Abstract
Much of the success of Plasmodium falciparum in establishing persistent infections is attributed to immune evasion through antigenic variation. This process involves periodically exchanging variants of the major surface antigen PfEMP1, a protein also responsible for parasite cytoadherence. PfEMP1 is encoded by genes of the 60-member var family, located at subtelomeric and internal chromosome loci. The active or silenced state of var genes is heritable, and its control by nonsequence information remains puzzling. Using FISH analysis, we demonstrate that both internal and subtelomeric var genes are positioned at the nuclear periphery in their repressed state. Upon activation, the same var genes are still found in the periphery, indicating that this zone can be transcriptionally competent, rather than uniformly silenced. However, activation of a var gene is linked with altered positioning at the nuclear periphery, with subtelomeric var loci exiting chromosome end clusters and being relocated to distinct nuclear sites. Serial sectioning of parasite nuclei reveals areas of both condensed and noncondensed chromatin at the nuclear periphery. Our results demonstrate that regulation of antigenic variation is associated with subnuclear position effects and point to the existence of transcriptionally permissive perinuclear zones for var genes.
Keywords: malaria, epigenetic, telomere, nucleus
The subtelomeric positioning of genes involved in host–pathogen interactions is a feature common to many parasitic eukaryotes. The phenomenon is found in organisms as evolutionarily diverse as diplomonads (Giardia), pathogenic fungi (Pneumocystis, Candida, and others), trypanosomatids (Trypanosoma brucei), and throughout the phylum Apicomplexa (Plasmodium spp., Toxoplasma, and Theileria). The subtelomeric grouping of virulence factors in such widely divergent groups suggests a convergent evolution of a feature whose significance is still puzzling. Suggested functions include control of antigen expression through telomeric silencing effects (1), expansion of virulence factor repertoire through duplication, and gene conversion after recombination between heterogeneously paired telomeres (2). In some of these organisms, subtelomerically encoded antigens are expressed in a mutually exclusive manner, with only a single representative expressed in any given individual. This phenomenon often is interpreted as a means of limiting unnecessary exposure of antigens to the immune system to prolong the course of an infection, thereby maximizing transmission.
In Plasmodium falciparum, at least three distinct gene families are encoded at subtelomeric loci: the vars, rifins, and stevors (although very little is known about the role or behavior of rifins and stevors). The var gene family, consisting of ≈60 representatives per haploid genome, is normally found immediately adjacent to the subtelomeric repeats and in groups at internal chromosome positions. Expression of var genes is monoallelic: individuals within populations generally transcribe one dominant var gene with other alleles excluded from expression (3, 4). Very low expression of other var genes, often detectable only by RT-PCR, has been reported. However, in Northern blots, only a single dominant transcript is observed, and only one resultant protein is detectable (4, 5). Genetic rearrangements are not necessary for silencing (3), and var promoters artificially removed from chromosomal context are activated by default (6), both indicative of epigenetic var control mechanisms.
The subtelomeric positioning of many var genes is suggestive of a system of epigenetic silencing reminiscent of the telomere position effect (TPE) originally characterized for the genes inserted at the subtelomeres of the budding yeast Saccharomyces cerevisiae (7, 8) and since described in other eukaryotes. In yeast, a large multiprotein complex (consisting of Rap1p, the Ku complex, Sir proteins, and Rif proteins) anchors telomere ends to nuclear pores and, in an independent activity, initiates condensation of local chromatin through the enzymatic modification of exposed histone tails. The proximity of the subtelomeres to the nuclear periphery is thought to assist their silencing; nontelomeric regions with heterologous silencing elements artificially tethered to the nuclear periphery are silenced in S. cerevisiae (9), whereas active subtelomeric genes are more likely to leave the periphery (10). We previously identified homologues of several yeast telomere silencing factors in the P. falciparum genome (11) and have recently shown that one of these silencing factors, PfSir2, interacts with inactive subtelomeric var genes but not with active var genes (12). Other published experiments demonstrate relaxed transcription from subtelomeric loci after deletions of the subtelomeric repeat structures, indicating a role for these elements in the silencing process (13, 14). However, almost nothing is known about how internal var genes are silenced.
