2.
At the regional level, haplotypes observed in Asia and Australasia were distributed among all three clusters. Conversely, all haplotypes detected in Europe belonged exclusively to cluster A. Cluster B2 showed the most limited distribution, being revealed only in Southern Japan and Tasmania. The unique haplotype found at Santa Barbara (Up20) occupied a pivotal position as the closest B1 haplotype to the B2 cluster. At the population level, only four populations, located in Asia and Australasia, showed haplotypes belonging to more than one cluster: both New Zealand populations, Japanese-cultivated, and Shimoda (Japan) displayed both A and B1 haplotypes.
Discussion
Characterizing Genetic Diversity: Introduced Populations Show both Severe Founder Effects and Elevated Levels of Genetic Diversity. The two mitochondrial intergenic regions developed here revealed 25 mitochondrial haplotypes, distributed both among and within populations, and proved efficient for use for a gene genealogy study at the population level in a brown seaweed. Although two complete brown alga mitochondrial genomes are available (37), published mitochondrial markers have been limited to conserved coding regions used for interspecific hybridization and higher-order phylogenies (44–46) except for one study of Ectocarpus laboratory strains (47). The intraspecific sequence divergence (π = 0.0387, Table 1) revealed in our study was more than four times higher than sequence divergence reported for RuBisCo spacer, of comparable sequence length, among different species of Undaria (π = 0.008, 268 bp, ref. 48), making these markers useful for phylogeographic studies.
Unexpectedly high genetic structure was revealed among Asian populations, i.e., in the native range of the species. This striking population structure in the native region, as attested by generally low within-population genetic diversity and little sharing of haplotypes among the sampled Asian populations, clearly suggests that single-source introductions would generate very low genetic diversity within introduced populations, even with large effective numbers of founders. Therefore, only multiple independent introduction events from distinct native populations can explain the substantial within-population genetic diversity (see Fig. 1, Table 2) of most introduced U. pinnatifida populations. Monomorphism, the expected genetic signature of a severe population bottleneck arising from single-event introductions (49, 50), was observed in only three spontaneous introduced populations, namely Tinderbox (Australasia), Thau (France), and Santa Barbara (U.S.). Tinderbox and Thau appear to have resisted recurrent introduction events, because 15 and 30 years, respectively, have passed since the initial sighting of U. pinnatifida. Likewise, Santa Barbara also appears to arise from a single source; however, it was sampled in 2002, only 1 year after its reported introduction (26).
Different Patterns of Recurrent Introductions in Europe and Australasia. Although the greater average within-population genetic diversity in both Europe and Australasia clearly points to multiple introduction events, the highly contrasting patterns of sequence divergence in these two regions suggest different colonization dynamics. In Europe, the overall reduction in regional sequence divergence, coupled with weaker intraregion population structure, compared with Asia clearly indicates a low incidence of divergent multiple-source introductions in the northeast Atlantic Ocean and North Sea. On the other hand, the high regional He values, coupled with higher within-population nucleotide diversity in Australasia compared with Asia, can be explained only by recurrent introduction events from all over the native area.
From Crops to Harbors in Europe. All haplotypes observed in Europe showed low divergence and grouped together in one cluster (cluster A, Fig. 2). This pattern is largely explained by the presence of one haplotype, Up01 that is ubiquitous throughout Europe and makes up 63% of all observed haplotypes, reaching a frequency of unity in Thau. In Europe, U. pinnatifida was first observed in 1971 in the Thau Lagoon, a center of intense oyster farming. This introduction occurred by means of Pacific oyster imports (51, 52) originating from the Sendai region, Northern Honshu, Japan (53), where cultivation of U. pinnatifida is practiced (54). The original deliberately introduced individuals for pilot wakame farms in Brittany in 1983 (55) were derived from individuals originating in Thau, as are the current strains cultivated in the sampled French aquaculture farm. Indeed, the lone haplotype observed in the French crop population was identical to the one detected in Thau. Interestingly, haplotypes from cluster A were extremely rare in Asian populations, except in the cultivated samples, where Up01 was shared by both Korean and Japanese crops. Up01 appears thus to be prevalent in an intensely cultivated strain and to be the origin of primary introduction in Europe.
