Abstract
Gnas is a complex gene with multiple imprinted promoters. The upstream Nesp and Nespas/Gnasxl promoters are paternally and maternally methylated, respectively. The downstream promoter for the stimulatory G protein α-subunit (Gsα) is unmethylated, although in some tissues (e.g., renal proximal tubules), Gsα is poorly expressed from the paternal allele. Just upstream of the Gsα promoter is a primary imprint mark (1A region) where maternal-specific methylation is established during oogenesis. Pseudohypoparathyroidism type 1B, a disorder of renal parathyroid hormone resistance, is associated with loss of 1A methylation. Analysis of embryos of Dnmt3L–/– mothers (which cannot methylate maternal imprint marks) showed that Nesp, Nespas/Gnasxl, and 1A imprinting depend on one or more maternal primary imprint marks. We generated mice with deletion of the 1A differentially methylated region. These mice had normal Nesp-Nespas/Gnasxl imprinting, indicating that the Gnas locus contains two independent imprinting domains (Nespas-Nespas/Gnasxl and 1A-Gsα) controlled by distinct maternal primary imprint marks. Paternal, but not maternal, 1A deletion resulted in Gsα overexpression in proximal tubules and evidence for increased parathyroid hormone sensitivity but had no effect on Gsα expression in other tissues where Gsα is normally not imprinted. The 1A region is a maternal imprint mark that contains one or more methylation-sensitive cis-acting elements that suppress Gsα expression from the paternal allele in a tissue-specific manner.
Keywords: genomic imprinting, pseudohypoparathyroidism, DNA methylation, guanine nucleotide binding protein
Genomic imprinting is an epigenetic phenomenon involving a small number of genes in which the two parental alleles have distinct epigenetic marks (e.g., DNA methylation and histone modification) that usually lead to allele-specific differences in gene expression (1). These marks are erased within primordial germ cells and reestablished during gametogenesis. The most well established examples of primary imprint marks are regions in which DNA methylation is established on one parental allele during gametogenesis and maintained throughout development. These primary imprint marks can then produce other allele-specific epigenetic changes within neighboring regions to generate large domains including multiple imprinted genes.
Gnas is a complex imprinted locus on mouse chromosome 2 that generates multiple gene products by the use of alternative promoters and first exons that splice onto a common set of downstream exons (exons 2–12, Fig. 1A) (2). The human ortholog GNAS at 20q13 has a similar overall structure and imprinting pattern. The major Gnas product, the stimulatory G protein α-subunit (Gsα), which mediates receptor-stimulated cAMP production, is generated from the most downstream promoter and first exon. Promoters for the chromogranin-like protein NESP55 (Nesp) and Gsα isoform XLαs (Gnasxl) are located 45 and 30 kb upstream of Gsα exon 1, respectively (3). Nesp and Gnasxl are oppositely imprinted; the Nesp promoter is DNA methylated on the paternal allele and transcriptionally active on the maternal allele, whereas Gnasxl is methylated on the maternal allele and active on the paternal allele (4, 5). Nesp is not a primary imprinting center because its imprinting is not established until after implantation (6). Paternal-specific antisense transcripts (Nespas) generated from the Gnasxl promoter region may be important for Nesp imprinting (7, 8).
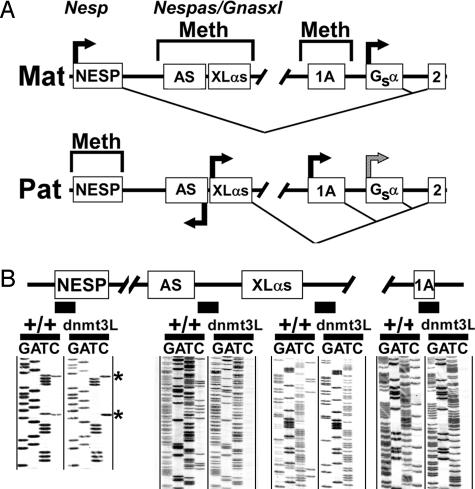
Fig. 1.
Gnas methylation analysis in embryos derived from Dnmt3L–/– mothers. (A) The maternal (Mat) and paternal (Pat) alleles of Gnas are depicted with alternative first exons NESP (Nesp), XLαs (Gnasxl), 1A, and Gsα splicing to common exon 2 (exons 3–12 are not shown). DNA methylation (Meth) and splicing patterns are shown above and below each image, respectively. Horizontal arrows indicate active promoters and the direction of transcription. The paternal Gsα promoter is suppressed in some tissues (gray arrow). The Gnasxl promoter also generates paternal-specific antisense transcripts (Nespas), whose first exon (AS) is shown. The diagram is not drawn to scale. (B) Bisulfite-modified genomic sequencing of +/+ embryos (8.5 days postcoitus) and embryos derived from Dnmt3L–/– mothers (Dnmt3L) within the Nesp, Nespas, Gnasxl, and 1A DMRs, respectively (black boxes indicate examined regions). Asterisks indicate positions of CpG cytosines in Nesp.
