Abstract
Gene 33 (Mig6, ERRFI1) is an adaptor protein with multiple cellular functions. We recently reported that depletion of this protein promotes lung epithelial cell transformation induced by hexavalent chromium [Cr(VI)]. However, the early molecular events that mediate this process are not clear. In the present study, we used single-cell RNA sequencing to compare gene expression profiles between BEAS-2B lung epithelial cells chronically exposed to a sublethal dose of Cr(VI) with or without CRISPR/cas9-mediated deletion of Gene 33. Our data reveal 83 differentially expressed genes. The most notable changes are genes associated with cell adhesion, oxidative stresses, protein ubiquitination, epithelial-mesenchymal transition/metastasis, and WNT signaling. Up-regulation of some neuro-specific genes is also evident, particularly ubiquitin carboxyl-terminal hydrolase L1 (UC HL1), a deubiquitinase and potential biomarker for lung cancer. Gene 33 deletion and/or Cr(VI) exposure did not cause discernable changes in cell morphology. However, Gene 33 deletion led to a modest but significant reduction of cells in the G2/M phase of the cell cycle regardless of Cr(VI) exposure. Gene 33 deletion also significantly reduced cell proliferation. Interestingly, Cr(VI) exposure eliminated the difference in cell proliferation between the two genotypes. Gene 33 deletion also significantly elevated cell migration. Our data indicate that combined Gene 33 deletion and chronic Cr(VI) exposure produces a gene expression pattern and a phenotype resemble those of the transformed lung epithelial cells. Given the known association of UCHL1 with lung cancer, we propose that UCHL1 is an important player in the early stage of lung epithelial cell transformation and tumorigenesis.
Keywords: Gene 33/Mig6, RNA-seq, Single cell, carcinogenesis, lung, Chromium
INTRODUCTION
Gene 33 is an inducible adaptor protein containing multiple domains for protein-protein interaction and signal transduction (Makkinje et al., 2000; Xu et al., 2005). This protein has a broad functional profile, which is reflected by its interaction with a number of important intracellular proteins (Fiorentino et al., 2000; Makkinje et al., 2000; Tsunoda et al., 2002; Xu et al., 2005; Zhang et al., 2007a; Hopkins et al., 2012). The most prominent function of Gene 33 identified to date is its inhibition of tyrosine kinases of the EGF receptor family thereby inhibiting the biological events mediated by these receptors (Xu et al., 2005; Anastasi et al., 2007; Zhang et al., 2007a). Because Gene 33 can also be induced by activation of the EGFR family receptors, it is considered a feedback inhibitor of these receptors (Anastasi et al., 2003; Zhang and Vande Woude, 2007). In the past decade or so, an increasing number of additional functions of this protein have been identified. Gene 33 binds to and activates the protein tyrosine kinase c-Abl thereby promoting apoptosis in a p73-dependent manner (Hopkins et al., 2012). Gene 33 appears to be involved in replicative and oncogene-induced cell senescence through its inhibition of EGFR signaling (Xie et al., 2013). Gene 33 has also been shown to regulate the function of NF-κB (Tsunoda et al., 2002). Accumulating evidence has linked Gene 33 with tumor cell migration and metastasis, likely by regulating the EGFR and HGF/c-Met signaling pathways, and the small G protein CDC42 (Pante et al., 2005; Jiang et al., 2016). Our recent work showed that Gene 33 may also be involved in the DNA damage response and genomic instability induced by the genotoxic carcinogen hexavalent chromium [Cr(VI)] (Park et al., 2016).
Current evidence supports a role of Gene 33 as a tumor suppressor in the lung. The evidence includes the existence of nonsense or missense mutations of gene 33 (errfi1) in lung cancers (Zhang et al., 2007b), reduced or loss of Gene 33 expression in human lung cancer samples and cell lines (Zhang et al., 2007b; Li et al.), and high incidence of spontaneous lung adenoma and adenocarcinoma in gene 33 null mice (Ferby et al., 2006; Zhang et al., 2007b). In addition, frequent loss of heterozygosity (LOH) of chromosome 1p36.32 (where gene 33 is located) occurs in lung cancer and is associated with tobacco smoking (Tseng et al., 2005). Despite the clear association of Gene 33 with lung cancer, the molecular mechanism underlying the involvement of Gene 33 in lung carcinogenesis is not fully understood, particularly at the early stage of cell transformation.
Cr(VI) is a well-established human lung carcinogen (Braver et al., 1985; Gibb et al., 2000; Park et al., 2004; Costa and Klein, 2006; Holmes et al., 2008; Halasova et al., 2009). Despite the consensus that Cr(VI) induces carcinogenesis by generating oxidative stresses during its intracellular reduction process which leads to DNA damage and genomic instability (Cheng et al., 1998; Cheng et al., 2000; Ding and Shi, 2002; Ha et al., 2003; Hu et al., 2011), the molecular mediators of this process remain incompletely understood. Our recent study demonstrated a link between Gene 33 and Cr(VI)-induced lung epithelial cell transformation (Park et al., 2016). We found that Gene 33 depletion promotes Cr(VI)-induced cell transformation in lung epithelial cells by elevating genomic instability (Park et al., 2016). To further understand how Gene 33 may regulate Cr(VI)-induced cell transformation at the molecular level, we studied the change in the global gene expression profile as a result of combined Gene 33 deletion and chronic Cr(VI) exposure in BEAS-2B lung epithelial cells using single-cell RNA-sequencing (scRNA-seq). We identified significant alteration in gene expression of 83 genes. These genes are associated with a wide array of cellular functions, including cell adhesion, oxidative stresses, transcriptional regulation, cell migration/metastasis, and protein ubiquitination. Of particular interest is the up-regulation of several neuro-specific genes, including ubiquitin carboxyl-terminal hydrolase L1 (UCHL1), a neuro-specific deubiquitinase whose overexpression is common in lung cancer. Our results suggest a potentially prominent role of UCHL1 in the early process of lung epithelial cell transformation and lung carcinogenesis.
MATERIALS AND METHODS
Cell lines and cell culture
BEAS-2B human lung bronchial epithelial cells were obtained from American Type Culture Collection. Cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) and antibiotics (100 unit/ml of penicillin and 100 μg/ml of streptomycin sulfate) under 37°C with 5% CO2. Cells were maintained by passing before reaching confluency.
Antibodies and siRNA
The antibody to Gene 33 has been described previously (Xu et al., 2005; Xu et al., 2006). The antibody to γH2AX was purchased from Biolegend. The antibody to GAPDH was from Santa Cruz Biotechnology. Antibodies to UCHL1 and H2AX were obtained from Cell Signaling Technology. The antibody for KRBOX1 was purchased from Sigma-Aldrich. The siRNA oligo targeting KRBOX1 was obtained from Dharmacon.
Western bolt
Standard Western blot procedure was used throughout the study. SDS-PAGE was carried out using the mini gel system from Bio-Rad after harvesting cells with 1X sample buffer. Proteins in gels were then transferred to PVDF membrane. After blocking with TBST containing 5% non-fat dry milk, the membrane was incubated overnight with primary antibodies diluted with TBST containing 5% BSA using dilutions suggested by the manufacturers. After thorough wash with TBST, the membrane was further incubated with horse radish peroxidase-conjugated secondary antibodies for 1 hour at room temperature followed by thorough washing with TBST buffer. The signals were developed with an ECL system (Pierce).
CRISPR/cas9-mediated Gene 33 deletion
A double-stranded sgRNA oligo (5′-CACCGCAGTCTGAACTCTTCTGCTC-3′/5′-AAACGAGCAGAAGAGTTCAGACTGC-3′) was cloned to pX330 vector (Addgene) and transfected to BEAS-2B cells along with a vector expressing GFP (pcDNA3-GFP, at 1/10 of the amount of pX330). Transfected cells expressing high levels of GFP (top 3%) were isolated 24 hours post-transfection using fluorescence-activated cell sorting (FACS) and seeded individually to 96-well plates. After clonal expansion, the expression levels of Gene 33 in cell clones were determined using WB. Clones with dramatically reduced level of Gene 33 were selected and tested for indel formation using the T7E1 assay according to the standard procedure and the primer pair 5′-GAGCCATGGGGAATATGAGG-3′/5′-GAAGATCCCACGTCCATGAA-3′. The same procedure was carried out using the empty pX330 vector to produce WT clones.
Flow cytometry
Cells were cultured in 6-well culture plates in triplicates, detached 24 hours later using Trypsin-EDTA fixed with 1% formaldehyde, stained with DAPI, and analyzed by flow cytometry using a MoFlo XDP high-speed flow cytometer.
Cell proliferation assay
BEAS-2B cells were seeded at 4×104/well in DMEM with 10% FBS in 12 well-plates in triplicates. At 24 hours, 48 hours, and 72 hours after seeding, cells were detached using Trypsin-EDTA and counted under a microscope using hemocytometer. Growth curves were then plotted. Data are shown as mean ± SD. Statistical significance was determined by the Student’s t test.