Both deliberate introduction for cultivation purposes (followed by escape of individuals outside the cultivation area) and local maritime transport have probably contributed to the spread of the Up01 haplotype from harbor to harbor (56), resulting in overall weak genetic structure in this region. Nevertheless, eight other haplotypes were detected in Europe. The close evolutionary relationship among these haplotypes suggests two scenarios: (i) rapid evolution of mitochondrial genome in Europe, with Up01 as a recent ancestor or (ii) recurrent introductions, also from cultivated strains. However, it is unlikely that, in the 30 years since the original introduction of Up01, recent sequence evolution could account for the presence of the other A haplotypes in Europe. Indeed, the oldest population in Europe, Thau, is still monomorphic. Furthermore, no signature of recent expansion was detected, as exemplified by the absence of significant departure from mutation-drift equilibrium in the European populations (Tajima's D =–0.04, P = 0.50; analysis not shown). On the other hand, as the result of Asian breeding programs (see ref. 57), haplotypes closely related to Up01 might also be associated with cultivated lines. For instance, Up06 present in at least one spontaneous European population (Brest, Fig. 1) was revealed in the Korean cultivated sample. Because of the close proximity of Pacific oyster farms and U. pinnatifida cultivation sites, importations of oysters may have served as vectors of introduction of cultivated strains of U. pinnatifida (52, 58). In addition, deliberate introductions of cultivated lines of U. pinnatifida, through official or unofficial channels, have probably occurred. For example, in northern Spain, where U. pinnatifida is also cultivated (59), Cudillero shows dominance of an alternate A haplotype. Cultivation may be at least partly responsible for the successful establishment of U. pinnatifida in Europe. Breeding programs reduce genetic variability and select for strains adapted for vigorous growth on ropes or nets, characteristics that may promote growth on artificial floating structures, typical of anthropogenic environments (ports, marinas, boat hulls, etc.), where spontaneous European populations of U. pinnatifida are usually found. Taken together, these pieces of evidence suggest that aquaculture was the source of both primary and secondary introductions of U. pinnatifida and its successful adaptation in Europe.
Multiple Ships and Divergent Sources in Australasia. All three haplotype clusters were well represented in Australasia, resulting in the highest values of sequence divergence observed at both the regional and population levels in this study, particularly in New Zealand. Based on morphological data and precise records of ship activities, it was suggested that two to three introductions to New Zealand from Asia by way of maritime traffic occurred independently in two distant ports (Wellington, in 1987 and Timaru, before 1988), with subsequent spread to intermediate points (27, 34). Our genetic data, however, suggest a greater number of recurrent (local) introductions all over New Zealand: of 12 haplotypes detected in New Zealand samples, only two were shared between Otago and Diamond Harbors. The heavy proliferation on natural substrates in New Zealand may be strongly linked to the redistribution of genetic variation from among (native) populations to within (introduced) populations, thereby stimulating novel, locally adaptive, genetic combinations.