Gsα is imprinted in a tissue-specific manner, being expressed primarily from the maternal allele in renal proximal tubules, pituitary, thyroid, and ovaries but biallelically expressed in most other tissues (9–13). (Throughout this article, we use the term Gsα imprinting to refer to allele-specific differences in Gsα expression in specific tissues, not the underlying epigenetic marks, which are presumably not tissue-specific.) Gsα imprinting is not associated with DNA methylation of its promoter but is associated with allele-specific differences in histone methylation (6, 14). Heterozygous Gsα-inactivating mutations in Albright hereditary osteodystrophy also lead to parathyroid hormone (PTH), TSH, and gonadotropin resistance (pseudohypoparathyroidism type 1A, PHP1A) when present on the maternal allele (2) because of the loss of Gsα expression from the active maternal allele in hormone target tissues.
Just upstream of the Gsα promoter is a differentially methylated region (DMR) that is methylated on the maternal allele and contains a promoter and first exon (exon 1A), which generates paternal-specific mRNAs of unknown function (6, 15) (Fig. 1 A). This DMR is a primary imprint mark, because its methylation is established during oogenesis and maintained throughout development (6). This region also has allele-specific differences in histone modifications (14). In PHP1B, in which patients develop renal PTH resistance but not Albright hereditary osteodystrophy, maternal imprinting (methylation) of the 1A DMR is lost (15). We have suggested that the 1A DMR has cis-acting negative regulatory elements (e.g., silencers or insulators) for the Gsα promoter that are both methylation-sensitive (and, hence, suppress Gsα only on the paternal allele) and tissue-specific (perhaps because of tissue-specific expression of trans-acting factors) (2, 15). This model predicts that loss of maternal 1A methylation in PHP1B leads to biallelic loss of Gsα expression in renal proximal tubules and renal PTH resistance, but it has little effect in most other tissues where normally the cis-acting elements do not affect Gsα promoter activity.
In this study, we directly tested the role of the 1A DMR on Gnas and tissue-specific Gsα imprinting by generating mice with deletion of the 1A DMR. Studies in these mice, as well as in mouse embryos that are unable to establish maternal primary methylation imprints, show that Gnas consists of two independent imprinting domains and that tissue-specific Gsα imprinting is controlled by elements with the 1A DMR.
Materials and Methods
Generation of Targeting Construct. A clone containing Gnas exons 1 and 1A was isolated from a 129SvEv mouse genomic DNA library (Stratagene). A 4.2-kb BamHI–SspI fragment (–1,601/+2,602; base pair position relative to Gsα translational start site) was subcloned into the BamHI and EcoRI sites downstream of the loxP–neomycin resistance gene (Neo)-loxP cassette within pLoxpneo (16) (Fig. 2A). Next, a 5.3-kb SacI fragment (–11,600/–6,267) was inserted into a HpaI site located upstream of the loxP–Neo cassette, and the targeting construct was isolated after NotI digestion.
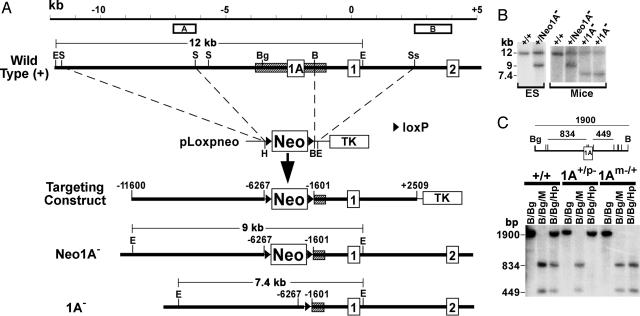
Fig. 2.
Generation of 1A– mice. (A) The upstream portion of the wild-type Gnas allele including exons 1A, 1, and 2 is shown at the top. The 1A DMR is shown by a hatched rectangle, and positions of probes A and B for Southern blot analysis are shown above. The scale at the top is given in kb, with 0 kb being the Gsα translational start site. Upstream SacI and downstream BamHI–SstI fragments were inserted into pLoxpneo to generate the targeting construct. Neo1A– and 1A– alleles are shown below. E, EcoRI; S, SacI; Bg, BglII; B, BamHI; Ss, SstI; H, HpaI; Neo, neomycin resistance gene; TK, thymidine kinase gene; triangle, loxP site. (B) Southern blot analysis of DNA from ES cells or mice after EcoRI digestion and hybridization with probe A. Genotypes are indicated above each lane (parental assignment unknown). EcoRI fragments derived from +, Neo1A–, and 1A– alleles are shown in A. (C) Southern blot analysis of liver DNA from +/+, 1A+/p–, and 1Am–/+ mice performed by using a BglII (Bg)–BamHI (B) probe within the 1A DMR after digestion with enzymes indicated above each lane (M, MspI; Hp, HpaII). A restriction map is shown, with M/Hp sites indicated as vertical lines and lengths of digestion products (in bp) shown above.
Generation of 1A– Mice. TC1 ES cells (17) were transfected with the targeting construct and selected with G418 and FIAU. Doubly resistant ES colonies were analyzed by Southern blot analysis for homologous recombination. ES cells heterozygous for the targeted mutation (Neo1A–, Fig. 2 A) were microinjected into C57/B6 blastocysts, and resulting male chimeras were mated with NIH Black Swiss females (Taconic Farms) to achieve germline transmission. Neo1A– mice were mated with EIIa promoter-cre mice (18) to excise Neo (1A– allele, Fig. 2 A; referred to as Δ–6267/–1601 in ref. 14). Mice were maintained on standard chow diet (National Institutes of Health, Bethesda) with 12:12-h light/dark cycle. E1– mice with deletion of adjoining downstream sequence from –1,601 to +419 (referred to as Δ–1601/+419 in ref. 14) have been described (M.C., unpublished data). Embryonic day 8.5 embryos were derived from Dnmt3L–/– mothers as described (19). Mouse studies were approved by the National Institute of Diabetes and Digestive and Kidney Diseases Animal Care and Use Committee.
