Abstract
In humans, purine de novo synthesis pathway consists of multi-functional enzymes. Nucleotide metabolism enzymes are potential drug targets for treating cancer and autoimmune diseases. Glycinamide ribonucleotide transformylase (GART) is one of the most important trifunctional enzymes involved in purine synthesis. Previous studies have demonstrated the role of folate inhibitors against tumor activity. In this present study, three components of GART enzyme were targeted as receptor dataset and in silico analysis was carried out with folate ligand dataset. To accomplish the task, Autodock 4.2 was used for determining the docking compatibilities of ligand and receptor dataset. Taken together, it has been suggested that folate ligands could be potentially used as inhibitors of GART.
Abbreviations: GAR, glycinamide ribonucleotide; GART, glycinamide ribonucleotide transformylase; DIA, 5-((4-carboxy-4-(4-(((2,4-diaminopyrido[3,2-d]pyrimidine-6-yl)methyl)amino)benzamido)butyl)carbamoyl)-isophthalic acid; DHFR, dihydrofolate reductase; PY899, 2,4-diamino-6-(3,4,5-trimethoxybenzyl)-5,6,7,8-tetrahydro-quinazoline; PY873, 2,4-diamino-6-(3,4,5-trimethoxyanilino)-methylpyrido[3,2-d]pyrimidine; HsGART, human GART tri-functional enzyme; GARS, glycinamide ribonucleotide synthetase; AIRS, aminoimidazole ribonucleotide synthetase; GARTfase, glycinamide ribonucleotide transformylase
Keywords: In silico, Inhibition, PY873, PY899, Isophthalic acid
1. Introduction
Nucleotide biosynthesis is an imperative phenomenon in the synthesis of nucleic acids and metabolic pathways. Nucleotide biosynthesis follows synergistic pathway involving coordination between de novo and salvage pathways. Since nucleotides play a pivotal role in human cells, the enzymes involved in nucleotide metabolism could be exploited as potential anti-proliferative drug targets against cancer and autoimmune diseases (Welin et al., 2010). During de novo purine synthesis, conversion of phosphoribosyl pyrophosphate to inosine monophosphate involves 10 individual steps (Hartman et al., 1959). However, six different multifunctional enzymes (three monofunctional, two bifunctional and one trifunctional enzymes) are engaged in catalysis of above-mentioned steps (Kappock et al., 2000).
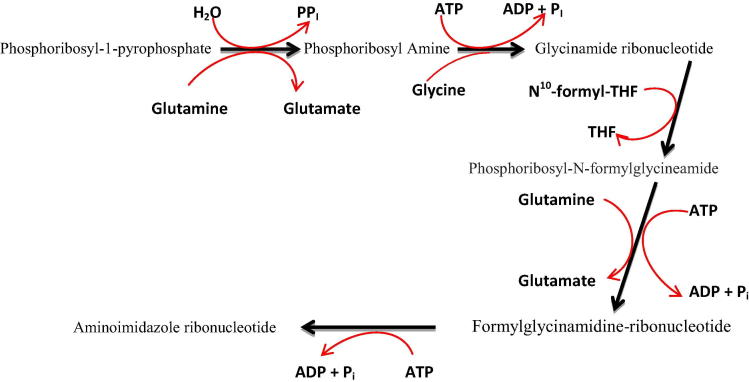
Glycinamide ribonucleotide transformylase (GART) is one of the most important trifunctional enzymes involved in purine synthesis. Human GART (HsGART) is composed of three units: glycinamide ribonucleotide synthetase (GARS), glycinamide ribonucleotide transformylase (GARTfase), aminoimidazole ribonucleotide synthetase (AIRS) and all of which work in a synchronized manner to facilitate purine synthesis. These three units of human GART (HsGART) catalyze steps 2, 3 and 5 of the de novo purine synthesis pathway. The second step of purine synthesis is dependent on GARS (N-terminal enzyme unit) that results in the generation of glycinamide ribonucleotide (GAR), adenosine diphosphate and phosphate ion. The third step is catalyzed by GARTfase (C-terminal enzyme unit) resulting in conversion of GAR to N-formylglycinamide ribonucleotide using 10-formyltetrahydrofolate as a cofactor. AIRS (the middle enzymatic domain of HsGART) is important for the conversion of formylglycinamidine ribonucleotide and adenosine triphosphate to aminoimidazole ribonucleotide (AIR), adenosine diphosphate and phosphate ion (Welin et al., 2010). This whole process is shown in Fig. 1. The core fourth step of the purine pathway is performed by phosphoribosyl formylglycinamidine amidotransferase, encoded by a separate gene (purL). Interestingly, phosphoribosylamine, the substrate of GARS, is quite unstable and quickly hydrolyzes to ribose 5-phosphate within few seconds at physiological temperatures (Rudolph et al., 1995). Because of its transient nature, phosphoribosylamine might be transferred from phosphoribosyl pyrophosphate amidotransferase to GARS (Wang et al., 1998).