Cell migration assay
Confluent monolayer of BEAS-2B cells cultured in DMEM with 10% FBS under 37°C with 5% CO2 were scratched with p1000 pipet tips. Cells were then washed with PBS and incubated in DMEM containing 1% FBS. Images were acquired under 40X magnification from three fields of each cell culture at 0 hour, 16 hours, and 24 hours after scratching. Gap distances were measured on the images using ImageJ software. Differences in gap distances between 0 hour and 16 or 24 hours were calculated and plotted. Data are shown as mean ± SD. Statistical significance was determined by the Student’s t test.
scRNA-seq
The protocol for scRNA-seq was adopted and modified from a previously published protocol (Picelli et al., 2014). Briefly, after trypsinization, single cells were mouth pipetted under a microscope into a 96-well plate with 6.5 μl of lysis buffer in each well. We picked 24 WT and 24 KO single cells, respectively. The lysis buffer composed of 4.5 μl of 0.2% Triton X-100 (Sigma-Aldrich), 2 U RNaseOUT, 1.0 μl of 10 mM dNTP (Life Technologies), and 1.0 μl of 10 μM oligo-dT30VN primer (Exiqon). Single cells were lysed at 72°C for 5 min and transferred on ice. Reverse transcription (RT) mix was then added, which contained 2.0 μl of 5× first-strand buffer, 0.5 μl of reverse transcriptase III, 0.5 μl of 100 mM DTT, 0.25 μl of RNaseOUT (Life Technologies), 2.0 μl of 5M betaine, 0.06 μl of 1M MgCl2 (Sigma-Aldrich), and 0.1 μl of 100μM template-switching oligo (TSO, Exiqon). The RT reaction was carried out with the initial incubation at 42°C for 90 min and 10 cycles of 50°C for 2 minutes and 42°C for 2 minutes. PCR mix of 12.5 μl of Q5 2× Hot-Start HiFi mix (New England Bio-labs) and 0.25 μl of 10μM In-Site PCR primers (Exiqon). The PCR reaction was as follows: 98°C incubation for 30 seconds; 20 cycles of 98°C for 20 seconds, 67°C for 15 seconds, 72°C for 6 minutes; followed by 5 minutes incubation at 72°C. The PCR products were purified using 40 μl of Ampure XP beads (Beckman Coulter). The quality of double-stranded cDNA (dsDNA) was assessed by Bioanalyzer 2100 (Agilent Technologies). We selected five WT and five KO cDNA samples in good quality and quantity for the NGS. The dsDNA (1 ng each) was subjected to library preparation using the Nextera XT DNA sample preparation kit (Illumina) and individualized index, as per the manufacturer’s instruction. The resulting library was assessed by Bioanalyzer 2100 and quantitated by Qubit dsDNA HS Assay with Qubit 2.0 Fluorometer (Invitrogen). After normalization of each sample, 10 libraries were pooled together and loaded into NextSeq 550 sequencing system (Illumina). Paired-end sequencing, 75 base pairs (bp) ×2, was performed using NextSeq Mid-Throughput Reagent v2 (Illumina), receiving from 11.5 to 14.3 million short-reads per sample. The data analyses were conducted using the TopHat-DESeq2 pipeline in the BaseSpace (Illumina) with the hg19 human genome as reference. From 89.83% to 91.73% short-reads were aligned to hg19.
RESULTS
Gene 33 deletion and/or chronic sublethal Cr(VI) exposure does not cause visible morphological changes of BEAS-2B cells
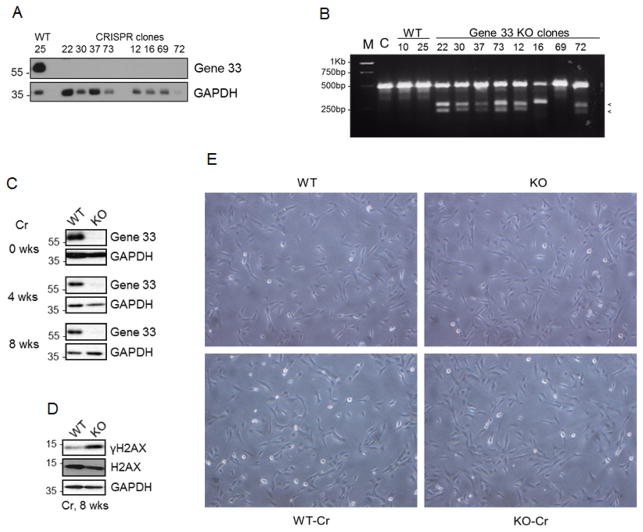
We used CRISPR/cas9 system to delete Gene 33 from BEAS-2B cells as described in Materials and Methods. We isolated 8 cell clones (KO) with dramatically reduced Gene 33 expression from cells transfected with the vector containing the sgRNA targeting gene 33 and two clones (WT) from cells transfected with the empty vector (Figure 1A). T7E1 assay confirmed the existence of indels, indicated by appearance of two smaller DNA fragments after enzymatic digestion, in most KO cells (Figure 1B). We chose a WT clone (#25) and a KO clone (#22) and treated them with a sublethal dose of Cr(VI) (0.5μM, Na2CrO4) continuously for 8 weeks. As shown in Figure 1C, KO cells maintained low Gene 33 expression levels throughout the treatment period compared to those of WT cells. Cr(VI) exposure led to a higher level of γH2AX in KO cells than in WT cells (Figure 1D), consistent with our previous observation that Gene 33 depletion with RNAi enhances the Cr(VI)-induced DNA damage response (Park et al., 2016). WT or KO cells with or without Cr(VI) treatment showed little visible difference in morphology at the population level (Figure 1E). This result agrees with a previous observation that long-term treatments with potassium dichromate (K2Cr2O7) at concentrations of 0.5μM or below does not affect the morphology of BEAS-2B cells (Costa et al., 2010).
FIGURE 1. CRISPR/cas9-mediated Gene 33 deletion and chronic sublethal Cr(VI) exposure did not cause visible morphological changes of BEAS-2B cells.
A. CRISPR/cas9-mediated Gene 33 deletion was carried out in BEAS-2B cells as described in Materials and Methods. Clones of control (WT) and Gene 33 deletion (KO) cells were isolated and expanded. Total cellular proteins of these cells were collected and subjected to Western blot (WB) to detect Gene 33. GAPDH was used a loading control. B. The T7E1 assay was performed using primers described in Materials and Methods on WT and KO cell clones. The appearance of two small DNA fragments indicates indel formation. M: DNA marker, C: the parental BEAS-2B cells. C. WB showing expression of Gene 33 at different time points after Cr(VI) treatment at 0.5 μM in WT and KO cells. D. WB showing the levels of γH2AX in WT and KO cells after 8 week treatment with Cr(VI) at 0.5 μM. E. Morphology of WT and KO cells with or without Cr(VI) treatment at 0.5 μM for 8 weeks. Pictures were taken at 60X.
scRNA-seq reveals differentially expressed genes between Cr(VI)-treated WT and KO cells
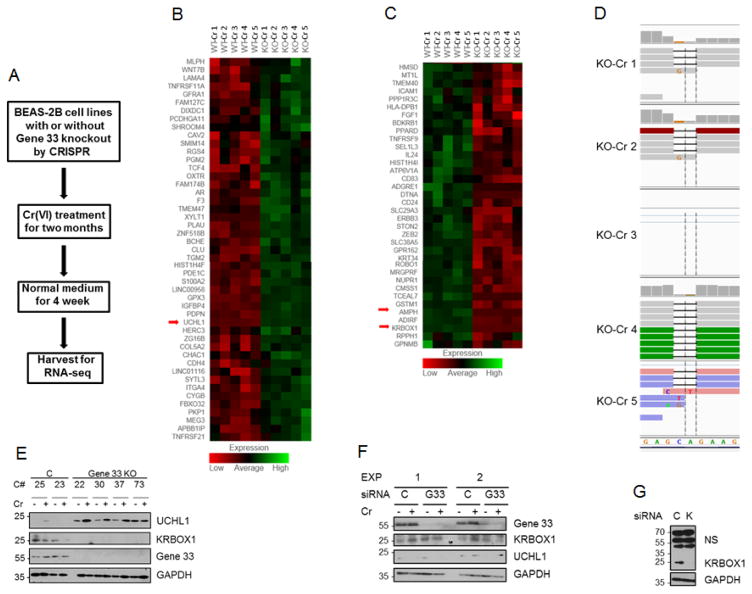
To investigate the effect of Gene 33 deletion on the global gene expression profile of BEAS-2B cells after chronically exposed with Cr(VI), we performed scRNA-seq on WT and KO cells treated with Cr(VI) for 8 weeks (named WT-Cr and KO-Cr, respectively). The experimental scheme for scRNA-seq is illustrated in Figure 2A. We carried out RNA-seq on 5 single cells from each genotype as detailed in Materials and Methods. The sequencing results were analyzed using DEseq software package to identify differentially expressed genes across the five single cells between the genotypes. We identified 83 genes that were differentially expressed between WT-Cr and KO-Cr cells at a statistically significant level (p<0.05), among which 47 were upregulated and 36 were downregulated in KO-Cr cells (Table 1, Figure 2B&C). As shown in the heat map, the expression levels of many of these genes were remarkably consistent between individual cells of each genotype (Figure 2B&C). The sequencing result also revealed the frameshifting deletion of 2 nucleotides at the expected sgRNA targeting sequence (Figure 2D), which was obviously responsible for the disappearance of the Gene 33 protein. In contrast, transcription of gene 33 was not affected as Gene 33 mRNA transcripts were detected in both genotypes and exhibited no significant difference (data not shown).
FIGURE 2. scRNA-seq reveals differentially expressed mRNA transcripts as a result of Gene 33 deletion and chronic Cr(VI) exposure.
A. An illustration of the experimental scheme. Normal medium: medium without Cr(VI). B&C. Heat maps showing the 83 differentially expressed genes between WT and KO cells chronically treated with Cr(VI). D. Sequence alignments showing the frameshift deletion of two nucleotides as a result of CRISPR-mediated indel formation in KO cells. Note that cell number 3 (KO 3) has no representation. E. Clones of WT and KO BEAS-2B cells with or without chronic Cr(VI) treatment as described in Materials and Methods were subjected to WB with the indicated antibodies. C#: cell clone number, C: WT clones, Gene 33 KO: KO clones. F. BEAS-2B cells were transfected with either a scrambled siRNA oligo (C) or an siRNA oligo targeting Gene 33 (G33) for 48 hours. Cells were either left untreated or treated with Cr(VI) at 5μM for 6 hours as indicated. Cells were then harvested for WB to detect the indicated proteins. G. BEAS-2B cells were transfected with either a scrambled (C) siRNA oligo or an siRNA targeting KRBOX1 (K). Cells were harvested 72 hours post-transfection for WB to detect the indicated proteins.