Genotyping. Genomic DNA was isolated from ES cells or tissues by using the Genomic DNA buffer set (Qiagen, Valencia, CA). Samples were digested with BglII and hybridized with a labeled 1.3-kb SspI–NotI genomic fragment (probe B, Fig. 2 A) or were digested with EcoRI and hybridized with an 878-bp KpnI–SacI genomic fragment (probe A, Fig. 2 A and B). Subsequent genotyping was performed by PCR of mouse tail DNA using the following cycling profile: 94°C for 5 min, followed by 35 cycles of 94°C for 45 s, 64°C for 45 s, and 72°C for 60 s. The following primers were used: 5′-GCCGATTTTT TGCGCGTCCCCTTC-3′ and 5′-GCTTCTTCTCCATCTTCTTGCG-3′, which generates a 530-bp band from the wild-type allele; and 5′-GATAGCCTTTCACCCAGTAG-3′ and 5′-TTTGCGGGCGGCACATCGCG-3′, which generates a 380-bp band from the 1A– allele.
Southern Blot Analysis with Methylation-Sensitive Restriction Enzymes. DNA samples (20 μg) were digested with the indicated restriction enzymes (New England Biolabs). Genomic probes were generated by PCR using the following sets of upstream and downstream primers: Nesp upstream region, 5′-TAGCCATGATGCTCTTGCTGAC-3′ and 5′-GGGTGAGATACAGTAGGTGC-3′; Gnasxl upstream region, 5′-GACGGAGCCGAACGTCCCTAC-3′ and 5′-GAGAAGTGAGGATATGCTTAGC-3′; and Nespas/Gnasxl promoter region, 5′-GTTGTAGTCAGGGCTTGCTCTG-3′ and 5′-TGCAGCTTCTTTGTGCACAGC-3′.
Bisulfite-Modified Genomic Sequencing. Bisulfite-modified genomic sequencing was performed on genomic DNA samples as described (6). Primers for the 1A DMR, Nesp, and Gnasxl first exon have been reported (6). For the Nespas/Gnasxl promoter region, the following initial upstream and downstream primers were used, respectively: 5′-GTAATTTTATAGGGTTTTATTG-3′ and 5′-ATCCATTCTCTTAAATACTCACC-3′; and the following nested upstream and downstream primers were used, respectively: 5′-GAGAGGATTAGTGGAGGTATTTTT-3′ and 5′-ACTCACCCTCTAACTCTACAAAAAAT-3′. Amplified fragments were gel-purified and sequenced with a nested primer by using the Thermo Sequenase kit (United States Biochemical).
Immunoblot Analysis. Proximal tubules were isolated from renal cortex as described (10). Tissues were homogenized in ice-cold lysis buffer (50 mM Tris·HCl/1% Nonidet P-40/0.5% Na deoxycholate/150 mM NaCl, 1 mM PMSF/1 mM Na3VO4/1 mM NaF, containing protease inhibitor mixture) (Roche) with a Teflon pestle (3 ml of buffer per g of tissue) and incubated for 60 min at 4°C. Homogenates were centrifuged at 4°C in a microcentrifuge for 10 min at 16,000 × g, and supernatants were used for immunoblotting. Protein concentrations were determined by the dye method (Bio-Rad). Immunoblotting was performed as described (10, 20). To normalize for loading, the relative amounts of protein in each sample were determined by Simply Blue SafeStain (Invitrogen) staining and quantified with alphaease fc software (Alpha Innotech, San Leandro, CA).
Real-Time Quantitative RT-PCR. Total RNA was isolated by using TRIzol reagent and treated with DNase I (Invitrogen). Quantitative RT-PCR was performed on an MxP3000 real-time PCR system (Stratagene) by using SYBR Green PCR master mix and RT-PCR kit (Applied Biosystems). The PCR cycling profile was 95°C for 10 min, followed by 40 amplification cycles of 95°C for 30 s, 58°C for 60 s, and 72°C for 30 s, and a final cycle of 95°C for 60 s and 55°C for 30 s to generate dissociation curves. A standard curve was generated in each experiment and used to determine the relative abundance of Gsα mRNA in each sample. Gsα expression was normalized to β-actin mRNA levels, which were determined in a similar fashion. Standard curves demonstrated efficiencies of >90% in all experiments. Generation of multiple RT-PCR products was ruled out by both dissociation curves and acrylamide gel electrophoresis. The following upstream and downstream primers were used: Gsα, 5′-CTCCGTTAAACCCATTAACATGCA-3′ and 5′-ACAAGCAGGTCTACCGGGCC-3′; and β-actin, 5′-GACCTCTATGCCAACACAGT-3′ and 5′-TAGGAGCCAGAGCAGTAATC-3′. Nontemplate controls were included to rule out nonspecific amplification. Nesp and Gnasxl transcripts were amplified by using the upstream primers 5′-CACTAATGGGTGACTCCGTCCA-3′ and 5′-GGACTACATGTGTACACACCG-3′, respectively, and the same downstream primer as described for Gsα.