An equally puzzling question in epigenetic var control is how one var gene can be highly expressed, whereas the others are silenced. Recent work on African trypanosomes (T. brucei) has put forward a model that might explain how activity could be restricted to one gene at a time. In this hypothesis, active genes transcribing variable surface antigen enjoy a privileged subnuclear location from which silenced variable surface glycoprotein genes are excluded, precluding their transcription (15). In this work, we investigated the nuclear position of var genes located in subtelomeric and chromosome internal positions. Both subtelomeric and internal var genes appear to be under a default epigenetic repression because of their position in the nuclear periphery, which is the preferential location of heterochromatin and silencing factors such as Sir2 (12). Upon activation, we observed that a subtelomere-associated var gene, although remaining in a perinuclear position, moves out of the telomeric cluster. Reconstruction of serially sectioned parasite nuclei revealed that the Plasmodium nuclear periphery consists of condensed chromatin material with one or more gaps of noncondensed chromatin. Such heterogeneity may be associated with zones with differential competency for transcription.
Materials and Methods
Parasites. Parasites were cultured by using the method established by Trager and Jensen (16) and gassed with a mixture of 5% CO2/1% O2/94% N2. Parasite strains used were the FCR3-C1 clone, a parasite from the FCR3 background with a disruption in the var1csa gene (17). This clone was panned on CHO cells expressing chondroitin sulfate A (CSA) by using the method described in ref. 3. Parasites were assayed (by FISH analysis or Northern blotting) within four to eight generations of panning. Dd2 parasites were a gift from Tom Wellems (National Institutes of Health, Bethesda).
Northern Blotting. Total RNA was prepared from synchronized parasites 12–16 h after invasion (ring) and 22–30 h after invasion (trophozoites). RNA was extracted with TRIzol (Invitrogen), and Northern blots were prepared as described in ref. 18. The var2csa probe consisted of the first 2 kb of the var2csa ORF from the FCR3 strain. The primers used to amplify the probe were 5′-AGCTGATCCTAGTGAAGTGG-3′ and 5′-TGAAGTATCTTGTTCAGCGG-3′. The exon 2 probe corresponds to bases 7930–9147 of varT11-1 (GenBank accession no. U67959). The var7 probe consisted of the first 2 kb of the var7 ORF from the parental Dd2 strain. The primers used to amplify the probe were 5′-TGTAAAAGAATATTATGAGCGTG-3′ and 5′-AGGTCAAGAACTCCATAGGGAC-3′.
FISH. FISH was conducted on air-dried infected red blood cells fixed with 4% paraformaldehyde. Red blood cells were washed in PBS, air-dried on a slide for >30 min, and then fixed in 4% paraformaldehyde solution for 15 min. Preparations were washed again in PBS and then hybridized with heat-denatured probe under a sealed rubber frame at 92°C for 3 min and at 37°C for 12 h. The probes for the var2csa and var7 genes were derived from genomic DNA by using the same primers detailed above. The probe adjacent (3 kb upstream) to the central var cluster (referred to as var-pfl0935c) was produced by using the primers 5′-TCCATGAATTTTCATCACATG-3′ and 5′-TGAAATTATTTTGTGGAGGC-3′. The internal non-var probe pfl0330c was produced by using the primers 5′-TGGTAATAAAAGATTAGAATTAGCTG-3′ and 5′-TTTTCCTACTGTCATACGAG-3′. The hybridization solution contained 50% formamide, 10% dextran sulfate, 1× SSC, 250 μg/ml herring sperm DNA, and ≈100 ng of dsDNA probe labeled by using fluorescein high-prime(Roche Applied Sciences). After hybridization, parasites were washed twice in 50% formamide/2× SSC at 50°C, once in 2× SSC at 37°C, and once in 4× SSC at room temperature. Parasites were finally washed in a solution of 100 mM Tris·HCl/150 mM NaCl/0.5% (vol/vol) Tween 20, mounted in Vectashield (Vector Laboratories), and imaged. Images were analyzed for intranuclear position of signals as described in ref. 13.