Figure 1.
Presentation of step 1–5 of the purine de novo bio-synthesis pathway: Phosphoribosyl-1-pyrophosphate (PRPP) to aminoimidazole ribonucleotide (AIR).
HsGART gene is localized on chromosome 21 and might be linked with trisomy disorders (Down syndrome) (Chadefaux et al., 1984). In addition, elevated serum purine levels associated with Down’s syndrome may possibly be due to overexpression of HsGART (Brodsky et al., 1997). In-vivo studies have suggested that inhibitors of folate-dependent enzymes play a crucial role in anti-tumor activity. The C-terminal GARTfase domain uses folate cofactor and this has been associated with anti-tumor activity (Costi and Ferrari, 2001).
The compound (6R)-dideazatetrahydrofolate (lometrexol) belongs to the class of anti-folates that are specific inhibitors of de novo purine synthesis due to potent inhibition of GART (Bronder and Moran, 2002). A study of the activity of pemetrexed (a commercially available chemotherapy drug) against several recombinant mouse and human enzymes in vitro led to the conclusion that both GART and dihydrofolate reductase (DHFR) were potential secondary targets for the polyglutamate forms of pemetrexed (Shih et al., 1997, Chattopadhyay et al., 2006).
Based on these studies, we designed an in silico study involving docking of three enzyme units of HsGART: GARS, AIRS and GARTfase against three 2,4-Diamino analogs of folic acid important for cancer chemotherapy (Sant et al., 1992). 2,4-Diamino analogs of folic acid have been important in cancer chemotherapy. Despite its complexity, the underlying basis of cell growth inhibition by these compounds relies on their ability to block de novo synthesis of the purine nucleotides, i.e., precursors of DNA (Schoettle et al., 1997). This work is an extension of our previous study where we reported inhibition of amido phosphoribosyltransferase using 2,4-Diamino analogs of folic acid (Batool et al., 2013).
2. Materials and methods
2.1. Receptor dataset
Since HsGART is composed of three enzyme units, PDB ids for its three components were gathered from the literature, (i) GARS (PDB id: 2QK4) (Welin et al., 2010), (ii) GARTfase (PDB id: 1ZLY) (Dahms et al., 2005) and (iii) AIRS (PDB id: 2V9Y) (Welin et al., 2010). Binding residue information of substrate binding for these PDB entries were collected separately. 3D structures for the binding sites of GARS and AIRS were taken from the previous studies (Welin et al., 2010, Connelly et al., 2013, Zhang et al., 2003). The binding site interacting residues are shown in Table 1.
Table 1.
Binding site residues for (i) 2QK4, (ii) 1ZLY and (iii) 2V9Y.