Table 1.
Differentially expressed Genes as a result of CRISPR-mediated gene 33 knockout after 8 weeks of Cr(VI) treatment
| genes up regulated with gene 33 KO | genes down regulated with gene 33 KO | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Symbol | Gene name | FC | p | Symbol | Gene name | FC | p |
| MLPH | Melanophilin | 2.5 | 0.040 | HMSD | histocompatibility minor serpin domain containing | 2.6 | 0.009 |
| WNT7B | Wnt family member 7B | 2.8 | 0.011 | MT1L | metallothionein 1L | 2.1 | 0.007 |
| LAMA4 | laminin subunit alpha 4 | 3.2 | <0.001 | TMEM40 | transmembrane protein 40 | 3.0 | 0.001 |
| TNFRSF11A | tumor necrosis factor receptor superfamily member 11a | 2.5 | 0.034 | ICAM1 | intercellular adhesion molecule 1 | 2.3 | 0.012 |
| GFRA1 | GDNF family receptor alpha 1 | 2.1 | 0.019 | PPP1R3C | protein phosphatase 1 regulatory subunit 3C | 2.3 | 0.040 |
| FAM127C | family with sequence similarity 127 member C | 5.3 | <0.001 | HLA-DPB1 | major histocompatibility complex, class II, DP beta 1 | 2.1 | 0.036 |
| DIXDC1 | DIX domain containing 1 | 2.6 | 0.021 | FGF1 | fibroblast growth factor 1 | 2.0 | 0.032 |
| PCDHGA11 | protocadherin gamma subfamily A, 11 | 4.0 | <0.001 | BDKRB1 | bradykinin receptor, beta 1 | 3.0 | 0.001 |
| SHROOM4 | shroom family member 4 | 3.0 | 0.002 | PPARD | peroxisome proliferator activated receptor delta | 2.1 | 0.012 |
| CAV2 | caveolin 2 | 1.9 | 0.045 | TNFRSF9 | tumor necrosis factor receptor superfamily, member 9 | 3.0 | 0.004 |
| SMIM14 | small integral membrane protein 14 | 2.6 | 0.004 | SEL1L3 | SEL1L family member 3 | 2.6 | 0.029 |
| RGS4 | regulator of G-protein signaling 4 | 2.6 | <0.001 | IL24 | interleukin 24 | 4.0 | <0.001 |
| PGM2 | phosphoglucomutase 2 | 2.6 | 0.009 | HIST1H4I | histone cluster 1, H4i | 2.3 | <0.001 |
| TCF4 | transcription factor 4 | 3.2 | 0.002 | ATP6V1A | ATPase H+ transporting V1 subunit A | 1.9 | 0.049 |
| OXTR | oxytocin receptor | 2.6 | 0.028 | CD83 | CD83 molecule | 3.2 | <0.001 |
| FAM174B | family with sequence similarity 174 member B | 2.6 | 0.021 | ADGRE1 | adhesion G protein-coupled receptor E1 | 9.1 | <0.001 |
| AR | androgen receptor | 4.3 | <0.001 | DTNA | dystrobrevin alpha | 4.3 | <0.001 |
| F3 | coagulation factor III | 1.9 | 0.021 | CD24 | CD24 molecule | 1.9 | 0.022 |
| TMEM47 | transmembrane protein 47 | 3.0 | 0.003 | SLC29A3 | solute carrier family 29 member 3 | 2.4 | 0.041 |
| XYLT1 | xylosyltransferase 1 | 3.7 | <0.001 | ERBB3 | erb-b2 receptor tyrosine kinase 3 | 2.6 | 0.028 |
| PLAU | plasminogen activator, urokinase | 2.0 | 0.003 | STON2 | stonin 2 | 2.6 | 0.034 |
| ZNF518B | zinc finger protein 518B | 3.0 | 0.004 | ZEB2 | zinc finger E-box binding homeobox 2 | 2.4 | <0.001 |
| BCHE | butyrylcholinesterase | 4.3 | <0.001 | SLC38A5 | solute carrier family 38 member 5 | 7.7 | <0.001 |
| CLU | clusterin | 2.1 | 0.002 | GPR162 | G protein-coupled receptor 162 | 3.2 | 0.002 |
| TGM2 | transglutaminase 2 | 3.0 | <0.001 | KRT34 | keratin 34 | 3.0 | 0.005 |
| HIST1H4F | histone cluster 1, H4f | 3.5 | <0.001 | ROBO1 | roundabout guidance receptor 1 | 2.9 | 0.011 |
| PDE1C | phosphodiesterase 1C | 4.0 | <0.001 | MRGPRF | MAS related GPR family member F | 2.6 | 0.019 |
| S100A2 | S100 calcium binding protein A2 | 2.6 | <0.001 | NUPR1 | nuclear protein transcription regulator 1 | 4.0 | <0.001 |
| LINC00958 | long intergenic non-protein coding RNA 958 | 5.7 | <0.001 | CMSS1 | cms1 ribosomal small subunit homolog | 1.9 | 0.017 |
| GPX3 | glutathione peroxidase 3 | 3.7 | <0.001 | TCEAL7 | transcription elongation factor A like 7 | 6.3 | <0.001 |
| IGFBP4 | insulin like growth factor binding protein 4 | 2.5 | <0.001 | GSTM1 | glutathione S-transferase mu 1 | 6.3 | <0.001 |
| PDPN | podoplanin | 7.5 | <0.001 | AMPH | amphiphysin | 25.0 | <0.001 |
| UCHL1 | ubiquitin C-terminal hydrolase L1 | 147.0 | <0.001 | ADIRF | adipogenesis regulatory factor | 3.4 | <0.001 |
| HERC3 | HECT and RLD domain containing E3 ubiquitin protein ligase 3 | 2.3 | 0.035 | KRBOX1 | KRAB box domain containing 1 | 100.0 | <0.001 |
| ZG16B | zymogen granule protein 16B | 2.8 | 0.017 | RPPH1 | ribonuclease P RNA component H1 | 1.9 | 0.037 |
| COL5A2 | collagen type V alpha 2 chain | 2.6 | <0.001 | GPNMB | glycoprotein nmb | 2.6 | 0.034 |
| CHAC1 | ChaC glutathione specific gamma-glutamylcyclotransferase 1 | 2.3 | 0.035 | ||||
| CDH4 | cadherin 4 | 2.6 | 0.011 | ||||
| LINC01116 | long intergenic non-protein coding RNA 1116 | 2.0 | 0.037 | ||||
| SYTL3 | synaptotagmin like 3 | 2.6 | 0.031 | ||||
| ITGA4 | integrin subunit alpha 4 | 2.6 | 0.035 | ||||
| CYGB | cytoglobin | 3.7 | <0.001 | ||||
| FBXO32 | F-box protein 32 | 3.0 | 0.005 | ||||
| PKP1 | plakophilin 1 | 2.6 | 0.032 | ||||
| MEG3 | maternally expressed 3 | 4.3 | <0.001 | ||||
| APBB1IP | amyloid beta precursor protein binding family B member 1 interacting protein | 4.9 | <0.001 | ||||
| TNFRSF21 | tumor necrosis factor receptor superfamily member 21 | 2.5 | 0.023 | ||||
FC = fold change, p = adjusted p-value
The differentially expressed genes are associated with a spectrum of cellular functions (Table 2). Of particular interest is the up-regulation of genes associated with cell adhesion and the oxidative stress. Eight adhesion-associated genes were up-regulated in KO-Cr cells: ITGA4, CAV2, CDH4, LAMA4, COL5A2, PKP1, PDPN, APBB1IP, and PCDHGA11. Two adhesion-associated genes were down regulated in KO-Cr cells: ICAM1 and ROBO1. These genes are associated with both cell-extra cellular matrix interaction (ITGA4, CAV2, LAMA4, COL5A2, ROBO1) and cell-cell adhesion (CDH4, PKP1, PDPN, PCDHGA11, ICAM1). Several genes associated with the oxidative stress were up-regulated in KO-Cr cells compared to WT-Cr cells: GSTM1, CTGB, GPX3, CHAC1, and CLU. In contrast, PPARD, was down-regulated.
Table 2.