Nevertheless, in contrast to New Zealand populations, those in Tasmania and mainland Australia showed relatively low within-population diversity, suggesting few locally successful, yet highly divergent, introduction events. Furthermore, although close geographically and connected via shipping traffic, Tasmania (first record of U. pinnatifida 1988, ref. 60) and Melbourne (1996, ref. 28) presented haplotypes that belonged to two different subclusters (B2 and B1, respectively; Figs. 1 and 2), suggesting independent primary introduction events (see ref. 34). Melbourne possesses haplotypes that are found in New Zealand, possibly representing a case of secondary relay from previously introduced populations, although a primary (perhaps cultivated) Asian origin cannot be excluded. Interestingly, haplotype composition in Argentina (first record of U. pinnatifida 1992, ref. 29) was highly similar to Melbourne, suggesting a single common origin for both introductions. Tasmania, on the other hand, appears to have arisen from an isolated and independent primary introduction. It was the only introduced area to show B2 haplotypes, separated by at least eight mutational steps from any other haplotype found in introduced populations. Furthermore, contrary to all other introduced areas, the initial populations in Tasmania were not found on floating structures in sheltered sites but on natural substrates in exposed sites (27, 60). In addition, the two Tasmanian populations have none of the four haplotypes found in the crop samples. The unusual Tasmanian haplotypes and habit could be interpreted as a cryptic (sub)species introduction. Given the highly localized distribution of B2 in Asia, the close phylogenetic relationship among the three Undaria species (61) and the great morphological plasticity noted within and among species in this genus (62, 63), cluster B2 may in fact represent an alternate Undaria subspecies. Likewise, specific ecological adaptations appear to have promoted other cryptic invasions, such as in the polychaete Marenzelleria sp. (64) or the suggested derived incipient species status of invasive Caulerpa taxifolia (65). Testing the hypothesis of reproductive isolation between mitochondrial haplotype clusters, assuming they correspond to divergent genetic strains, in U. pinnatifida calls for the use of nuclear markers with analysis of cytonuclear disequilibrium (66).
Conclusion
Genetic Characteristics and Invasibility. Genetic characteristics associated with invasion success are typically classified into two categories according to species: either (pre)adapted but relatively reduced genetic diversity resulting from population bottlenecks (e.g., ant, ref. 67; reed, ref. 68) or rapid evolution associated with enhanced levels of diversity due to recurrent introduction events (e.g., lizard, ref. 24; mollusks, refs. 69 and 70). In contrast, we uncovered a multiplicity of genetic characteristics in introduced U. pinnatifida populations. In particular, contrary to the well studied worldwide introduced alga C. taxifolia (65, 71), the pervasiveness of U. pinnatifida does not stem from the (rapid) evolution of a particular invasive type(s) with secondary relay around the world. We have effectively shown that the successful establishment of U. pinnatifida varies from (i) introduction of cultivated strains preadapted to anthropogenic habitats (i.e., floating structures and artificial substrata in Europe); (ii) divergent multiple-source introductions resulting in high genetic diversity promoting rapid local adaptation (New Zealand); (iii) putative secondary relay from these locally adapted invasive types (Melbourne and Argentina); and, finally, (iv) single-source, but cryptic, introductions (Tasmania). These different types of introduction processes have occurred independently around the world and promoted the settlement of the species over a large spectrum of environments.
We have demonstrated that a variety of microevolutionary processes can account for the success of a single worldwide invasive species. It is thus clearly difficult, based on localized genetic architecture only, to predict the invasive potential of a given species, and studies over the global range of introduction are essential. The multiplicity of processes and genetic signatures associated with the successful invasion of a given species confirms that multiple facets of global change, e.g., aquaculture practices, alteration of habitats, and increased traffic, act in synergy at the worldwide level, facilitating successful pandemic introductions.
Supplementary Material
Acknowledgments
This study would not have been possible without generous help from our colleagues from around the world (listed in Table 2) who sampled (or helped us sample) Undaria populations, for which we are extremely grateful. We thank C. Daguin, D. Jollivet, C. Lemaire, C. Maggs, and M. Valero for helpful comments on the manuscript. We are indebted to M. Perennou for help in using the facilities provided by the Génopole Ouest/Génomer sequencing platform. This study was part of the European Network of Excellence “Marine Genomics” (Contract 505403). This work was also supported by a Ph.D. fellowship (“Renouvellement des Compétences” Program) from the Région Bretagne (to M.V.) and the Centre National de la Recherche Scientifique through an Action Thématique et Incitative sur Programme (ATIP) grant (to F.V.) and a postdoctoral fellowship (to C.R.E.).
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY821889–AY821906).
References
- 1.Chapin, F. S. I., Zavaleta, E. S., Eviner, V. T., Naylor, R. L., Vitousek, P. M., Reynolds, H. L., Hopper, D. U., Lavorel, S., Sala, O. E., Hobbie, S. E., et al. (2000) Nature 405, 234–242. [DOI] [PubMed] [Google Scholar]
- 2.Carlton, J. T. (2000) in Invasive Species in a Changing World, eds. Mooney, H. A. & Hobbs, R. (Island Press, Washington, DC), pp. 31–53.