| No | PDB id | Substrate binding residues |
|---|---|---|
| 1 | 2QK4 | Lys148, Val160, Lys162, Glu190, Glu191, Leu192, Leu193, Glu197, Arg220, Asn229, Leu287 |
| 2 | 1ZLY | Ile91, Ala140, Glu141, Asp144, Val143, Leu85 |
| 3 | 2V9Y | Thr498, Gly499, Gly623, His620, Asn622, Met519, His680, Tyr537, Glu570 |
2.2. Ligand dataset
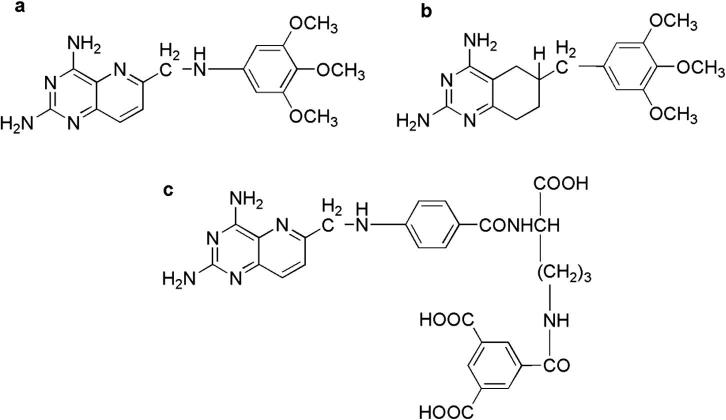
The ligand dataset was prepared from three antifolate diamino inhibitors including (a) 2,4-diamino-6-(3,4,5-trimethoxybenzyl)-5,6,7,8-tetrahydro-quinazoline (PY899), (b) 5-((4-carboxy-4-(4-(((2,4-diaminopyrido[3,2-d]pyrimidine-6-yl)methyl)amino)benzamido)butyl)carbamoyl) isophthalic acid (DIA), and (c) 2,4-diamino-6-(3,4,5-trimethoxyanilino)-methylpyrido[3,2-d]pyrimidine (PY873). The structures of PY873, PY899 and DIA were drawn using Chemoffice. Fig. 2 shows the 2D chemical structures of these ligands.
Figure 2.
2D structures of (a) PY873, (b) PY899 and (c) DIA.
2.3. Docking studies
Using automated docking program AutoDock4®, molecular docking was performed to monitor possible binding interactions of the three antifolate inhibitors with all three components of HsGART (Morris et al., 1998). In brief, polar hydrogen atoms and Kollman charges were assigned to the receptor proteins. For ligands, Gasteiger partial charges were designated and non-polar hydrogen atoms were merged. All torsions for ligands were allowed to rotate during docking procedure. The program AutoGrid® was used to generate the grid maps. Each grid was centered at the structure of the corresponding receptor. The grid dimensions were 80 * 80 * 80 Å3 with points separated by 0.375 Å having random starting positions, orientations and torsions. The translation, quaternion and torsion steps were taken from default values indicated in AutoDock. The Lamarckian genetic algorithm method was used for minimization using default parameters. The standard docking protocol for rigid and flexible ligand docking consisted of 100 runs, using an initial population of 150 randomly placed individuals, with 2.5 × 106 energy evaluations, a maximum number of 27,000 iterations, a mutation rate of 0.02, a crossover rate of 0.80 and an elitism value of 1. Cluster analysis using an RMS tolerance of 1.0 Å was performed on the docked results. The binding energy of each cluster is the mean binding energy of all the conformations present within the cluster; the cluster with lowest binding energy and higher number of conformations within it was selected as the docked pose of that particular ligand.
3. Results
Three components of HsGART were subjected to docking studies using Autodock 4.2. Binding information pertaining to these components was gathered and used for a binding analysis of ligand dataset. Each docked pose was monitored individually in order to analyze the binding interactions. Table 2 shows binding energy, intermolecular energy, van der Waals + hydrogen-bonding + desolvation energies, electrostatic and final internal energy, torsional energy and system’s unbound energies along with inhibition constant values for the docking studies.
Table 2.
Docking Results for (i) glycinamide ribonucleotide synthetase (2QK4), (ii) glycinamide ribonucleotide transformylase (1ZLY) and (iii) aminoimidazole ribonucleotide synthetase (2V9Y).