Serum Chemistries. Serum calcium, albumin, and phosphorus were measured by colorimetric assays (Stanbio, Boeme, TX). Serum PTH was measured by using a rat PTH immunoradiometric assay (Immutopics, San Clemente, CA).
Results
Gnas Imprinting Depends on Maternal Primary Methylation Imprint Marks. To test the role of maternal primary imprint marks in Gnas imprinting, we examined methylation of Gnas DMRs in embryos derived from Dnmt3L–/– mothers (Fig. 1B). Dnmt3L, although it is not a DNA methylase, has sequence similarity to active DNA methyltransferases of the Dnmt3 family. Heterozygous embryos derived from Dnmt3L–/– mothers specifically fail to establish methylation at maternal primary imprint marks during oogenesis (19). Gnas methylation was determined by bisulfite-modified genomic sequencing, a method by which DNA is chemically modified so that unmethylated cytosines are converted to uracil before PCR amplification and direct sequencing. In the final sequence, methylated cytosines remain as cytosines, whereas unmethylated cytosines are converted to thymines. In +/+ embryos CpG cytosines were partially converted to thymines at all Gnas DMRs, consistent with the presence of one methylated and one unmethylated allele. In embryos from Dnmt3L–/– mothers, all cytosines within the Nespas/Gnasxl promoter, a region within the same DMR located just downstream of the XLαs first exon, and 1A DMR were fully converted to thymidine, consistent with both parental alleles being unmethylated (the paternal epigenotype). In contrast, there was no CpG cytosine-to-thymidine conversion within Nesp, consistent with both alleles being methylated (the paternal epigenotype). Consistent with these methylation changes, quantitative real-time PCR experiments showed these embryos to have undetectable levels of Nesp mRNA and a 2.3-fold increase in the level of Gnasxl mRNA (data not shown). Therefore, embryos from Dnmt3L–/– mothers have a paternal epigenotype in both alleles at all Gnas DMRs, indicating that Gnas imprinting is determined by one or more maternal primary imprint marks. The findings at Nesp are consistent with prior results showing that Nesp methylation is not established until after implantation development and, therefore, depends on a primary imprint mark within another Gnas region (6).
Generation of Mice with a 1A DMR Deletion. The 1A DMR has been shown to be a maternal primary imprint mark (6). To determine its role in Gnas imprinting, we generated mice with a 1A DMR deletion by targeted mutagenesis (Fig. 2 A), in which the region from –6,267 to –1,601 (base pair positions relative to the Gsα translational start site) was replaced by a single loxP site (1A– allele). Correct gene targeting, germline transmission, and generation of the 1A– allele were confirmed by Southern blot analysis using internal probe A (Fig. 2 A and B) and 3′ probe B (Fig. 2 A and data not shown).
Homozygous 1A–/– mice or mice heterozygous for 1A– on the maternal (1Am–/+) or paternal (1A+/p–) allele had no obvious phenotype. To confirm that the deletion did not affect 1A imprinting in the opposite allele, liver DNA was subjected to Southern blot analysis with methylation-sensitive restriction enzymes and hybridized to a 1,900-bp BglII–BamHI genomic probe within the 1A DMR (Fig. 2C). MspI, which is insensitive to CpG methylation at MspI/HpaII sites, completely digested all samples. In +/+ samples, HpaII, which digests the same sites only when they are unmethylated, generated both the undigested 1,900-bp band and smaller digestion products, consistent with the presence of a methylated maternal and an unmethylated paternal allele (6). HpaII digestion of 1A+/p– samples produced only the undigested band, consistent with deletion of the unmethylated paternal 1A DMR and normal maternal 1A methylation, whereas 1Am–/+ samples showed only the digested bands, consistent with deletion of the methylated maternal 1A DMR and no methylation of the remaining paternal allele. Therefore, heterozygous 1A deletion does not have a transacting effect on the imprinting status of opposite allele. Similar results were obtained with spleen, lung, and kidney DNA (data not shown).
The 1A deletion extends beyond the 5′ end of the DMR, because the most upstream differentially HpaII site is at –2,754 (data not shown). However, the 1A deletion does not remove the 3′ end of the DMR, because we have shown that differential methylation extends a further ≈600 bp downstream of the 1A deletion (6). Southern blot analysis showed that 1A deletion had little effect on methylation of the remaining ≈600-bp downstream region. Conversely, removal of this ≈600-bp downstream region in E1– mice with a –1,601/+419 deletion had no effect on methylation of the major upstream portion of the DMR removed in the 1A deletion (Fig. 6, which is published as supporting information on the PNAS web site). Whether this result indicates that the cis elements required for 1A DMR methylation lie outside of the DMR is unknown.