Differentially expressed genes as a result of CRISPR-mediated gene 33 knockout and 8 weeks of Cr(VI) treatment: major functional groups
| Function | Gene symbol | Gene name | FC |
|---|---|---|---|
|
| |||
| EMT and Metastasis | ZEB2 | zinc finger E-box binding homeobox 2 | −2.4 |
| NUPR1 | nuclear protein transcription regulator 1 | −4.0 | |
| ErbB3 | erb-b2 receptor tyrosine kinase 3 | −2.6 | |
| GPNMB | glycoprotein nmb | −2.6 | |
| Cell adhesion | CDH4 | cadherin 4 | +2.6 |
| ITGA4 | integrin subunit alpha 4 | +2.6 | |
| CAV2 | caveolin 2 | +1.9 | |
| APBB1IP | amyloid beta precursor protein binding family B member 1 interacting protein | +4.9 | |
| LAMA4 | laminin subunit alpha 4 | +3.2 | |
| PCDHGA11 | protocadherin gamma subfamily A, 11 | +4.0 | |
| COL5A2 | collagen type V alpha 2 chain | +2.6 | |
| PKP1 | plakophilin 1 | +2.6 | |
| PDPN | podoplanin | +7.5 | |
| ICAM1 | intercellular adhesion molecule 1 | −2.3 | |
| ROBO1 | roundabout guidance receptor 1 | −2.9 | |
| Oxidative stress | GSTM1 | glutathione S-transferase mu 1 | +6.3 |
| PPARD | peroxisome proliferator activated receptor delta | −2.1 | |
| CYGB | cytoglobin | +3.7 | |
| GPX3 | glutathione peroxidase 3 | +3.7 | |
| CHAC1 | ChaC glutathione specific gamma-glutamylcyclotransferase 1 | +2.3 | |
| CLU | Clusterin | +2.1 | |
| GPCR signaling | ADGRE1 | adhesion G protein-coupled receptor E1 | −9.1 |
| BDKRB1 | bradykinin receptor, beta 1 | −3.0 | |
| RGS4 | regulator of G-protein signaling 4 | +2.6 | |
| MRGPRF | MAS related GPR family member F | −2.6 | |
| GPR162 | G protein-coupled receptor 162 | −3.2 | |
| RTK signaling | ErbB3 | erb-b2 receptor tyrosine kinase 3 | −2.6 |
| FGF1 | fibroblast growth factor 1 | −2.0 | |
| GFRA1 | GDNF family receptor alpha 1 | +2.1 | |
| IGFBP4 | insulin like growth factor binding protein 4 | +2.5 | |
| Protein phosphatase | PPP1R3C | protein phosphatase 1 regulatory subunit 3C | −2.3 |
| WNT signaling | WNT7B | Wnt family member 7B | +2.8 |
| DIXDC1 | DIX domain containing 1 | +2.6 | |
| TCF4 | transcription factor 4 | +3.2 | |
| TLR signaling | ZG16B | zymogen granule protein 16B | +2.8 |
| TNF signaling | TNFRSF11A | tumor necrosis factor receptor superfamily member 11a | +2.5 |
| TNFRSF21 | tumor necrosis factor receptor superfamily member 21 | +2.5 | |
| TNFRSF9 | tumor necrosis factor receptor superfamily, member 9 | −3.0 | |
| Vesicle trafficking | CAV2 | caveolin 2 | +1.9 |
| SYTL3 | synaptotagmin like 3 | +2.6 | |
| STON2 | stonin 2 | −2.6 | |
| AMPH | amphiphysin | −25.0 | |
| histone | HIST1H4F | histone cluster 1, H4f | +3.5 |
| HIST1H4I | histone cluster 1, H4i | −2.3 | |
| Calcium signaling | PDE1C | phosphodiesterase 1C | +4.0 |
| S100A2 | S100 calcium binding protein A2 | +2.6 | |
| Long non-coding RNA | MEG3 | maternally expressed 3 | +4.3 |
| LINC01116 | long intergenic non-protein coding RNA 1116 | +2.0 | |
| LINC00958 | long intergenic non-protein coding RNA 958 | +5.7 | |
| Cytoskeleton | KRT34 | keratin 34 | −3.0 |
| SHROOM4 | shroom family member 4 | +3.0 | |
| Transcriptional regulator | KRABOX1 | KRAB box domain containing 1 | −100.0 |
| ZNF518B | zinc finger protein 518B | +3.0 | |
| NUPR1 | nuclear protein transcription regulator 1 | −4.0 | |
| TCF4 | transcription factor 4 | +3.2 | |
| Ubiquitination | HERC3 | HECT and RLD domain containing E3 ubiquitin protein ligase 3 | +2.3 |
| UCHL1 | ubiquitin C-terminal hydrolase L1 | +147.0 | |
| FBXO32 | F-box protein 32 | +3.0 | |
| Carrier protein | MLPH | Melanophilin | +2.5 |
| SLC38A5 | solute carrier family 38 member 5 | −7.7 | |
Three long non-coding RNAs were also significantly upregulated, among which MEG3 (up 4.3 folds) is the best studied. MEG3 has been shown to activate p53 by enhancing its stability and transcriptional activity (Zhang et al., 2003; Zhou et al., 2007; Zhou et al., 2012; Zhu et al., 2015). It has been implicated in pituitary tumor and suppresses growth of cancer cells as a tumor suppressor (Zhang et al., 2003; Zhou et al., 2007; Zhou et al., 2012; Zhu et al., 2015). A recent study showed that MEG3 can modulate TGF-β genes by binding to their distal regulatory elements (Mondal et al., 2015).
Other interesting changes include down regulation of genes associated with epithelial-mesenchymal transition (EMT) and metastasis (ZEB2, NUPR1, ErbB3, and GPNMB) as well as up regulation of genes for WNT signaling (WNT7B, DIXDC1, TCF4). NUPR1 is a transcription regulator that mediates diverse cellular functions, including cell proliferation, hypotrophy, apoptosis, autophagy, DNA repair, and EMT (Gironella et al., 2009; Goruppi and Iovanna, 2010; Cano et al., 2011). NUPR1 has been implicated in a number of cancers (Cano et al., 2011; Grasso et al., 2015; Emma et al., 2016). Of interest, NUPR1 has recently been shown to be induced by Cr(VI) and to suppress acetylation of histone H4 at lysine 16 (Chen et al., 2016). ZEB2 is considered one of the master regulators of EMT (Vandewalle et al., 2005; Gregory et al., 2011). Significant changes also occurred in genes associated with TNF signaling, GPCR signaling, vesicle trafficking, cytoskeleton, transcription regulation, and ubiquitination. Another notable finding is the change in expression of a number of neuro-specific genes, particularly up-regulation of UCHL1, CDH4, TCF4, GFRA1, and PCDHGA11 (Table 1, highlighted).
The three genes that showed most dramatic changes are ubiquitin C-terminal hydrolase L1 (UCHL1, up 147 folds in KO-Cr cells), KRAB box domain containing 1 (KRBOX1, down 100 folds in KO-Cr cells), and amphiphysin (AMPH, down 25 folds in KO-Cr cells). Expression of these three genes showed remarkable consistency cross the 5 WT clones and 5 KO clones (Figure 2B&C). UCHL1 is a deubiquitinase that catalyzes the hydrolytic removal of ubiquitin from its substrates and is strongly associated with lung carcinogenesis/progression (Hibi et al., 1998; Hibi et al., 1999; Luchansky et al., 2006; Orr et al., 2011). KRBOX1 is a member of a large family of KRAB-associated zinc finger proteins, which are believed to be DNA-binding repressors of transcription (Huntley et al., 2006; Imbeault et al., 2017). A closer examination of the raw sequencing data revealed that although KRBOX1 mRNA expression was reduced 100 folds in KO-Cr cells statistically, the transcript of this gene was actually not detected in KO-Cr cells. This could either be a result of insufficient depth of the sequencing or the loss of KRBOX1 gene in these cells. AMPH is a neuro-specific protein associated with synaptic vesicles (Sulzer et al., 1995). Elevated expression of AMPH has been shown in breast cancer cells (Brennan et al., 2012).
The dramatic up-regulation of UCHL1 and down regulation of KRBOX1 in KO-Cr cells prompted us to choose these proteins for further validation. We used WB to examine the expression levels of these two proteins in cell clones used for RNA-seq (clones 25 and 22) along with another WT clone (#23) and three additional KO clones (#30, 37, and 73), with or without chronic Cr(VI) treatments (same treatment scheme as for clones 25 and 22). As shown in Figure 2E, WT clones (25 and 23) had very low levels while KO clones (22, 30, 37, and 73) showed much higher levels of the UCHL1 protein. In contrast, expression levels of the KRBOX1 protein were high in WT clones but undetectable in KO clones. Note that Cr(VI) treatment appears to have a tendency of further elevating the levels of UCHL1 and suppressing the levels of KRBOX1 (Figure 2E). As expected, the Gene 33 protein in KO cells was undetectable (Figure 2E). These results agree with those from RNA-seq. These data also confirm that the changes in expression of UCHL1 and KRBOX1 were indeed a result of Gene 33 deletion and Cr(VI) treatment rather than a clonal effect. Surprisingly, transient knockdown of Gene 33 using siRNA did not significantly affect the levels of UCHL1 and KRBOX1 proteins in the parental BEAS-2B cells (Figure 2F), suggesting that the altered expression of these genes was a result of long-term transcriptional reprograming after Gene 33 deletion rather than an acute and direct result of Gene 33 deletion.
KRBOX1 is a protein with a calculated molecular weight of about 14 kD. According to the information provided by the manufacture (Sigma-Aldrich), the antibody to KRBOX1 (Cat. SAB1305246) should detect the endogenous KRBOX1 at an apparent molecular weight of around 35 kD. We found that the antibody detects several dominant bands on WB with the lysate of BEAS-2B cells (Figure 2G). We used RNAi targeting KRBOX1 to further characterize this antibody. As shown in Figure 2G, the band at 25 kD disappeared after transfection of an siRNA oligo to KRBOX1 into BEAS-2B cells, suggesting that this band mostly likely represents KRBOX1. Furthermore, WB also showed that this band disappears as a result of Gene 33 deletion (Figure 2E). These data confirm that KRBOX1 has an apparent molecular weight of about 25 kD in these cells.