| PDB IDS | 2QK4 |
1ZLY |
2V9Y |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligand/properties | PY873 | PY899 | DIA | PY873 | PY899 | DIA | PY873 | PY899 | DIA |
| Binding energy (kcal/mol) | −6.03 | −6.83 | −5.52 | −7.06 | −6.22 | −5.97 | −6.57 | −6.11 | −5.53 |
| Ki (μM) | 37.1 | 9.85 | 90.16 | 6.7 | 27.54 | 42.18 | 15.4 | 33.43 | 87.97 |
| Intermolecular energy (kcal/mol) | −8.43 | −8.32 | −9.99 | −9.45 | −7.71 | 11.34 | −8.95 | −7.6 | −10.9 |
| vdW + Hbond + desolv energy (kcal/mol) | −7.53 | −7.49 | −7.54 | −8.9 | −7.44 | −8.39 | −7.66 | −7.13 | −7.6 |
| Electrostatic energy (kcal/mol) | −0.85 | −0.83 | −2.45 | −0.55 | −0.27 | −2.95 | −1.29 | −0.47 | −3.3 |
| Final total internal energy (kcal/mol) | −0.77 | −0.11 | 164.57 | −0.2 | −0.18 | −0.57 | −0.86 | −0.02 | −1.08 |
| Torsional free energy (kcal/mol) | 2.39 | 1.49 | 4.47 | 2.39 | 1.49 | 5.37 | 2.39 | 1.49 | 5.37 |
| Unbound system’s energy (kcal/mol) | −0.77 | −0.11 | 164.57 | −0.2 | −0.18 | −0.57 | −0.86 | −0.02 | −1.08 |
| Temperature (K) | 298.5 | 298.5 | 298.5 | 298.5 | 298.5 | 298.5 | 298.5 | 298.5 | 298.5 |
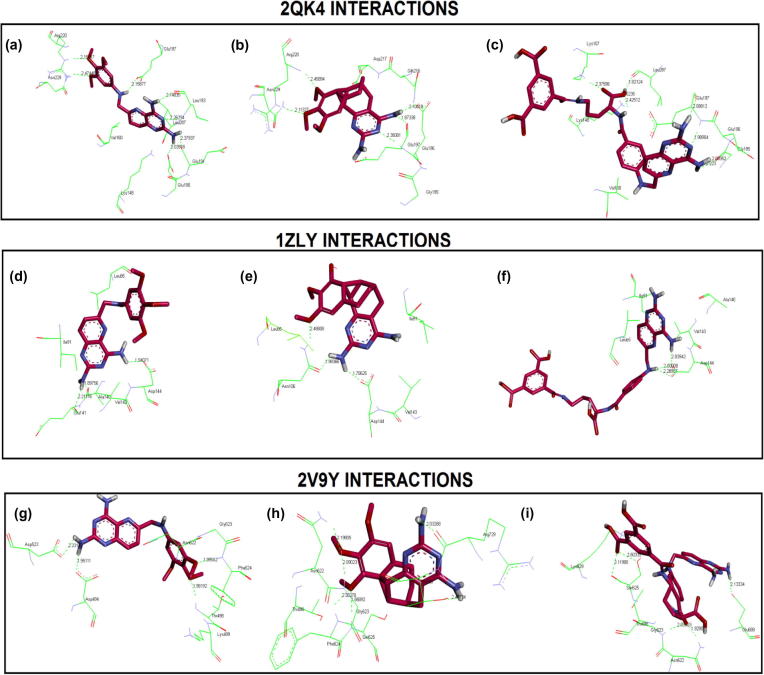
Docking analysis revealed that the ligand dataset showed interactions with the binding site residues of all the receptors included in the study. Graphically interactions are shown in Fig. 3.
Figure 3.
Glycinamide ribonucleotide synthetase (2QK4) interactions with, (a) PY873, (b) PY899, (c) DIA, glycinamide ribonucleotide transformylase (1ZLY) interactions, (d) PY873, (e) PY899, (f) DIA, (iii) aminoimidazole ribonucleotide synthetase (2V9Y) interactions, (g) PY873, (h) PY899 and (i) DIA.
3.1. 2QK4 results
The docking results show clearly that PY873, PY899 and DIA interact to the binding site of GARS. PY873 makes the following seven hydrogen bonds: two bonds with Leu193 through its oxygen atom at a distance of 2.26 and 2.14 Å (−O…H, distance; 2.26 Å, 2.12 Å); third hydrogen bond with Glu190 (−O…NH, distance; 2.03 Å); fourth hydrogen bond with Glu191 (−O…NH, distance; 2.37); fifth hydrogen bond with Glu197 (−O…NH, distance; 2.158 Å); sixth hydrogen bond with Arg220 (−H…O, distance; 2.158 Å) and a seventh hydrogen bond with Asn229 (−H…O, distance; 2.47 Å). PY873 exhibits hydrophobic interactions with Lys148 and Val160 as shown in Fig. 3a.