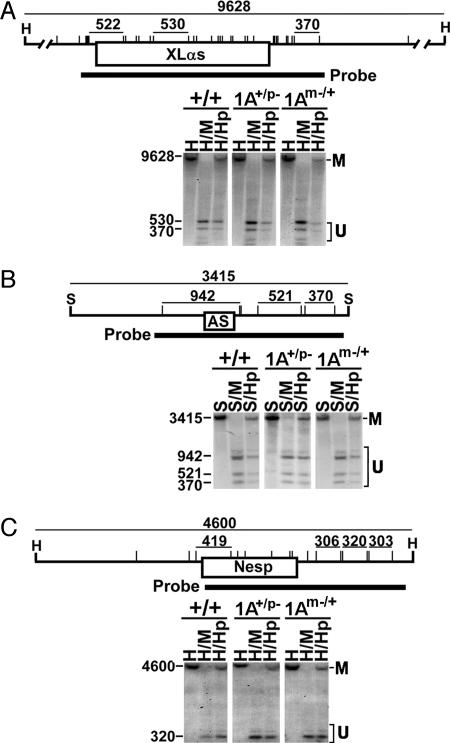
The 1A Deletion Has No Effect on Nesp or Nespas/Gnasxl Imprinting. To determine whether Nesp and Nespas-Gnasxl imprinting depend on the 1A DMR, we performed Southern blot analysis on liver DNA digested with MspI or HpaII and hybridized with Nesp-, Nespas/Gnasxl-, and Gnasxl first exon-specific genomic probes (Fig. 3). MspI and HpaII digestion showed similar results for +/+, 1A+/p–, and 1Am–/+ samples in all three regions, which in each case was consistent with the presence of one methylated and one unmethylated allele. Therefore, 1A deletion on either allele had no effect on Nesp/Nespas/Gnasxl imprinting. Identical results were obtained also with spleen, lung, and kidney DNA, and they were confirmed by bisulfite-modified genomic sequencing (data not shown). The same studies performed with E1– heterozygotes produced identical results (Fig. 7, which is published as supporting information on the PNAS web site), indicating that neither the 1A DMR nor Gsα promoter regions are required to establish or maintain Nesp and Nespas/Gnasxl imprinting.
Fig. 3.
Effect of 1A deletion on Nesp and Nespas/Gnasxl methylation. (A) A restriction map of a 9,628-bp HindIII (H) fragment including XLαs exon 1 is shown, with MspI (M)/HpaII (Hp) sites indicated by vertical lines. The size of restriction fragments in bp is shown at the top, and the position of the hybridization probe is shown at the bottom. Lower shows Southern blot analysis of liver DNA from +/+, 1A+/p–, and 1Am–/+ mice after digestion with enzymes indicated above each lane. (B) Southern blot analysis of a 3,415-bp SacI (S) fragment including the Nespas first exon (AS) and promoter region. (C) Southern blot analysis of a 4,600-bp HindIII fragment from the Nesp region. In each image, the fragments derived from the methylated and unmethylated alleles are designated as M and U, respectively.
Paternal 1A Deletion Reverses Tissue-Specific Gsα Imprinting. We have proposed that tissue-specific Gsα imprinting results from cis-acting negative regulatory elements within the 1A DMR that are both tissue-specific and DNA methylation-sensitive (and, therefore, do not suppress Gsα expression from the maternal allele) (2, 15). Our model predicts that maternal 1A deletion would not affect Gsα expression, whereas paternal 1A deletion would relieve paternal Gsα imprinting leading to Gsα overexpression in tissues, such as proximal tubules, where Gsα is normally imprinted. Consistent with this prediction, Gsα mRNA and protein expression in proximal tubules were unaffected in 1Am–/+ mice but were increased significantly by 72% and 57%, respectively, in 1A+/p– mice (Fig. 4 A and B). Because ≈70% of Gsα expressed in proximal tubules is derived from the maternal allele (10), the increase in Gsα expression observed in 1A+/p– mice is within the range expected from complete loss of paternal Gsα imprinting. Gsα expression was increased >2-fold by the paternal 1A deletion when Gsα expression from the maternal allele was disrupted (E1m–/+:1A+/p– vs. E1m–/+, data not shown), providing further evidence that the observed increase in Gsα expression is from the paternal allele. Along with Gsα overexpression in proximal tubules, 1A+/p– mice also had evidence for increased PTH sensitivity, with lower serum PTH levels, normal serum calcium, and a trend toward lower serum phosphorus (Table 1). In contrast, serum PTH, calcium, and phosphorus were unaffected in 1Am–/+ mice. Neither maternal nor paternal 1A deletion affected Gsα expression in renal inner medulla or liver (Fig. 4 A and B), two tissues where Gsα is normally expressed equally from both parental alleles (10, 14).
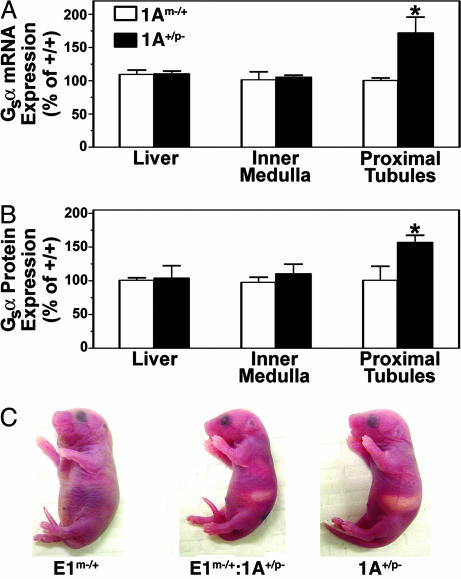
Fig. 4.