EM. Red blood cells containing nonsynchronized parasites were fixed in 1% glutaraldehyde in RPMI medium 1640/Hepes for 1 h at 4°C. Parasites were embedded in LR Gold resin (Electron Microscopy Sciences, Fort Washington, PA) and series of ultrathin sections were cut. Sections were poststained with uranyl acetate and lead citrate and observed by using a transmission electron microscope (CM120 BioTWIN, Philips) at 80 kV.
Mapping. To confirm that no large-scale rearrangements occurred that effected the analyzed var clusters, we mapped the probe for the var2csa and the probe adjacent to the central var cluster (var-pfl0935c) by hybridizing to pulsed-field gel electrophoresis-separated chromosomes. This hybridization confirmed that the target sequences remain on chromosome 12 (data not shown). The var2csa gene is highly conserved between isolates and is assumed to retain its subtelomeric position (19). var7 has been previously mapped in Dd2 (20). The control housekeeping gene pfl0330c (for RNA polymerase III subunit) is assumed to retain the same position as in 3D7. Map positions are given as in 3D7 but may be slightly modified in FCR3 subtelomeric positions.
Results
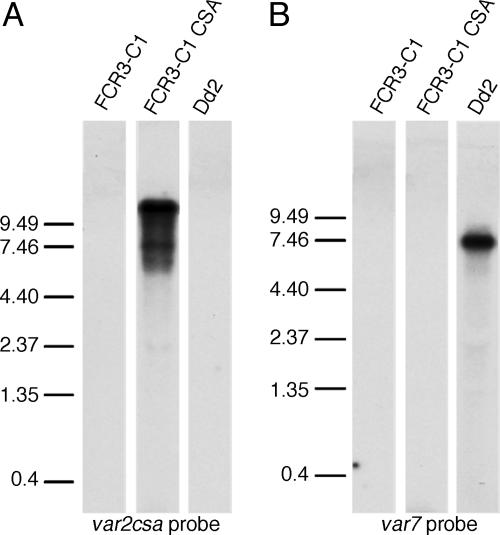
var Gene Transcription from Subtelomeric and Chromosome-Internal Loci. To examine var genes in “on” and “off” states, we used P. falciparum parasite lines in which the var2csa gene was differentially active. To avoid potential interference from the var1csa (previously called varCSA) gene, which appears to be constitutively transcribed among different populations (18), we worked with FCR3 parasites with a disrupted var1csa, called FCR3 C1 (17). These parasites do not initially adhere to CSA, but after repeated panning, they are able to adhere specifically to CSA. This clone is known as FCR3 C1CSA. In FCR3 C1CSA parasites, Northern blots of total RNA using a universal var gene probe detected transcriptional activation of one dominant var gene, and a single PfEMP1 is detected by surface iodination (17). Wild-type parasites have been reported to up-regulate the subtelomeric var2csa gene in CSA-binding parasites in a mutually exclusive fashion (21). We hypothesized that this same var gene (designated PFL0030c in the fully sequenced 3D7 strain) was up-regulated in FCR3 C1CSA. To test this hypothesis, we made a probe specific to the start of the var2csa gene. This probe recognized a strongly transcribed band at 9.2 kb in the FCR3 C1CSA clone, corresponding to the expected size of the var2csa transcript but no product in the FCR3 C1 clone (Fig. 1). Consistent with the typical profile of expressed var genes, this gene was up-regulated in ring-stage parasites, then almost disappeared in trophozoite-stage parasites (data not shown). The same membrane also was hybridized with a probe made against the exon 2 of varT11-1, which contains sequence semiconserved between var genes. This probe detected additional var bands in the unpanned parasites but no additional var genes to the var2csa-specific probe in the FCR3 C1CSA parasites (data not shown). These data demonstrate that var2csa is actively transcribed in FCR3 C1CSA parasites but silenced in the genetically identical FCR3 C1 parasites. It is possible that additional genes may be transcribed in the FCR3 C1CSA parasites below levels detectable by Northern blotting.