PY899 makes five of the following hydrogen bonds with the binding residues: two hydrogen bonds with Glu197 (−O…H, distance; 1.9 Å and −O…N, distance; 2.4 Å); two hydrogen bonds with Arg220 (–H…O, distance; 2.46 Å and –H…O, distance; 2.11 Å) and one hydrogen bond with Glu196 (–O…H, distance; 2.11 Å). PY899 exhibits hydrophobic interactions with Asn229, Asp217, Gln216, and Gly195 residues (Fig. 3b).
Fig. 3c shows the binding interactions for DIA which makes the following eight hydrogen bonds with the binding residues: Two hydrogen bonds with Glu197 (–O…H, distance; 2.088 Å and –O…NH, distance; 1.98 Å); Two H Bonds with Gly195 (–O…NH, distance; 2.083 Å and –O…NH, distance; 2.223 Å); two hydrogen bonds with Lys107 (−O…NH, distance; 2.4 Å and –O…NH, distance; 2.2 Å); one hydrogen bond with Leu287 (–H…O, distance; 1.8 Å) and one hydrogen bond with Val160 (–O…NH, distance; 2.3 Å). DIA exhibits hydrophobic interactions with Lys148.
3.2. 1ZLY results
PY873 makes the following three hydrogen bonds with the binding residues: first hydrogen bond is observed with Ala140 (H…O, distance; 1.89 Å); second hydrogen bond with Glu141 (H…O, distance; 2.2 Å) and the third hydrogen bond with Asp144 (H…O, distance; 1.9 Å). Hydrophobic interactions are observed with Leu85, Ile91 and Val143 (Fig. 3d).
PY899 makes the following three hydrogen bonds with the binding residues: First hydrogen bond with Asp144 (H…O, distance; 1.7 Å) and the other two hydrogen bonds with Asn106 (H…O, distance; 1.98 Å and H…N, distance; 2.48 Å). Hydrophobic interactions are noted for Leu85, Ile95 and Val143 residues (Fig. 3e).
DIA makes three hydrogen bonds with Asp144 (H…O, distance; 2 Å, H…O, distance; 2 Å and H…O, distance; 2 Å) while hydrophobic interactions are observed with Leu85, Ile91, Ala140 and Val143 (Fig. 3f).
3.3. 2V9Y results
PY873 makes four hydrogen bonds with the following binding residues: (1) Thr498 (O…HN, distance; 1.98 Å); (2) Gly623 (O…HN, distance; 1.9 Å); (3) Asp494 (H…O, distance; 1.95 Å); (4) Asp523 (H…O, distance; 2.2 Å). Hydrophobic interactions are observed with Lys499, Asn622 and Phe624 (Fig. 3g).
PY899 makes the following six hydrogen bonds: First hydrogen bond with Asn622 (H…O, distance; 2); second hydrogen bond with Gly623 (HN…O, distance; 2 Å); third hydrogen bond with Ser625 (HN…O, distance;1.86 Å); fourth hydrogen bond found with Arg729 (O….H, distance; 2 Å), fifth hydrogen bond noticed with Phe624 (HN…O, distance; 2.3 Å) and sixth hydrogen bond seen with Thr730 (O…HN, distance; 2.49 Å). Hydrophobic interactions are exhibited with Thr498 (Fig. 3h).
DIA makes the following five hydrogen bonds: First hydrogen bond with Lys629 (H…O, distance; 3 Å); second hydrogen bond with Glu688 (O…H, distance; 2.1 Å); third hydrogen bond with Asn622 (H…O, distance; 1.9 Å); fourth hydrogen bond with Gly623 (H…O, distance; 2.03 Å); fifth hydrogen bond with Ser625 (H…O, distance; 2.6 Å) while hydrophobic interactions are seen with Thr498 (Fig. 3i).