Paternal 1A deletion reverses Gsα imprinting and the E1m–/+ neonatal phenotype. (A) Gsα mRNA expression in liver, renal inner medulla, and renal proximal tubules of 1Am–/+ and 1A+/p– mice relative to paired +/+ littermates (n = 5 pairs for all groups, except n = 4 for 1Am–/+ inner medulla). (B) Relative Gsα protein expression in same tissues. (n = 5 pairs for inner medulla and proximal tubules, n = 4 for 1Am–/+ liver, and n = 3 for 1A+/p– liver). Results are mean ± SEM. *P < 0.05 vs. 100% by t test. (C) Representative E1m–/+ (Left), E1m–/+:1A+/p– (Middle), and 1A+/p– (Right) neonates are shown.
Table 1. Serum Ca, phosphorus (Ph), and PTH.
| Genotype | Ca, mg/dl | Ph, mg/dl | PTH, pg/ml |
|---|---|---|---|
| +/+ | 10.1 ± 0.2 | 9.5 ± 0.4 | 157 ± 21 |
| 1A+/p- | 10.0 ± 0.4 | 8.5 ± 0.3 | 51 ± 22* |
| 1Am-/+ | 9.4 ± 0.5 | 9.2 ± 0.5 | 151 ± 50 |
Measurements were done in 3-month-old male mice (n = 10 for +/+, n = 5 for 1A+/p- and 1Am-/+). *, P < 0.05 vs. wild type.
Loss of the maternal, but not paternal, Gnas allele produces a severe phenotype with large bodies and s.c. edema at birth (10, 21, 22). Mice with a maternal Gsα exon 1 deletion (E1m–/+) have the same phenotype (Fig. 4C, 13 of 13 pups), providing further evidence that this phenotype results from loss of Gsα expression from the maternal allele (M.C., unpublished data). This phenotype is absent in 1A– homozygotes and heterozygotes (Fig. 4C). Paternal 1A deletion reversed the early E1m–/+ phenotype in E1m–/+:1A+/p– mice (11 of 11 pups), suggesting that loss of paternal Gsα imprinting in one or more tissues can correct the Gsα deficiency caused by maternal Gsα deletion.
Discussion
Gnas is a unique gene that contains multiple oppositely imprinted promoters within a single transcriptional unit. Here, we show that embryos derived from oocytes lacking Dnmt3L (and, therefore, unable to establish primary maternal methylation imprints) had a paternal-specific epigenotype on both parental alleles at all Gnas DMRs, confirming that Gnas imprinting depends on one or more maternal primary imprint marks. A similar effect on GNAS imprinting was also found in human biparental complete hydatidiform moles (23), in which primary maternal methylation imprints fail to be established by other unknown mechanisms (24). However, unlike the mouse embryos, the moles had a biparental pattern of random and partial methylation of the Gnasxl first exon, reminiscent of the pattern observed in this region in normal mouse blastocysts (6). In contrast, the Nespas/Gnasxl promoter located within the same DMR is unmethylated in both parental alleles in both maternal imprinting defect models, suggesting that the Nespas/Gnasxl DMR undergoes a complicated imprinting process during development in which imprinting of the more upstream promoter is established earlier than the more downstream Gnasxl first exon. This nonuniform imprinting process may explain why the upstream and downstream portions of this DMR do not have the same imprinting pattern in some PHP1B patients (25).
Because the 1A DMR was shown to contain a maternal primary imprint mark (6), we examined the effect of deleting this region in mice. The 1A– homozygotes have no obvious phenotype, providing further evidence that exon 1A-specific mRNAs have no clear function. The 1A DMR deletion had no effect on Nesp or Nespas/Gnasxl imprinting, showing that imprinting of these latter regions does not require the 1A DMR. Similar findings were observed in another 1A DMR-deletion mouse model (26). That Nesp-Nespas/Gnasxl imprints independently of the 1A DMR is further supported by the fact that in many PHP1B patients 1A DMR imprinting is lost without any effect on imprinting of the upstream regions and that 1A DMR, but not Nesp-Nespas/Gnasxl imprinting, appears to depend on a cis-acting element within the closely linked STX16 gene (27). Methylation studies suggest the presence of a second maternal primary imprint mark within the Nespas/Gnasxl promoter (28). A likely scenario is that methylation is first established within this promoter and then spreads into the Gnasxl first exon. Nesp imprinting is established later in development either by the action of paternal antisense Nespas transcripts (29) or other mechanisms (30, 31).
Tissue-specific Gsα imprinting occurs even though its promoter is unmethylated and has histone modifications consistent with open chromatin on both parental alleles (6, 14). Loss of maternal 1A methylation in PHP1B (15) strongly suggests that the 1A DMR controls Gsα imprinting. Tissue-specific Gsα imprinting is not determined by exon 1A promoter activity or by its mRNA transcripts, as exon 1A- and Gsα-specific mRNAs have a similar tissue distribution pattern (6). It is more likely that the 1A DMR harbors cis-acting regulatory elements that are both tissue-specific and methylation-sensitive (Fig. 5). In the model shown in Fig. 5, the 1A DMR contains a silencer that in proximal tubules binds a tissue-specific repressor and inhibits Gsα expression on the paternal allele. DNA methylation on the maternal allele blocks repressor binding, allowing Gsα to be expressed from this allele. In most other tissues, the repressor is not expressed, and therefore, Gsα is expressed biallelically. In PHP1B, loss of maternal 1A DMR methylation allows the repressor to bind to both alleles in proximal tubules, leading to Gsα deficiency and PTH resistance. In most other tissues, the repressor is absent, and therefore, Gsα expression is unaffected. As predicted, 1Am–/+ mice had no changes in Gsα expression because the silencer is inactive on the maternal allele. In contrast, paternal 1A deletion resulted in Gsα overexpression in proximal tubules but not in other tissues where normally Gsα is biallelically expressed, consistent with deletion of a tissue-specific silencer or other negative regulatory element. Our results directly confirm that the 1A DMR controls tissue-specific Gsα imprinting.