- 3.Occhipinti-Ambrogi, A. & Savini, D. (2003) Marine Pollut. Bull. 46, 542–551. [DOI] [PubMed] [Google Scholar]
- 4.Stachowicz, J. J., Terwin, J. R., Whitlatch, R. B. & Osman, R. W. (2002) Proc. Natl. Acad. Sci. USA 99, 15497–15500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suarez, A. V., Bolger, D. T. & Case, T. J. (1998) Ecology 79, 2041–2056. [Google Scholar]
- 6.Sax, D. F. & Brown, J. H. (2000) Global Ecol. Biogeogr. 9, 363–371. [Google Scholar]
- 7.Sala, O. E., Chapin, F. S., III, Armesto, J. J., Berlow, E., Bloomfield, J., Dirzo, R., Huber-Sanwald, E., Huenneke, L. F., Jackson, R. B., Kinzig, A., et al. (2000) Science 287, 1770–1774. [DOI] [PubMed] [Google Scholar]
- 8.Simberloff, D. & Von Holle, B. (1999) Biol. Invasions 1, 21–32. [Google Scholar]
- 9.Dukes, J. S. & Mooney, H. A. (1999) Trends Ecol. Evol. 14, 135–139. [DOI] [PubMed] [Google Scholar]
- 10.Levine, J. M. & D'Antonio, C. M. (1999) Oikos 87, 15–26. [Google Scholar]
- 11.Bax, N., Williamson, A., Aguero, M., Gonzalez, E. & Geeves, W. (2003) Marine Polut. 27, 313–323. [Google Scholar]
- 12.Sax, D. F. & Gaines, S. D. (2003) Trends Ecol. Evol. 18, 561–566. [Google Scholar]
- 13.Roberts, C. M. & Hawkins, J. P. (1999) Trends Ecol. Evol. 14, 241–246. [DOI] [PubMed] [Google Scholar]
- 14.McKinney, M. L. & Lockwood, J. L. (1999) Trends Ecol. Evol. 14, 450–453. [DOI] [PubMed] [Google Scholar]
- 15.Myers, N. (1997) Science 278, 597–598. [Google Scholar]
- 16.Kolar, C. S. & Lodge, D. (2001) Trends Ecol. Evol. 16, 199–204. [DOI] [PubMed] [Google Scholar]
- 17.Williamson, M. (1996) Biological Invasions (Chapman & Hall, London).
- 18.Ruiz, G. M., Fofonoff, P., Carlton, J. T., Wonham, M. J. & Hines, A., H. (2000) Annu. Rev. Ecol. Syst. 31, 481–531. [Google Scholar]
- 19.Goulletquer, P., Bachelet, G., Sauriau, P. G. & Noel, P. (2002) in Invasive Aquatic Species of Europe, eds. Leppäkoski, E., Gollash, S. & Olenin, S. (Kluwer, Dordrecht, The Netherlands), pp. 276–290.