Gene 33 deletion reduces cell proliferation and Cr(VI) treatment reverses this effect
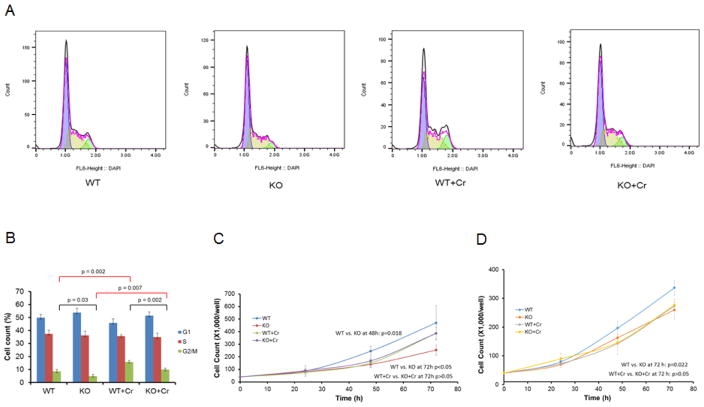
As an inhibitor of EGFR signaling, Gene 33 has been shown to inhibit EGF induced cell cycle entry under serum-starvation (Xu et al., 2005). However, our recent study showed that transient knockdown of Gene 33 with siRNA had no significant effect on cell cycle progression of BEAS-2B cells under normal cell culture condition (without serum-starvation) (Park et al., 2016). To check the effect of CRISPR/cas9-mediated Gene 33 deletion on cell cycle progression in BEAS-2B cells, we compared cell cycle profiles of WT and KO cells with or without chronic Cr(VI) treatment. Interestingly, we did detect statistically significant difference in cell cycle profiles between WT and KO cells: KO cells had a reduced number of cells in the G2/M phase of the cell cycle compared to WT cells regardless the Cr(VI) treatment (Figure 3A&B). This result is in contrast with our previous result that transient knockdown of Gene 33 has no significant effect on cell cycle distribution. This discrepancy may be a result of the long-term adaption process occurred in the KO cells, as apparently also occurred in UCHL1 and KRBOX1 expression (Figure 2F). We also observed a significant increase of cells in the G2/M phase of the cell cycle with Cr(VI)-treated cells, which agrees with the previous finding that Cr(VI) induces G2/M phase arrest as a result of Cr(VI)-induced DNA damage and repair (Chiu et al., 2010).
FIGURE 3. Effect of Gene 33 deletion and chronic Cr(VI) exposure on cell cycle progression and cell proliferation.
A. Representative histograms showing cell cycle analysis of WT and KO BEAS-2B cells with or without chronic Cr(VI) treatment. B. Cell cycle distribution of WT and KO BEAS-2B cells with or without chronic Cr(VI) treatment. C&D. Growth curves of two pairs of WT and KO BEAS-2B cell clones with or without chronic Cr(VI) treatment.
We next compared the growth curves of two WT clones and two Gene 33-deleted clones. We found that Gene 33 deletion significantly reduced the increase in cell number over time, indicating a potential slowdown of cell proliferation (Figure 3C&D). These results are unexpected as transient Gene 33 depletion with RNAi did not affect cell cycle profile in our previous study (Park et al., 2016). However, these results confirm that the phenotype produced by stable deletion of Gene 33 is not identical to that produced by transient Gene 33 knockdown in BEAS-2B cells. Interestingly, Cr(VI) treatment eliminated the difference in cell proliferation between WT and Gene 33-deleted cells (Figure 3C&D). These data indicate that suppression of cell proliferation may not be the main mechanism underlying the tumor suppressive function of Gene 33, particularly in response to chronic Cr(VI) exposure.
Deletion of Gene 33 promotes cell migration
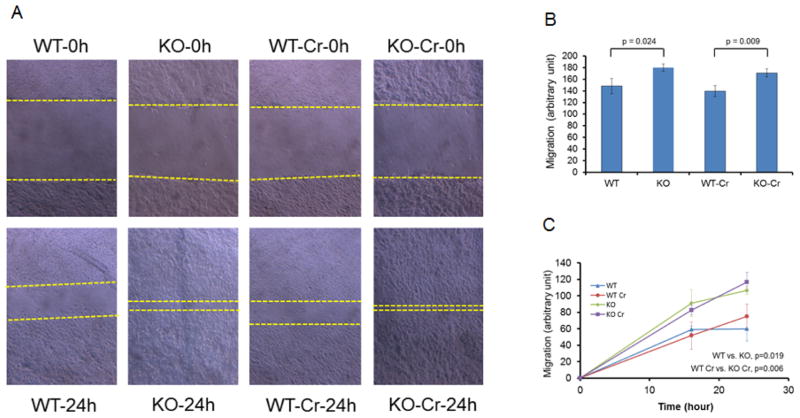
Gene 33 has been shown to regulate cell migration and tumor metastasis (Pante et al., 2005; Jiang et al., 2016). We thus compared cell migration between KO (#22) and WT (#25) cells with or without Cr(VI) treatment using the scratch assay. Our results showed that Gene 33 deletion led to a significant increase in cell migration with or without Cr(VI) exposure (Figure 4A&B). To confirm this result, we repeated the experiment with another pair of KO and WT cells (#30, KO and 23, WT, respectively). Again, KO cells exhibited significantly quicker migration than the WT cells (Figure 4C). These results confirm the previous finding that Gene 33 negatively regulates cell migration (Pante et al., 2005; Jiang et al., 2016). Intriguingly however, Cr(VI) treatment alone had limited effect on cell migration under our experimental conditions (Figure 4).
FIGURE 4. Effects of Gene 33 deletion and chronic Cr(VI) treatment on cell migration.
A. Representative images from the scratch assay. B&C. Migration rates of two pairs of WT and KO BEAS-2B cell clones with or without chronic treatment with Cr(VI).
DISCUSSION
In the present study we analyzed gene expression profiles o f BEAS-2B lung epithelial cells chronically treated with Cr(VI), with or without CRISPR/cas9-mediated deletion of Gene 33, using scRNA-seq. We identified 83 genes with significantly altered expression. Some of these genes are known to be associated transformed lung epithelial cells and lung cancer, consistent with the previous observations that Gene 33 depletion and/or chronic Cr(VI) treatments induce transformation of BEAS-2B cells (Park et al., 2016). In particular, we observed dramatic up-regulation of UCHL1 in KO cells. We also observed a significantly lowered expression of KRBOX1, a KRAB-associated zinc finger protein with unknown function. We found that KO cells have higher levels of cell migration but lower levels of cell proliferation compared to WT cells. Chronic Cr(VI) treatment eliminated the difference in cell proliferation but had limited effect on cell migration between these two genotypes. The enhanced cell migration is consistent with a transformed phenotype of BEAS-2B cells (Cartularo et al., 2016; Pratheeshkumar et al., 2016).
UCHL1 is a neuro-specific gene known to be associated with the progression of Alzheimer disease (Tramutola et al., 2016). It has limited expression in normal lungs but is dramatically induced in lung cancer (Hibi et al., 1998; Hibi et al., 1999). In fact, most lung cancer cells and primary lung tumors (both small and non-small cell lung cancers) express high levels of UCHL1 (Hibi et al., 1999). Elevated UCHL1 expression has also been shown in the human airway epithelium of cigarette smokers (Carolan et al., 2006). This protein is therefore considered a potential biomarker for lung tumorigenesis (Hibi et al., 1999; Orr et al., 2011). UCHL1 has been shown to be a key regulator of lung tumor cell invasion and metastasis (Kim et al., 2009; Goto et al., 2015; Kim et al., 2015).
As a deubiquitinase, UCHL1 functions to re move ubiquitin from its substrates thereby preventing them from proteasome-mediated degradation (Luchansky et al., 2006). At the molecular level, the potential tumor promoting functions of UCHL1 include to stabilize HIF-1α by mediating its deubiquitination thereby promoting the hypoxic response (Goto et al., 2015), to prevent degradation of β-catenin thereby enhancing β-catenin-mediated gene transcription (Bheda et al., 2009), to inhibit EGFR degradation thereby promoting breast cancer progression (Jin et al., 2015), and to enhance p53 degradation and elevate the AKT pathway thereby reducing cell death (Hussain et al., 2010). UCHL1 has also been shown to interact with CDK1, CDK4, and CDK5 thereby promoting their activity independent of its catalytic activity (Kabuta et al., 2013). However, UCHL1 has also been shown to possess a tumor suppressing function in certain tumors (Hurst-Kennedy et al., 2012). Accordingly, UCHL1 may stabilize p53 thereby triggering cell cycle arrest (Li et al., 2010), stabilize NOXA thereby promoting cell death (Brinkmann et al., 2013), prevent degradation of p27 by interacting with JAB1 and p27 (Caballero et al., 2002). Thus, the overall role of UCHL1 in a particular tumor is likely dependent on the specific targets with which it interacts in a tumor type-specific fashion. Interestingly, a ubiquitin E3 ligase activity has also been reported for UCHL1 (Liu et al., 2002).
Our results also show up-regulation of the transcription factor TCF4 and some components of the WNT pathway (WNT7B and DIXDC1) in KO cells. Given the reported roles of TCF4 and β-catenin, a key downstream target of the WNT pathway, in promoting UCHL1 transcription (Bheda et al., 2009), the elevated expression of UCHL1 was probably a result of the up-regulation of these genes. Of note, UCHL1 also promotes the activity of the WNT pathway by stabilizing β-catenin (Bheda et al., 2009). The WNT pathway has been well-established to play a critical role in lung tumorigenesis (Stewart, 2014). Thus, the interplay of these proteins may play a significant role in lung epithelial cell transformation.
KRBOX1 belongs to a large family (close to 300) of KRAB-associated zinc finger proteins (Huntley et al., 2006; Imbeault et al., 2017). These proteins are believed to have DNA binding capability and serve as transcription repressors (Huntley et al., 2006; Imbeault et al., 2017). The regulatory targets of most of these proteins, including KRBOX1, remain unknown. The expression pattern and specific function of KRBOX1 in lung cancer have not been documented either. KRBOX1 gene is located at chromosome 3p22.1 where LOH is one of the early genetic events occurring in lung carcinogenesis (Zabarovsky et al., 2002). In fact, our RNA-seq did not detect any transcript of KRBOX1 in KO-Cr cells, suggesting a possible loss of KRBOX1 gene in these cells. Further genetic verification is needed to confirm this.