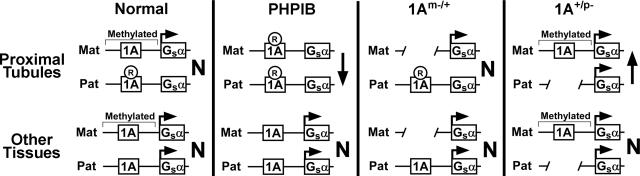
Fig. 5.
Model for role of 1A DMR in tissue-specific Gsα imprinting. The 1A DMR is shown to have a methylation-sensitive silencer that binds a tissue-specific repressor (R). (Upper) In proximal tubules, the repressor normally binds to and suppresses the Gsα promoter on the paternal allele, but it is unable to bind to or suppress Gsα promoter on the methylated maternal allele. (Lower) In most other tissues, the repressor is absent, and therefore, 1A imprinting does not lead affect Gsα expression. In PHP1B, loss of maternal 1A methylation allows the repressor to bind to and suppress Gsα expression from both alleles in proximal tubules, leading to Gsα deficiency (down arrow) and PTH resistance. Gsα expression is unaffected (N) in most other tissues where the repressor is absent. Maternal 1A deletion (1Am–/+) has no effect on Gsα expression. Paternal 1A deletion (1A+/p–) leads to Gsα overexpression in proximal tubules (up arrow) and increased PTH sensitivity but has no effect on Gsα expression in other tissues.
The exact nature of the putative regulatory element within the 1A DMR is unknown. Although methylation-sensitive insulators are important control elements in other imprinted genes (30–32), they are not tissue-specific. Tissue-specific silencers are a strong candidate, because they have been identified in other imprinted genes, including Igf2 and Igf2r (33). Another recent 1A DMR knockout model also provides evidence that this region controls tissue-specific Gsα imprinting (26). Based on the minimal overlap between the two deletions, the elements that regulate Gsα imprinting lie within a 2-kb region (–1.6 to –3.6 kb upstream of the Gsα coding region).
Although heterotrimeric G proteins are not generally considered to be rate-limiting signaling components, Gsα expression and PTH signaling were similarly reduced in proximal tubules by ≈70% in mice with a maternal Gnas mutation (10), suggesting that Gsα is rate-limiting for PTH action in this tissue. Consistent with Gsα being rate-limiting, a modest increase in Gsα expression in proximal tubules of 1A+/p– mice had a significant effect on circulating PTH levels consistent with increased PTH signaling. Gsα may be rate-limiting because it is poorly expressed in these cells (10). 1A+/p– mice will be useful to further examine PTH-Gsα signaling in proximal tubules.
Supplementary Material
Author contributions: J.L., M.C., C.D., J.G.N., B.E., and L.S.W. designed research; J.L., M.C., C.D., J.G.N., and B.E. performed research; C.D., D.B., and T.H.B. contributed new reagents/analytic tools; J.L., D.B., T.H.B., and L.S.W. analyzed data; and J.L. and L.S.W. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Gsα, stimulatory G protein α-subunit; PHP1A/B, pseudohypoparathyroidism types 1A and 1B; PTH, parathyroid hormone; DMR, differentially methylated region.