- 20.Ricciardi, A. & MacIsaac, H. J. (2000) Trends Ecol. Evol. 15, 62–65. [DOI] [PubMed] [Google Scholar]
- 21.Drake, J. M. & Lodge, D. M. (2004) Proc. R. Soc. London 271, 575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, C. E. (2002) Trends Ecol. Evol. 17, 386–391. [Google Scholar]
- 23.Sakai, A. K., Allendorf, F. W., Holt, J. S., Lodge, M., Molofsky, J., With, K. A., Baughman, S., Cabin, R. J., Cohen, J. E., Ellstrand, N. C., McCauley, D. E., et al. (2001) Annu. Rev. Ecol. Syst. 32, 305–332. [Google Scholar]
- 24.Kolbe, J. J., Glor, R. E., Schettino, L. S., Lara, A. C., Larson, A. & Losos, J. B. (2004) Nature 431, 177–181. [DOI] [PubMed] [Google Scholar]
- 25.Carlton, J. T. (1996) Biol. Conserv. 78, 97–106. [Google Scholar]
- 26.Silva, P. C., Woodfield, A. R., Cohen, A. N., Harris, L. H. & Goddard, J. H. R. (2002) Biol. Invasions 4, 333–338. [Google Scholar]
- 27.Hay, C. H. (1990) Br. Phycol. J. 25, 301–313. [Google Scholar]
- 28.Campbell, S. J., Bité, J. S. & Burridge, T. R. (1999) Bot. Mar. 42, 231–241. [Google Scholar]
- 29.Casas, G. N. & Piriz, M. L. (1996) Hydrobiologia 326/327, 213–215. [Google Scholar]
- 30.Aguilar-Rosas, R., Aguilar-Rosas, L. E., Avila-Serrano, G. & Marcos-Ramírez, R. (2004) Bot. Mar. 47, 255–258. [Google Scholar]
- 31.Thornber, C. S., Kinlan, B. P., Graham, M. H. & Stachowicz, J. J. (2004) Mar. Ecol. Prog. Ser. 268, 69–80. [Google Scholar]
- 32.Floc'h, J.-Y., Pajot, R. & Mouret, V. (1996) Hydrobiologia 326/327, 217–222. [Google Scholar]
- 33.Valentine, J. P. & Johnson, C. R. (2003) J. Exp. Mar. Biol. Ecol. 295, 63–90. [Google Scholar]
- 34.Hay, C. H. & Sanderson, J. C. (1999) in Islands in the Pacific Century: Pacific Science Inter-Congress (University of the South Pacific, Suva, Fiji).
- 35.Orensanz, J. M. L., Schwindt, E., Pastorino, G., Bortolus, A., Casas, G., Darrigran, G., Elías, R., Lopez Gappa, J. J., Obenat, S., Pascual, M., et al. (2002) Biol. Invasions 4, 115–143. [Google Scholar]
- 36.Grime, J. P. (1977) Am. Nat. 111, 1169–1194. [Google Scholar]
- 37.Oudot-Le Secq, M. P., Kloareg, B. & Loiseaux de Goër, S. (2002) J. Phycol. 37, 163–172. [Google Scholar]
- 38.Sunnucks, P., Wilson, A. C. C., Beheregaray, L. B. & Zenger, K. (2000) Mol. Ecol. 9, 1699–1710. [DOI] [PubMed] [Google Scholar]
- 39.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hall, T. A. (1999) Nuclear Acids Symp. Ser. 41, 95–98. [Google Scholar]
- 41.Coyer, J. A., Hoarau, G., Stam, W. T. & Olsen, J. L. (2004) Mol. Ecol. 13, 1323–1326. [DOI] [PubMed] [Google Scholar]
- 42.Excoffier, L., Smouse, P. E. & Quattro, J. M. (1992) Mol. Mar. Biol. Biotechnol. 5, 295–298. [Google Scholar]
- 43.Posada, D. & Crandall, K. A. (2001) Trends Ecol. Evol. 16, 37–45. [DOI] [PubMed] [Google Scholar]
- 44.Coyer, J. A., Peters, A. F., Hoarau, G., Stam, W. T. & Olsen, J. L. (2002) Eur. J. Phycol. 37, 173–178. [Google Scholar]
- 45.Coyer, J. A., Peters, A. F., Hoarau, G., Stam, W. T. & Olsen, J. L. (2002) Proc. R. Soc. London 269, 1829–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ehara, M., Kitayama, T., Watanabe, K. I., Inagaki, Y., Hayashi-Ishimaru, Y. & Ohama, T. (1999) Phycol. Res. 47, 225–231. [Google Scholar]
- 47.Peters, A. F., Scornet, D., Müller, D. G., Kloareg, B. & Cock, J. M. (2004) Eur. J. Phycol. 39, 235–242. [Google Scholar]
- 48.Yoon, H. S. & Boo, S. M. (1999) Hydrobiologia 398/399, 47–55. [Google Scholar]
- 49.Allendorf, F. W. & Lundquist, L. L. (2003) Conserv. Biol. 17, 24–30. [Google Scholar]
- 50.Davies, N., Villanblanca, F. X. & Roderick, G. K. (1999) Genetics 153, 351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perez, R., Lee, J. Y. & Juge, C. (1981) Science Pêche 315, 1–12. [Google Scholar]
- 52.Wolff, W. J. & Reise, K. (2002) in Invasive Aquatic Species of Europe, eds. Leppäkoski, E., Gollasch, S. & Olenin, S. (Kluwer, Dordrecht, The Netherlands), pp. 193–205.