Fig. 1.
Northern blots of var gene transcription. (A) Parasites were probed with a 2-kb fragment specific to the 5′ end of the var2csa gene. In the Dd2 strain and in unpanned FCR3 C1 parasites, no signal was detected even after overexposure. In the CSA-panned FCR3 C1CSA line, a strong 9.5- to 10-kb signal was detected, consistent with the predicted size of the mature var2csa transcript. (B) Parasites were probed with a 2-kb fragment specific to the 5′ end of the var7 gene. In both the CSA-panned and nonpanned C1 parasites, no signal was detected even after overexposure, whereas in the Dd2 strain, a strong 7.5- to 8-kb signal was detected, consistent with the predicted size of the var7 gene.
To characterize an expressed var gene encoded at a chromosomal internal position, we examined the var7 gene. This gene has previously been described as the dominant var gene transcribed in Dd2 parasites (6, 22). We used a probe specific to the 5′ end of the var7 ORF to probe Dd2 parasites and found a strong band of the expected size for var7, 7.3 kb (Fig. 1). Again, the profile of expression was that typical of active var genes, with strong expression in ring stages and almost no expression in trophozoite stages (data not shown). No var2csa transcript was detected in Dd2 parasites, nor was var7 detected in FCR3 C1 or FCR3 C1CSA parasites.
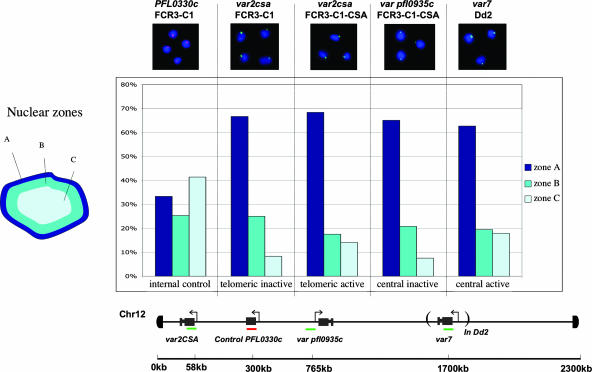
Subnuclear Localization of Internal and Subtelomeric var Genes. FISH using probes specific to var genes allowed their localization in asexual intraerythrocytic parasites. Synchronized late-ring-stage parasites were labeled with specific probes, and the location of the probe was scored in relation to the parasite nuclear periphery (Fig. 2) using the imagej pointpicker software (http://rsb.info.nih.gov/ij) tool. Fluorescent foci were scored for at least 50 cells and at least three independent hybridizations for each probe and were sorted as being localized into one of three concentric nuclear zones of equal area. A control gene from a chromosome internal position on chromosome 2 (which lacks internal var genes) was randomly distributed among the three zones (Fig. 2). However, the subtelomeric and chromosome internal var genes were found to be mainly localized at the nuclear periphery (zone A) in a distribution notably different from that expected from a random distribution (Fig. 2). These data indicate that subtelomeric var genes are specifically associated with the periphery of the nucleus, an unsurprising finding given that Plasmodium telomere ends have previously been shown to be nuclear peripheral (2). However, internal var genes are >500 kb away from the closest telomere, a distance sufficient to traverse the nucleus many times over in a noncompacted DNA configuration. This fact suggests that internal var gene loci are physically looped back to the nuclear periphery with intervening stretches probably found in more central nuclear regions.
Fig. 2.