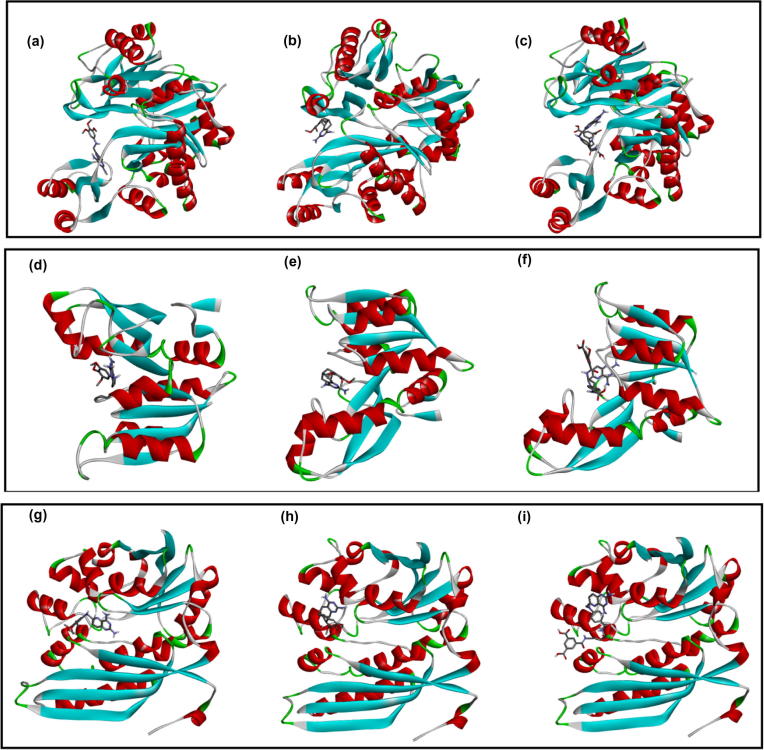
Fig. 4 shows binding orientation of the ligands with a binding pocket of three components of HsGART. This present study suggests that each of the three ligands binds to the same binding cavity with its respective receptors.
Figure 4.
Binding position for 2QK4 Interactions (a) PY873 (b) PY899, (c) DIA, Binding position for 1ZLY Interactions (d) PY873, (e) PY899, (f) DIA, Binding position for 2V9Y Interactions (g) PY873, (h) PY899 and (i) DIA. Receptors are shown in ribbon while ligands are shown in sticks.
4. Discussion
This study focuses on the mechanism by which certain antifolate inhibitors act against purine synthesis pathways. Previous studies have demonstrated that methotrexate polyglutamates retain a potent ability to inhibit DHFR and act as potent inhibitors of several folate-dependent enzymes, including thymidylate synthase and de novo purine synthesis enzymes (Baram et al., 1988, Allegra et al., 1985a, Allegra et al., 1985b, Baggott et al., 1986). Recent investigations regarding the mechanism of action of dihydrofolate polyglutamates have indicated that metabolic inhibition is a multifactorial event that includes folate substrate depletion and direct inhibition of several critical folate-dependent enzymes by interaction at multiple intracellular sites (Allegra et al., 1986, Baram et al., 1987, Matherly et al., 1987).
HsGART consists of three enzyme units: GARS, GARTfase and AIRS. HsGART is involved in catalyzing steps 2, 3 and 5 steps of de novo purine synthesis pathway. In this study, we analyzed the binding of three DHFR antifolates, PY873, PY899 and DIA, with human GART. Taken together, our study reveals that three studied inhibitors demonstrate interactions with the binding residues of receptor dataset along with favorable energy values. Based on the current study, we propose GART as a potential target for designing of new specifically potent and selective antifolate type inhibitors to find a suitable candidate for targeted chemotherapy.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
The authors (SB, MSN, FP, MAK) would like to thank the facilities provided by the Department of BioSciences, COMSATS Institute of Information Technology, Pakistan and the King Fahd Medical Research Center, King Abdulaziz University, Saudi Arabia. The authors are grateful to Miss Maleeha Waqar for her assistance in creating Fig. 1 in light of a template provided by MAK in this project. MAK would also like to thank the School of Molecular and Microbial Biosciences, University of Sydney, Australia for the award of a prestigious U2000 Postdoctoral Fellowship in 2000. This three-year highly competitive award funded his research on the “Inhibition of amido phosphoribosyltransferase by new antifolates: Design and mechanism of action of purine antagonists” and it is the basis of this current study.