The dramatic inverse correlation between UCHL1 and KRBOX1 expression patterns and the potential role of KRBOX1 as a suppressor of gene transcription intrigued us to investigate whether UCHL1 is a direct target of KRBOX1. However, we were unable to detect any change in UCHL1 protein expression upon transient depletion of KRBOX1 using siRNA in wild type BEAS-2B cells (data not shown). Nonetheless, it is conceivable that loss of KRBOX1 may lead to de-repression of some of the up-regulated genes identified herein, which contribute to the transformed phenotype of KO-Cr cells.
Neuroendocrine differentiation is a common feature of lung cancer, including most small cell lung carcinoma (SCLC) and a significant portion (10–30%) of non-small cell lung carcinoma (Berendsen et al., 1989; Carnaghi et al., 2001). While SCLC is considered neuroendocrine in nature, the clinical significance of neuroendocrine differentiation in NSCLC is unclear (Berendsen et al., 1989; Carnaghi et al., 2001). Our data s how that a number of neuro-specific genes were up-regulated in KO-Cr cells. However, the upregulated neuro-specific genes identified in the present study are different from the previously reported neuroendocrine genes (e.g. NSP, NCAM, SYN, CgA, Leu-7, and NF) associated with lung cancer (Carnaghi et al., 2001). The significance of this observation re mains to be determine d. Nonetheless, the expression of these genes appears to indicate a tendency of neural differentiation, which may be associated with early neoplastic transformation of these cells. Further investigation is clearly needed to interpret these findings.
The up-regulation of genes associated with the oxidative stress in KO-Cr cells is intriguing. Cr(VI) is known to produce oxidative stresses in the cell during its reduction process, which contribute to the toxicity and tumorigenicity of Cr(VI) (Cheng et al., 1998; Cheng et al., 2000; Ding and Shi, 2002; Ha et al., 2003; Hu et al., 2011). Elevated expression of oxidative stress-related genes may reflect an adaptive response to an increased oxidative stress in KO-Cr cells as BEAS-2B cells with depletion of Gene 33 show higher levels of DNA damage (Figure 1D) (Park et al., 2016). It will be important to determine how Gene 33 may regulate the intracellular oxidative response and the DNA damage response.
The increased cell mobility in KO cells is consistent with the elevated expression of genes associated with focal adhesion (Table 2). However, it is surprising that some EMT-associated genes, particularly ZEB2, were down-regulated. EMT is typically associated with greater cell mobility and reduced intercellular adhesion of epithelial cells (Kalluri and Weinberg, 2009). Accordingly, we did also observed up-regulation of several genes associated with intercellular adhesion (CDH4, PKP1, PDPN, and PCDHGA11). The significance of these changes remains to be determined. It is possible that these changes represent those occur in the very early phase of the cell transformation process when EMT is suppressed. Supporting this notion, no visible morphological changes occur after Gene 33 deletion and/or Cr(VI) treatments (Figure 1E).
It is interesting that Gene 33 deletion reduced rather than increased cell proliferation and that Cr(VI) treatment eliminated the difference (Figure 3C&D). We also observed a difference in cell cycle distribution between the two genotypes (Figure 3B). The significance of these observations in the context of cell transformation and Cr(VI) treatment remains to be determined. Given that stable depletion of Gene 33 with shRNA alone promotes transformation of BEAS-2B cells (Park et al., 2016), the change in cell migration is likely an indication of the early stage of cell transformation. As discussed, the up-regulation of neuro-specific genes in KO-Cr cells may also indicate neoplastic transformation of these cells. Our previous work showed that Cr(VI) suppresses the expression of the Gene 33 protein in BEAS-2B cells, which may contribute to elevated transformation of these cells (Park et al., 2016). The data presented herein reveal some early molecular events that may mediate this process. Given that Gene 33 deletion dramatically up-regulates UCHL1, Cr(VI) tends to promote UCHL1 expression, and that elevated UCHL1 is known to up-regulated in lung cancer, we propose that UCHL1 plays an important role in the early stage of lung epithelial cell transformation and lung tumorigenesis.
The present study identifies a number of genes that are potentially important for lung epithelial cell transformation. The most interesting ones are UCHL1 and KRBOX1, the two most altered. While the connection between UCHL1 and lung cancer has been well-established, its role in the early stage of lung epithelial cell transformation has not been studied. On the other hand, the function of KRBOX1 is completely unknown. It is also interesting that some components of the WNT pathway also significantly changed. Given the potential mutual regulatory relationship between UCHL1 and the WNT pathway (Bheda et al., 2009), it is highly likely that their changes were interconnected. The immediate focus of future investigations should be on whether and how these proteins and pathways contribute to lung epithelial cell neoplastic transformation and lung tumorigenesis, both in vitro and in vivo.
Highlights.
CRISPR/cas9-mediated deletion of Gene 33 (Mig6, ERRFI1) was carried out in BEAS-2B cells.
Single cell RNA-seq revealed 83 differentially expressed genes as a result of Gene 33 depletion and chronic Cr(VI) exposure.
UCHL1 and KRBOX1 are the most differentially expressed genes.
Gene 33-depleted cells showed a phenotype and a gene expression pattern indicating cell transformation.
UCHL1 may play a key role in lung epithelial cell transformation.
Acknowledgments
This work was supported in part by National Institutes of Health Grant R21ES023862 (DX), a startup fund from NYMC (DX), funds from the NYMC Castle-Krob Research Endowment Fund under the College’s Intramural Research Support Program (DX), and Philips Research North America (JTF).
Footnotes
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest with the contents of this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anastasi S, Baietti MF, Frosi Y, Alema S, Segatto O. The evolutionarily conserved EBR module of RALT/MIG6 mediates suppression of the EGFR catalytic activity. Oncogene. 2007;26:7833–7846. doi: 10.1038/sj.onc.1210590. [DOI] [PubMed] [Google Scholar]
- Anastasi S, Fiorentino L, Fiorini M, Fraioli R, Sala G, Castellani L, Alema S, Alimandi M, Segatto O. Feedback inhibition by RALT controls signal output by the ErbB network. Oncogene. 2003;22:4221–4234. doi: 10.1038/sj.onc.1206516. [DOI] [PubMed] [Google Scholar]
- Berendsen HH, de Leij L, Poppema S, Postmus PE, Boes A, Sluiter HJ, The H. Clinical characterization of non-small-cell lung cancer tumors showing neuroendocrine differentiation features. J Clin Oncol. 1989;7:1614–1620. doi: 10.1200/JCO.1989.7.11.1614. [DOI] [PubMed] [Google Scholar]
- Bheda A, Yue W, Gullapalli A, Whitehurst C, Liu R, Pagano JS, Shackelford J. Positive reciprocal regulation of ubiquitin C-terminal hydrolase L1 and beta-catenin/TCF signaling. PloS one. 2009;4:e5955. doi: 10.1371/journal.pone.0005955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braver ER, Infante P, Chu K. An analysis of lung cancer risk from exposure to hexavalent chromium. Teratog Carcinog Mutagen. 1985;5:365–378. doi: 10.1002/tcm.1770050507. [DOI] [PubMed] [Google Scholar]
- Brennan DJ, O’Connor DP, Laursen H, McGee SF, McCarthy S, Zagozdzon R, Rexhepaj E, Culhane AC, Martin FM, Duffy MJ, Landberg G, Ryden L, Hewitt SM, Kuhar MJ, Bernards R, Millikan RC, Crown JP, Jirstrom K, Gallagher WM. The cocaine- and amphetamine-regulated transcript mediates ligand-independent activation of ERalpha, and is an independent prognostic factor in node-negative breast cancer. Oncogene. 2012;31:3483–3494. doi: 10.1038/onc.2011.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann K, Zigrino P, Witt A, Schell M, Ackermann L, Broxtermann P, Schull S, Andree M, Coutelle O, Yazdanpanah B, Seeger JM, Klubertz D, Drebber U, Hacker UT, Kronke M, Mauch C, Hoppe T, Kashkar H. Ubiquitin C-terminal hydrolase-L1 potentiates cancer chemosensitivity by stabilizing NOXA. Cell Rep. 2013;3:881–891. doi: 10.1016/j.celrep.2013.02.014. [DOI] [PubMed] [Google Scholar]
- Caballero OL, Resto V, Patturajan M, Meerzaman D, Guo MZ, Engles J, Yochem R, Ratovitski E, Sidransky D, Jen J. Interaction and colocalization of PGP9.5 with JAB1 and p27(Kip1) Oncogene. 2002;21:3003–3010. doi: 10.1038/sj.onc.1205390. [DOI] [PubMed] [Google Scholar]