FISH localization of var genes. var genes positioned at subtelomeric and chromosome internal loci were localized both in parasites where they were actively transcribed and in parasites where they were silenced. Fluorescent foci were scored for localization in three concentric zones of equal area. An internally coded control is randomly distributed among all three zones, whereas var genes are preferentially positioned in the peripheral zone. Like subtelomeric var genes, internal var genes also associate with the nuclear periphery. No differences are observed in this assay between active and inactive var genes. Map distances from the start of the chromosome 12 are inferred from the 3D7 genome.
The distribution in zones A–C seen for active var genes was not distinguishable from that seen for the inactive var genes, i.e., exclusion from the nuclear core (zones B and C) and association with the nuclear periphery (zone A). These data indicate that repositioning to a nuclear-internal environment is not required for var gene transcription, and that sectors of the nuclear periphery are transcriptionally competent.
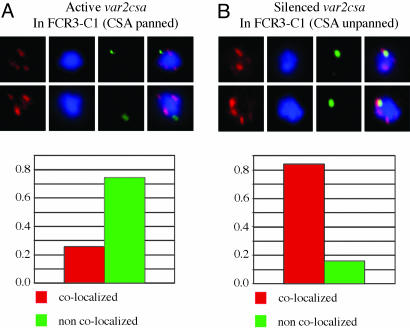
Colocalization Between var Genes and Telomeric Clusters. To test whether there was a relationship between the transcription state of var genes and their localization with telomeric clusters (which are silencing zones in yeast), we performed two-color FISH with different var gene probes and telomeric probes. Parasites were synchronized and processed at late ring stage, the stage with the peak of var gene expression. Two different probes were used for telomeric clusters, one to the telomeric repeat structure and the other to the telomeric proximal rep20 repeat region. Both probes gave the same localization results. Double labeling with the specific var probes allowed scoring of colocalization, with colocalization defined as any overlap between the fluorescent foci. In all cases, >50 nuclei were scored from experimental replicates. In the case of the unpanned FCR3 C1 parasites, a strong association was seen between the inactive var2csa and telomeric clusters, with colocalization occurring in 84% of parasites (Fig. 3), However, in FCR3 C1CSA parasites, the actively transcribed var2csa gene was dissociated from telomeric clusters, colocalizing in only 26% of parasites (Fig. 3). In many cases, the active var2sa was located 100–300 nm from the nearest telomeric cluster. Although the active var2csa dissociated from the telomeric clusters, var2csa probes still showed the gene to be associated with the nuclear periphery (Fig. 2). Both active and inactive central var genes also were scored for colocalization. These genes showed an infrequent, random association with the telomeric clusters. No difference was observed between the active and inactive central var genes (data not shown).
Fig. 3.
FISH colocalization between var genes and telomeric clusters. The localization of telomeric clusters (red probe) was compared with that of a var gene (green probe). (A) An actively transcribed var gene (var2csa) is compared with telomeric clusters. (B) The same var gene is in a silenced state. In parasites expressing var2csa, the gene is mostly physically separated from telomeric clusters, with distinct fluorescent foci clearly discernible. However, in genetically identical parasites with var2csa silenced, the var2csa gene colocalizes with the telomeric clusters.