- 53.Boudouresque, C. F., Gerbal, M. & Knoepffler-Peguy, M. (1985) Phycologia 24, 364–366. [Google Scholar]
- 54.Ohno, M. & Largo, D. B. (1998) in Seaweed Resources of the World, eds. Critchley, A. T. & Ohno, M. (Japanese International Cooperation Agency, Yokosura, Japan), pp. 1–14.
- 55.Perez, R., Kaas, R. & Barbaroux, O. (1984) Science Pêche 343, 3–15. [Google Scholar]
- 56.Fletcher, R. L. & Farrel, P. (1999) Helgolander Meeresuntersuchungen 52, 259–275. [Google Scholar]
- 57.Critchley, A. T. & Ohno, M. (1998) in Seaweed Resources of the World (Japanese International Cooperation Agency, Yokosura, Japan), p. 431.
- 58.International Council for the Exploration of the Sea (2001) ICES Coop. Res. Rep. 248, 58–59. [Google Scholar]
- 59.Juanes, J. A. & Sosa, P. A. (1998) in Seaweed Resources of the World, eds. Critchley, A. T. & Ohno, M. (Japan International Cooperation Agency, Yokosura, Japan), pp. 164–175.
- 60.Sanderson, J. C. (1990) Bot. Mar. 33, 153–157. [Google Scholar]
- 61.Yoon, H. S., Lee, J. Y., Boo, S. M. & Bhattacharya, D. (2001) Mol. Phylogenet. Evol. 21, 231–243. [DOI] [PubMed] [Google Scholar]
- 62.Saito, Y. (1972) in Contribution to the Systematics of the Benthic Marine Algae of the North Pacific, eds. Abbott, A. & Kurogi, M. (Japanese Society of Phycology, Kobe, Japan), pp. 117–132.
- 63.Stuart, M. D., Hurd, C. L. & Brown, M. T. (1999) Hydrobiologia 398/399, 191–199. [Google Scholar]
- 64.Bastrop, R., Jürss, K. & Sturmbauer, C. (1998) Mol. Biol. Evol. 15, 97–103. [DOI] [PubMed] [Google Scholar]
- 65.Meusnier, I., Valero, M., Olsen, J. L. & Stam, W. T. (2004) Eur. J. Phycol. 39, 83–92. [Google Scholar]
- 66.Basten, C. J. & Asmussen, M. A. (1997) Genetics 146, 1165–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tsutsui, N. D., Suarez, A. V., Holway, D. A. & Case, T. J. (2000) Proc. Natl. Acad. Sci. USA 97, 5948–5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Saltonstall, K. (2002) Proc. Natl. Acad. Sci. USA 99, 2445–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bachelet, G., Simon-Bouhet, B., Desclaux, C., Garcia-Meunier, P., Mairesse, G., de Montaudouin, X., Raigné, H., Randriambao, K., Sauriau, P.-G. & Viard, F. (2004) Mar. Ecol. Prog. Ser. 276, 147–159. [Google Scholar]
- 70.Dupont, L., Jollivet, D. & Viard, F. (2003) Mar. Ecol. Prog. Ser. 253, 183–195. [Google Scholar]
- 71.Jousson, O., Pawlowski, J., Zaninetti, L., Zechman, F., Dini, F., Di Guiseppe, G., Woodfield, G., Millar, R. & Meinesz, A. (2000) Nature 408, 157. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.