- Cano CE, Hamidi T, Sandi MJ, Iovanna JL. Nupr1: the Swiss-knife of cancer. J Cell Physiol. 2011;226:1439–1443. doi: 10.1002/jcp.22324. [DOI] [PubMed] [Google Scholar]
- Carnaghi C, Rimassa L, Garassino I, Santoro A. Clinical significance of neuroendocrine phenotype in non-small-cell lung cancer. Ann Oncol. 2001;12(Suppl 2):S119–123. doi: 10.1093/annonc/12.suppl_2.s119. [DOI] [PubMed] [Google Scholar]
- Carolan BJ, Heguy A, Harvey BG, Leopold PL, Ferris B, Crystal RG. Up-regulation of expression of the ubiquitin carboxyl-terminal hydrolase L1 gene in human airway epithelium of cigarette smokers. Cancer Res. 2006;66:10729–10740. doi: 10.1158/0008-5472.CAN-06-2224. [DOI] [PubMed] [Google Scholar]
- Cartularo L, Kluz T, Cohen L, Shen SS, Costa M. Molecular Mechanisms of Malignant Transformation by Low Dose Cadmium in Normal Human Bronchial Epithelial Cells. PloS one. 2016;11:e0155002. doi: 10.1371/journal.pone.0155002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Kluz T, Fang L, Zhang X, Sun H, Jin C, Costa M. Hexavalent Chromium (Cr(VI)) Down-Regulates Acetylation of Histone H4 at Lysine 16 through Induction of Stressor Protein Nupr1. PloS one. 2016;11:e0157317. doi: 10.1371/journal.pone.0157317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Liu S, Dixon K. Analysis of repair and mutagenesis of chromium-induced DNA damage in yeast, mammalian cells, and transgenic mice. Environ Health Perspect. 1998;106(Suppl 4):1027–1032. doi: 10.1289/ehp.98106s41027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Sonntag DM, de Boer J, Dixon K. Chromium(VI)-induced mutagenesis in the lungs of big blue transgenic mice. J Environ Pathol Toxicol Oncol. 2000;19:239–249. [PubMed] [Google Scholar]
- Chiu A, Shi XL, Lee WK, Hill R, Wakeman TP, Katz A, Xu B, Dalal NS, Robertson JD, Chen C, Chiu N, Donehower L. Review of chromium (VI) apoptosis, cell-cycle-arrest, and carcinogenesis. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2010;28:188–230. doi: 10.1080/10590501.2010.504980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa AN, Moreno V, Prieto MJ, Urbano AM, Alpoim MC. Induction of morphological changes in BEAS-2B human bronchial epithelial cells following chronic sub-cytotoxic and mildly cytotoxic hexavalent chromium exposures. Mol Carcinog. 2010;49:582–591. doi: 10.1002/mc.20624. [DOI] [PubMed] [Google Scholar]
- Costa M, Klein CB. Toxicity and carcinogenicity of chromium compounds in humans. Crit Rev Toxicol. 2006;36:155–163. doi: 10.1080/10408440500534032. [DOI] [PubMed] [Google Scholar]
- Ding M, Shi X. Molecular mechanisms of Cr(VI)-induced carcinogenesis. Mol Cell Biochem. 2002;234–235:293–300. [PubMed] [Google Scholar]
- Emma MR, Iovanna JL, Bachvarov D, Puleio R, Loria GR, Augello G, Candido S, Libra M, Gulino A, Cancila V, McCubrey JA, Montalto G, Cervello M. NUPR1, a new target in liver cancer: implication in controlling cell growth, migration, invasion and sorafenib resistance. Cell Death Dis. 2016;7:e2269. doi: 10.1038/cddis.2016.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferby I, Reschke M, Kudlacek O, Knyazev P, Pante G, Amann K, Sommergruber W, Kraut N, Ullrich A, Fassler R, Klein R. Mig6 is a negative regulator of EGF receptor-mediated skin morphogenesis and tumor formation. Nat Med. 2006;12:568–573. doi: 10.1038/nm1401. [DOI] [PubMed] [Google Scholar]
- Fiorentino L, Pertica C, Fiorini M, Talora C, Crescenzi M, Castellani L, Alema S, Benedetti P, Segatto O. Inhibition of ErbB-2 mitogenic and transforming activity by RALT, a mitogen-induced signal transducer which binds to the ErbB-2 kinase domain. Mol Cell Biol. 2000;20:7735–7750. doi: 10.1128/mcb.20.20.7735-7750.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb HJ, Lees PS, Pinsky PF, Rooney BC. Lung cancer among workers in chromium chemical production. Am J Ind Med. 2000;38:115–126. doi: 10.1002/1097-0274(200008)38:2<115::aid-ajim1>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Gironella M, Malicet C, Cano C, Sandi MJ, Hamidi T, Tauil RM, Baston M, Valaco P, Moreno S, Lopez F, Neira JL, Dagorn JC, Iovanna JL. p8/nupr1 regulates DNA-repair activity after double-strand gamma irradiation-induced DNA damage. J Cell Physiol. 2009;221:594–602. doi: 10.1002/jcp.21889. [DOI] [PubMed] [Google Scholar]
- Goruppi S, Iovanna JL. Stress-inducible protein p8 is involved in several physiological and pathological processes. J Biol Chem. 2010;285:1577–1581. doi: 10.1074/jbc.R109.080887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y, Zeng L, Yeom CJ, Zhu Y, Morinibu A, Shinomiya K, Kobayashi M, Hirota K, Itasaka S, Yoshimura M, Tanimoto K, Torii M, Sowa T, Menju T, Sonobe M, Kakeya H, Toi M, Date H, Hammond EM, Hiraoka M, Harada H. UCHL1 provides diagnostic and antimetastatic strategies due to its deubiquitinating effect on HIF-1alpha. Nat Commun. 2015;6:6153. doi: 10.1038/ncomms7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso D, Bintz J, Lomberk G, Molejon MI, Loncle C, Garcia MN, Lopez MB, Urrutia R, Iovanna JL. Pivotal Role of the Chromatin Protein Nupr1 in Kras-Induced Senescence and Transformation. Sci Rep. 2015;5:17549. doi: 10.1038/srep17549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory PA, Bracken CP, Smith E, Bert AG, Wright JA, Roslan S, Morris M, Wyatt L, Farshid G, Lim YY, Lindeman GJ, Shannon MF, Drew PA, Khew-Goodall Y, Goodall GJ. An autocrine TGF-beta/ZEB/miR-200 signaling network regulates establishment and maintenance of epithelial-mesenchymal transition. Mol Biol Cell. 2011;22:1686–1698. doi: 10.1091/mbc.E11-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha L, Ceryak S, Patierno SR. Chromium (VI) activates ataxia telangiectasia mutated (ATM) protein. Requirement of ATM for both apoptosis and recovery from terminal growth arrest. J Biol Chem. 2003;278:17885–17894. doi: 10.1074/jbc.M210560200. [DOI] [PubMed] [Google Scholar]
- Halasova E, Matakova T, Kavcova E, Musak L, Letkova L, Adamkov M, Ondrusova M, Bukovska E, Singliar A. Human lung cancer and hexavalent chromium exposure. Neuro Endocrinol Lett. 2009;30(Suppl 1):182–185. [PubMed] [Google Scholar]
- Hibi K, Liu Q, Beaudry GA, Madden SL, Westra WH, Wehage SL, Yang SC, Heitmiller RF, Bertelsen AH, Sidransky D, Jen J. Serial analysis of gene expression in non-small cell lung cancer. Cancer Res. 1998;58:5690–5694. [PubMed] [Google Scholar]
- Hibi K, Westra WH, Borges M, Goodman S, Sidransky D, Jen J. PGP9.5 as a candidate tumor marker for non-small-cell lung cancer. Am J Pathol. 1999;155:711–715. doi: 10.1016/S0002-9440(10)65169-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes AL, Wise SS, Wise JP., Sr Carcinogenicity of hexavalent chromium. Indian J Med Res. 2008;128:353–372. [PubMed] [Google Scholar]
- Hopkins S, Linderoth E, Hantschel O, Suarez-Henriques P, Pilia G, Kendrick H, Smalley MJ, Superti-Furga G, Ferby I. Mig6 is a sensor of EGF receptor inactivation that directly activates c-Abl to induce apoptosis during epithelial homeostasis. Dev Cell. 2012;23:547–559. doi: 10.1016/j.devcel.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Liu X, Chervona Y, Yang F, Tang MS, Darzynkiewicz Z, Dai W. Chromium induces chromosomal instability, which is partly due to deregulation of BubR1 and Emi1, two APC/C inhibitors. Cell Cycle. 2011;10:2373–2379. doi: 10.4161/cc.10.14.16310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntley S, Baggott DM, Hamilton AT, Tran-Gyamfi M, Yang S, Kim J, Gordon L, Branscomb E, Stubbs L. A comprehensive catalog of human KRAB-associated zinc finger genes: insights into the evolutionary history of a large family of transcriptional repressors. Genome Res. 2006;16:669–677. doi: 10.1101/gr.4842106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst-Kennedy J, Chin LS, Li L. Ubiquitin C-terminal hydrolase l1 in tumorigenesis. Biochem Res Int. 2012;2012:123706. doi: 10.1155/2012/123706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S, Foreman O, Perkins SL, Witzig TE, Miles RR, van Deursen J, Galardy PJ. The de-ubiquitinase UCH-L1 is an oncogene that drives the development of lymphoma in vivo by deregulating PHLPP1 and Akt signaling. Leukemia. 2010;24:1641–1655. doi: 10.1038/leu.2010.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbeault M, Helleboid PY, Trono D. KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature. 2017;543:550–554. doi: 10.1038/nature21683. [DOI] [PubMed] [Google Scholar]
- Jiang X, Niu M, Chen D, Chen J, Cao Y, Li X, Ying H, Bergholz J, Zhang Y, Xiao ZX. Inhibition of Cdc42 is essential for Mig-6 suppression of cell migration induced by EGF. Oncotarget. 2016;7:49180–49193. doi: 10.18632/oncotarget.10205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Zhang W, Xu J, Wang H, Zhang Z, Chu C, Liu X, Zou Q. UCH-L1 involved in regulating the degradation of EGFR and promoting malignant properties in drug-resistant breast cancer. Int J Clin Exp Pathol. 2015;8:12500–12508. [PMC free article] [PubMed] [Google Scholar]