References
- 1.Reik, W. & Walter, J. (2001) Nat. Rev. Genet. 2, 21–32. [DOI] [PubMed] [Google Scholar]
- 2.Weinstein, L. S., Yu, S., Warner, D. R. & Liu, J. (2001) Endocr. Rev. 22, 675–705. [DOI] [PubMed] [Google Scholar]
- 3.Kelsey, G., Bodle, D., Miller, H. J., Beechey, C. V., Coombes, C., Peters, J. & Williamson, C. M. (1999) Genomics 62, 129–138. [DOI] [PubMed] [Google Scholar]
- 4.Peters, J., Wroe, S. F., Wells, C. A., Miller, H. J., Bodle, D., Beechey, C. V., Williamson, C. M. & Kelsey, G. (1999) Proc. Natl. Acad. Sci. USA 96, 3830–3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayward, B. E., Moran, V., Strain, L. & Bonthron, D. T. (1998) Proc. Natl. Acad. Sci. USA 95, 15475–15480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu, J., Yu, S., Litman, D., Chen, W. & Weinstein, L. S. (2000) Mol. Cell. Biol. 20, 5808–5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wroe, S. F., Kelsey, G., Skinner, J. A., Bodle, D., Ball, S. T., Beechey, C. V., Peters, J. & Williamson, C. M. (2000) Proc. Natl. Acad. Sci. USA 97, 3342–3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayward, B. E. & Bonthron, D. T. (2000) Hum. Mol. Genet. 9, 835–841. [DOI] [PubMed] [Google Scholar]
- 9.Liu, J., Erlichman, B. & Weinstein, L. S. (2003) J. Clin. Endocrinol. Metab. 88, 4336–4341. [DOI] [PubMed] [Google Scholar]
- 10.Yu, S., Yu, D., Lee, E., Eckhaus, M., Lee, R., Corria, Z., Accili, D., Westphal, H. & Weinstein, L. S. (1998) Proc. Natl. Acad. Sci. USA 95, 8715–8720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mantovani, G., Ballare, E., Giammona, E., Beck-Peccoz, P. & Spada, A. (2002) J. Clin. Endocrinol. Metab. 87, 4736–4740. [DOI] [PubMed] [Google Scholar]
- 12.Hayward, B. E., Barlier, A., Korbonits, M., Grossman, A. B., Jacquet, P., Enjalbert, A. & Bonthron, D. T. (2001) J. Clin. Invest. 107, R31–R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Germain-Lee, E. L., Ding, C.-L., Deng, Z., Crane, J. L., Saji, M., Ringel, M. D. & Levine, M. A. (2002) Biochem. Biophys. Res. Commun. 296, 67–72. [DOI] [PubMed] [Google Scholar]
- 14.Sakamoto, A., Liu, J., Greene, A., Chen, M. & Weinstein, L. S. (2004) Hum. Mol. Genet. 15, 819–828. [DOI] [PubMed] [Google Scholar]
- 15.Liu, J., Litman, D., Rosenberg, M. J., Yu, S., Biesecker, L. G. & Weinstein, L. S. (2000) J. Clin. Invest. 106, 1167–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang, X., Li, C., Xu, X. & Deng, C. (1998) Proc. Natl. Acad. Sci. USA 95, 3667–3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng, C., Wynshaw-Boris, A., Zhou, F., Kuo, A. & Leder, P. (1996) Cell 84, 911–921. [DOI] [PubMed] [Google Scholar]
- 18.Xu, X., Li, C., Garrett-Beal, L., Larson, D., Wynshaw-Boris, A. & Deng, C. X. (2001) Genesis 30, 1–6. [DOI] [PubMed] [Google Scholar]
- 19.Bourc'his, D., Xu, G., Lin, C., Bollman, B. & Bestor, T. H. (2001) Science 294, 2536–2539. [DOI] [PubMed] [Google Scholar]
- 20.Simonds, W. F., Goldsmith, P. K., Woodard, C. J., Unson, C. G. & Spiegel, A. M. (1989) FEBS Lett. 249, 189–194. [DOI] [PubMed] [Google Scholar]
- 21.Williamson, C. M., Beechey, C. V., Papworth, D., Wroe, S. F., Wells, C. A., Cobb, L. & Peters, J. (1998) Genet. Res. 72, 255–265. [DOI] [PubMed] [Google Scholar]
- 22.Skinner, J. A., Cattanach, B. M. & Peters, J. (2002) Genomics 80, 373–375. [DOI] [PubMed] [Google Scholar]
- 23.Judson, H., Hayward, B. E., Sheridan, E. & Bonthron, D. T. (2002) Nature 416, 539–542. [DOI] [PubMed] [Google Scholar]
- 24.Hayward, B. E., De Vos, M., Judon, H., Hodge, D., Huntriss, J., Picton, H. M., Sheridan, E. & Bonthron, D. T. (2002) BMC Genet. 4, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, J., Nealon, J. G. & Weinstein, L. S. (2005) Hum. Mol. Genet. 14, 95–102. [DOI] [PubMed] [Google Scholar]
- 26.Williamson, C. M., Ball, S. T., Nottingham, W. T., Skinner, J. A., Plagge, A., Turner, M. D., Powles, N., Hough, T., Papworth, D., Fraser, W. D., et al. (2004) Nat. Genet. 36, 894–899. [DOI] [PubMed] [Google Scholar]
- 27.Bastepe, M., Frohlich, L. F., Hendy, G. N., Indridason, O. S., Josse, R. G., Koshiyama, H., Korkko, J., Nakamoto, J. M., Rosenbloom, A. L., Slyper, A. H., et al. (2003) J. Clin. Invest. 112, 1255–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coombes, C., Arnaud, P., Gordon, E., Dean, W., Coar, E. A., Williamson, C. M., Feil, R., Peters, J. & Kelsey, G. (2003) Mol. Cell. Biol. 23, 5475–5488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wutz, A. & Barlow, D. P. (1998) Nature 389, 745–749. [DOI] [PubMed] [Google Scholar]
- 30.Bell, A. C. & Felsenfeld, G. (2000) Nature 405, 482–485. [DOI] [PubMed] [Google Scholar]
- 31.Hark, A. T., Schoenherr, C. J., Katz, D. J., Ingram, R. S., Levorse, J. M. & Tilghman, S. M. (2000) Nature 405, 486–489. [DOI] [PubMed] [Google Scholar]
- 32.Chao, W., Huynh, K. D., Spencer, R. J., Davidow, L. S. & Lee, J. T. (2002) Science 295, 345–347. [DOI] [PubMed] [Google Scholar]
- 33.Ferguson-Smith, A. C. (2000) Curr. Biol. 10, R872–R875. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.