Footnotes
Peer review under responsibility of King Saud University.
References
- Allegra C.J., Chabner B.A., Drake J.C., Lutz R., Rodbard D., Jolivet J. Enhanced inhibition of thymidylate synthase by methotrexate polyglutamates. J. Biol. Chem. 1985;260:9720–9726. [PubMed] [Google Scholar]
- Allegra C.J., Drake J.C., Jolivet J., Chabner B.A. Inhibition of phosphoribosylaminoimidazolecarboxamide transformylase by methotrexate and dihydrofolic acid polyglutamates. Proc. Natl. Acad. Sci. U.S.A. 1985;82:4881–4885. doi: 10.1073/pnas.82.15.4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allegra C.J., Fine R.L., Drake J.C., Chabner B.A. The effect of methotrexate on intracellular folate pools in human MCF-7 breast cancer cells. Evidence for direct inhibition of purine synthesis. J. Biol. Chem. 1986;261:6478–6485. [PubMed] [Google Scholar]
- Baggott J.E., Vaughn W.H., Hudson B.B. Inhibition of 5-aminoimidazole-4-carboxamide ribotide transformylase, adenosine deaminase and 5′-adenylate deaminase by polyglutamates of methotrexate and oxidized folates and by 5-aminoimidazole-4-carboxamide riboside and ribotide. Biochem. J. 1986;236:193–200. doi: 10.1042/bj2360193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baram J., Allegra C.J., Fine R.L., Chabner B.A. Effect of methotrexate on intracellular folate pools in purified myeloid precursor cells from normal human bone marrow. J. Clin. Invest. 1987;79:692–697. doi: 10.1172/JCI112872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baram J., Chabner B.A., Drake J.C., Fitzhugh A.L., Sholar P.W., Allegra C.J. Identification and biochemical properties of 10-formyldihydrofolate, a novel folate found in methotrexate-treated cells. J. Biol. Chem. 1988;263:7105–7111. [PubMed] [Google Scholar]
- Batool S., Nawaz M.S., Kamal M.A. In silico analysis of the amido phosphoribosyltransferase inhibition by PY873, PY899 and a derivative of isophthalic acid. Invest. New Drugs. 2013;31:1355–1363. doi: 10.1007/s10637-013-9944-9. [DOI] [PubMed] [Google Scholar]
- Brodsky G., Barnes T., Bleskan J., Becker L., Cox M., Patterson D. The human GARS-AIRS-GART gene encodes two proteins which are differentially expressed during human brain development and temporally overexpressed in cerebellum of individuals with Down syndrome. Human Mol. Genet. 1997;6:2043–2050. doi: 10.1093/hmg/6.12.2043. [DOI] [PubMed] [Google Scholar]
- Bronder J.L., Moran R.G. Antifolates targeting purine synthesis allow entry of tumor cells into S phase regardless of p53 function. Cancer Res. 2002;62:5236–5241. [PubMed] [Google Scholar]
- Chadefaux B., Allard D., Rethore M.O., Raoul O., Poissonnier M., Gilgenkrantz S., Cheruy C., Jerome H. Assignment of human phosphoribosylglycinamide synthetase locus to region 21q221. Hum. Genet. 1984;66:190–192. doi: 10.1007/BF00286599. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S., Zhao R., Krupenko S.A., Krupenko N., Goldman I.D. The inverse relationship between reduced folate carrier function and pemetrexed activity in a human colon cancer cell line. Mol. Cancer Ther. 2006;5:438–449. doi: 10.1158/1535-7163.MCT-05-0243. [DOI] [PubMed] [Google Scholar]
- Connelly S., DeMartino J.K., Boger D.L., Wilson I.A. Biological and structural evaluation of 10R- and 10S-methylthio-DDACTHF reveals a new role for sulfur in inhibition of glycinamide ribonucleotide transformylase. Biochemistry. 2013;52:5133–5144. doi: 10.1021/bi4005182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costi M.P., Ferrari S. Update on antifolate drugs targets. Curr. Drug Targets. 2001;2:135–166. doi: 10.2174/1389450013348669. [DOI] [PubMed] [Google Scholar]