- Kabuta T, Mitsui T, Takahashi M, Fujiwara Y, Kabuta C, Konya C, Tsuchiya Y, Hatanaka Y, Uchida K, Hohjoh H, Wada K. Ubiquitin C-terminal hydrolase L1 (UCH-L1) acts as a novel potentiator of cyclin-dependent kinases to enhance cell proliferation independently of its hydrolase activity. J Biol Chem. 2013;288:12615–12626. doi: 10.1074/jbc.M112.435701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Kim YM, Lim S, Nam YK, Jeong J, Kim HJ, Lee KJ. Ubiquitin C-terminal hydrolase-L1 is a key regulator of tumor cell invasion and metastasis. Oncogene. 2009;28:117–127. doi: 10.1038/onc.2008.364. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Magesh V, Lee JJ, Kim S, Knaus UG, Lee KJ. Ubiquitin C-terminal hydrolase-L1 increases cancer cell invasion by modulating hydrogen peroxide generated via NADPH oxidase 4. Oncotarget. 2015;6:16287–16303. doi: 10.18632/oncotarget.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Tao Q, Jin H, van Hasselt A, Poon FF, Wang X, Zeng MS, Jia WH, Zeng YX, Chan AT, Cao Y. The tumor suppressor UCHL1 forms a complex with p53/MDM2/ARF to promote p53 signaling and is frequently silenced in nasopharyngeal carcinoma. Clin Cancer Res. 2010;16:2949–2958. doi: 10.1158/1078-0432.CCR-09-3178. [DOI] [PubMed] [Google Scholar]
- Li Z, Dong Q, Wang Y, Qu L, Qiu X, Wang E. Downregulation of Mig-6 in nonsmall-cell lung cancer is associated with EGFR signaling. Mol Carcinog. 2011;51:522–534. doi: 10.1002/mc.20815. [DOI] [PubMed] [Google Scholar]
- Liu Y, Fallon L, Lashuel HA, Liu Z, Lansbury PT., Jr The UCH-L1 gene encodes two opposing enzymatic activities that affect alpha-synuclein degradation and Parkinson’s disease susceptibility. Cell. 2002;111:209–218. doi: 10.1016/s0092-8674(02)01012-7. [DOI] [PubMed] [Google Scholar]
- Luchansky SJ, Lansbury PT, Jr, Stein RL. Substrate recognition and catalysis by UCH-L1. Biochemistry. 2006;45:14717–14725. doi: 10.1021/bi061406c. [DOI] [PubMed] [Google Scholar]
- Makkinje A, Quinn DA, Chen A, Cadilla CL, Force T, Bonventre JV, Kyriakis JM. Gene 33/Mig-6, a transcriptionally inducible adapter protein that binds GTP-Cdc42 and activates SAPK/JNK. A potential marker transcript for chronic pathologic conditions, such as diabetic nephropathy. Possible role in the response to persistent stress. J Biol Chem. 2000;275:17838–17847. doi: 10.1074/jbc.M909735199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal T, Subhash S, Vaid R, Enroth S, Uday S, Reinius B, Mitra S, Mohammed A, James AR, Hoberg E, Moustakas A, Gyllensten U, Jones SJ, Gustafsson CM, Sims AH, Westerlund F, Gorab E, Kanduri C. MEG3 long noncoding RNA regulates the TGF-beta pathway genes through formation of RNA-DNA triplex structures. Nat Commun. 2015;6:7743. doi: 10.1038/ncomms8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr KS, Shi Z, Brown WM, O’Hagan KA, Lappin TR, Maxwell P, Percy MJ. Potential prognostic marker ubiquitin carboxyl-terminal hydrolase-L1 does not predict patient survival in non-small cell lung carcinoma. J Exp Clin Cancer Res. 2011;30:79. doi: 10.1186/1756-9966-30-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pante G, Thompson J, Lamballe F, Iwata T, Ferby I, Barr FA, Davies AM, Maina F, Klein R. Mitogen-inducible gene 6 is an endogenous inhibitor of HGF/Met-induced cell migration and neurite growth. J Cell Biol. 2005;171:337–348. doi: 10.1083/jcb.200502013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park RM, Bena JF, Stayner LT, Smith RJ, Gibb HJ, Lees PS. Hexavalent chromium and lung cancer in the chromate industry: a quantitative risk assessment. Risk Anal. 2004;24:1099–1108. doi: 10.1111/j.0272-4332.2004.00512.x. [DOI] [PubMed] [Google Scholar]
- Park S, Li C, Zhao H, Darzynkiewicz Z, Xu D. Gene 33/Mig6 inhibits hexavalent chromium-induced DNA damage and cell transformation in human lung epithelial cells. Oncotarget. 2016;7:8916–8930. doi: 10.18632/oncotarget.6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, Faridani OR, Bjorklund AK, Winberg G, Sagasser S, Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- Pratheeshkumar P, Son YO, Divya SP, Turcios L, Roy RV, Hitron JA, Wang L, Kim D, Dai J, Asha P, Zhang Z, Shi X. Hexavalent chromium induces malignant transformation of human lung bronchial epithelial cells via ROS-dependent activation of miR-21-PDCD4 signaling. Oncotarget. 2016;7:51193–51210. doi: 10.18632/oncotarget.9967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart DJ. Wnt signaling pathway in non-small cell lung cancer. J Natl Cancer Inst. 2014;106:djt356. doi: 10.1093/jnci/djt356. [DOI] [PubMed] [Google Scholar]
- Sulzer D, Chen TK, Lau YY, Kristensen H, Rayport S, Ewing A. Amphetamine redistributes dopamine from synaptic vesicles to the cytosol and promotes reverse transport. J Neurosci. 1995;15:4102–4108. doi: 10.1523/JNEUROSCI.15-05-04102.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tramutola A, Di Domenico F, Barone E, Perluigi M, Butterfield DA. It Is All about (U)biquitin: Role of Altered Ubiquitin-Proteasome System and UCHL1 in Alzheimer Disease. Oxid Med Cell Longev. 2016;2016:2756068. doi: 10.1155/2016/2756068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng RC, Chang JW, Hsien FJ, Chang YH, Hsiao CF, Chen JT, Chen CY, Jou YS, Wang YC. Genomewide loss of heterozygosity and its clinical associations in non small cell lung cancer. Int J Cancer. 2005;117:241–247. doi: 10.1002/ijc.21178. [DOI] [PubMed] [Google Scholar]
- Tsunoda T, Inokuchi J, Baba I, Okumura K, Naito S, Sasazuki T, Shirasawa S. A novel mechanism of nuclear factor kappaB activation through the binding between inhibitor of nuclear factor-kappaBalpha and the processed NH(2)-terminal region of Mig-6. Cancer Res. 2002;62:5668–5671. [PubMed] [Google Scholar]
- Vandewalle C, Comijn J, De Craene B, Vermassen P, Bruyneel E, Andersen H, Tulchinsky E, Van Roy F, Berx G. SIP1/ZEB2 induces EMT by repressing genes of different epithelial cell-cell junctions. Nucleic Acids Res. 2005;33:6566–6578. doi: 10.1093/nar/gki965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie B, Zhao L, Chen H, Jin B, Mao Z, Yao Z. The mitogen-inducible gene-6 is involved in regulation of cellular senescence in normal diploid fibroblasts. Biology of the cell / under the auspices of the European Cell Biology Organization. 2013;105:488–499. doi: 10.1111/boc.201200052. [DOI] [PubMed] [Google Scholar]
- Xu D, Makkinje A, Kyriakis JM. Gene 33 is an endogenous inhibitor of epidermal growth factor (EGF) receptor signaling and mediates dexamethasone-induced suppression of EGF function. J Biol Chem. 2005;280:2924–2933. doi: 10.1074/jbc.M408907200. [DOI] [PubMed] [Google Scholar]
- Xu D, Patten R, Force T, Kyriakis JM. Gene 33/RALT is induced in cardiomyocytes by hypoxia where it promotes cell death by suppressing PI-3-kinase and ERK survival signaling. Mol Cell Biol. 2006;26:5043–5054. doi: 10.1128/MCB.02387-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabarovsky ER, Lerman MI, Minna JD. Tumor suppressor genes on chromosome 3p involved in the pathogenesis of lung and other cancers. Oncogene. 2002;21:6915–6935. doi: 10.1038/sj.onc.1205835. [DOI] [PubMed] [Google Scholar]
- Zhang X, Pickin KA, Bose R, Jura N, Cole PA, Kuriyan J. Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface. Nature. 2007a;450:741–744. doi: 10.1038/nature05998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhou Y, Mehta KR, Danila DC, Scolavino S, Johnson SR, Klibanski A. A pituitary-derived MEG3 isoform functions as a growth suppressor in tumor cells. J Clin Endocrinol Metab. 2003;88:5119–5126. doi: 10.1210/jc.2003-030222. [DOI] [PubMed] [Google Scholar]
- Zhang YW, Staal B, Su Y, Swiatek P, Zhao P, Cao B, Resau J, Sigler R, Bronson R, Vande Woude GF. Evidence that MIG-6 is a tumor-suppressor gene. Oncogene. 2007b;26:269–276. doi: 10.1038/sj.onc.1209790. [DOI] [PubMed] [Google Scholar]
- Zhang YW, Vande Woude GF. Mig-6, signal transduction, stress response and cancer. Cell Cycle. 2007;6:507–513. doi: 10.4161/cc.6.5.3928. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhang X, Klibanski A. MEG3 noncoding RNA: a tumor suppressor. J Mol Endocrinol. 2012;48:R45–53. doi: 10.1530/JME-12-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R, Ansell PJ, Zhao J, Weng C, Klibanski A. Activation of p53 by MEG3 non-coding RNA. J Biol Chem. 2007;282:24731–24742. doi: 10.1074/jbc.M702029200. [DOI] [PubMed] [Google Scholar]
- Zhu J, Liu S, Ye F, Shen Y, Tie Y, Zhu J, Wei L, Jin Y, Fu H, Wu Y, Zheng X. Long Noncoding RNA MEG3 Interacts with p53 Protein and Regulates Partial p53 Target Genes in Hepatoma Cells. PloS one. 2015;10:e0139790. doi: 10.1371/journal.pone.0139790. [DOI] [PMC free article] [PubMed] [Google Scholar]