- Dahms T.E., Sainz G., Giroux E.L., Caperelli C.A., Smith J.L. The apo and ternary complex structures of a chemotherapeutic target: human glycinamide ribonucleotide transformylase. Biochemistry. 2005;44:9841–9850. doi: 10.1021/bi050307g. [DOI] [PubMed] [Google Scholar]
- Hartman S.C., Buchanan J.M. Nucleic acids, purines, pyrimidines (nucleotide synthesis) Ann. Rev. Biochem. 1959;28:365–410. doi: 10.1146/annurev.bi.28.070159.002053. [DOI] [PubMed] [Google Scholar]
- Kappock T.J., Ealick S.E., Stubbe J. Modular evolution of the purine biosynthetic pathway. Curr. Opin. Chem. Biol. 2000;4:567–572. doi: 10.1016/s1367-5931(00)00133-2. [DOI] [PubMed] [Google Scholar]
- Matherly L.H., Barlowe C.K., Phillips V.M., Goldman I.D. The effects on 4-aminoantifolates on 5-formyltetrahydrofolate metabolism in L1210 cells. A biochemical basis of the selectivity of leucovorin rescue. J. Biol. Chem. 1987;262:710–717. [PubMed] [Google Scholar]
- Morris G.M., Goodsell D.S., Halliday R.S., Huey R., Hart W.E., Belew R.K., Olson A.J. Automated docking using a Lamarckian genetic algorithm and empirical binding free energy function. J. Comput. Chem. 1998;19:1639–1662. [Google Scholar]
- Rudolph J., Stubbe J. Investigation of the mechanism of phosphoribosylamine transfer from glutamine phosphoribosylpyrophosphate amidotransferase to glycinamide ribonucleotide synthetase. Biochemistry. 1995;34:2241–2250. doi: 10.1021/bi00007a019. [DOI] [PubMed] [Google Scholar]
- Sant M.E., Lyons S.D., Phillips L., Christopherson R.I. Antifolates induce inhibition of amido phosphoribosyltransferase in leukemia cells. J. Biol. Chem. 1992;267:11038–11045. [PubMed] [Google Scholar]
- Schoettle S.L., Crisp L.B., Szabados E., Christopherson R.I. Mechanisms of inhibition of amido phosphoribosyltransferase from mouse L1210 leukemia cells. Biochemistry. 1997;36:6377–6383. doi: 10.1021/bi962598m. [DOI] [PubMed] [Google Scholar]
- Shih C., Chen V.J., Gossett L.S., Gates S.B., MacKellar W.C., Habeck L.L., Shackelford K.A., Mendelsohn L.G., Soose D.J., Patel V.F., Andis S.L., Bewley J.R., Rayl E.A., Moroson B.A., Beardsley G.P., Kohler W., Ratnam M., Schultz R.M. LY231514, a pyrrolo[2,3-d]pyrimidine-based antifolate that inhibits multiple folate-requiring enzymes. Cancer Res. 1997;57:1116–1123. [PubMed] [Google Scholar]
- Wang W., Kappock T.J., Stubbe J., Ealick S.E. X-ray crystal structure of glycinamide ribonucleotide synthetase from Escherichia coli. Biochemistry. 1998;37:15647–15662. doi: 10.1021/bi981405n. [DOI] [PubMed] [Google Scholar]
- Welin M., Grossmann J.G., Flodin S., Nyman T., Stenmark P., Tresaugues L., Kotenyova T., Johansson I., Nordlund P., Lehtio L. Structural studies of tri-functional human GART. Nucleic Acids Res. 2010;38:7308–7319. doi: 10.1093/nar/gkq595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Desharnais J., Marsilje T.H., Li C., Hedrick M.P., Gooljarsingh L.T., Tavassoli A., Benkovic S.J., Olson A.J., Boger D.L., Wilson I.A. Rational design, synthesis, evaluation, and crystal structure of a potent inhibitor of human GAR Tfase: 10-(trifluoroacetyl)-5,10-dideazaacyclic-5,6,7,8-tetrahydrofolic acid. Biochemistry. 2003;42:6043–6056. doi: 10.1021/bi034219c. [DOI] [PubMed] [Google Scholar]