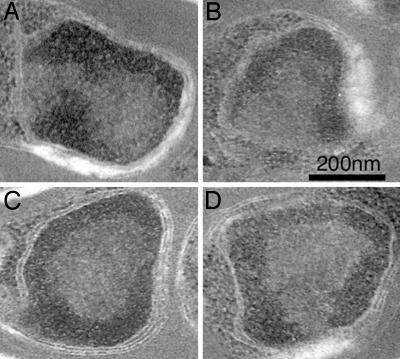
EM of Nuclear Ultrastructure. Because both active and inactive var genes are found in the nuclear periphery, we sought to characterize the ultrastructure of this subnuclear region by using EM. Mixed-stage preparations of P. falciparum were fixed, serially sectioned, and poststained. EM revealed that the periphery of P. falciparum nuclei consists of electron-dense, heterochromatin-like material. In ring-stage parasites, this zone is variegated in appearance, becoming more distinct and forming well defined boundaries in schizonts and merozoites. This material extended ≈50 nm from the nuclear membrane and was of approximately the same electron density throughout. Although the condensed region sometimes extended inwards toward the center of the nucleus (Fig. 4 A and B), it was generally of a uniform diameter (Fig. 4 C and D). The density also was consistent within each nucleus, although the region was denser in postsegmented schizonts than in other stages. It is unclear whether differences between early- and late-stage parasites reflect a biological difference or are attributable to differential fixation. Reconstruction after serial sectioning showed that some surface of the inner nuclear membrane was always bordered by an electron-sparse region, consistent with less condensed genetic material or euchromatin. The patches varied in size from ≈100 to 300 nm in length, but at least one was present in of all of the >20 parasite nuclei examined by serial sectioning.
Fig. 4.
Nuclear ultrastructure. Regions of condensed and relaxed genetic material at the nuclear periphery. Parasite nuclei from mixed intraerythrocytic cultures were serial-sectioned to examine patterns of nuclear condensation. Nuclei shown here are from segmented schizonts. Nuclei contained a region of electron-dense material at the periphery, consistent with heterochromatic material. Although this region spread over most of the internal surface of the nuclear membrane, in each nucleus examined, there was at least one clear region, free of heterochromatin somewhere at the nuclear periphery. This observation is consistent with the presence of transcriptionally active and inactive zones at the nuclear periphery.
Discussion
The var family, responsible for antigenic variation in P. falciparum, is concentrated in groups at chromosomally central and subtelomeric positions. Only one var gene in an individual is strongly transcribed, whereas most var genes are silenced, evocative of TPE silencing. TPE is best characterized in S. cerevisiae, but variants have been described in Drosophila (23), humans (24), and Giardia (25), implying a very ancient eukaryotic origin. In S. cerevisiae, TPE is associated with localization to the nuclear periphery and also with telomeric clustering. Whereas individual telomeres can bind some of the silencing factors, clusters appear to create a threshold concentration of the silent information regulator (SIR) complex that allows effective silencing (26, 27). Although it has been best described in modified chromosome systems, TPE is likely involved the silencing of several multigene families at yeast subtelomeres. Plasmodium telomeres have previously been shown to form clusters (2), and probably all 28 P. falciparum subtelomeres in natural isolates encode var genes. However, coordinated regulation of mutually exclusive expression of var genes by using only TPE is inconceivable. In this study, we have examined whether silencing of var genes is determined by their relationship with telomeric clusters. For subtelomeric var genes, we have shown that a silenced locus is physically associated with the telomeric cluster, whereas the same locus in an active state dissociates from the cluster (Fig. 3). TPE has previously been proposed as a means of repressing transcription of subtelomeric virulence factors (1, 28); we show that derepression is linked to exit from telomere clusters (Fig. 3).
Movement of active var gene loci away from telomere clusters could serve a twofold purpose. First, dissociation from the cluster physically separates the locus from the local concentration of silencing proteins found at telomeric clusters. Consistent with this interpretation, we have recently shown that telomeres and inactive subtelomeric var gene promoters associate with the important PfSir2 silencing protein, whereas the promoters of active var genes are devoid of PfSir2 (12). This absence would very likely have implications for the local chromatin structure, and indeed we have shown that the histones of inactive and active var genes are differentially acetylated (12). Experiments in yeast (29, 30), Drosophila (31), and mammalian systems (32) demonstrate that genes can be recruited from one area of the nucleus to another to be repressed. These recruitments are sometimes linked to chromatin modification, and this also may be the case with var genes. The second purpose of var locus movement may be to bring the promoter into closer association with factories of transcriptional machinery (33) that could be immobilized to nucleoskeletal fibers (34, 35).
The concept of var gene activation through association with factories of transcription is congruent with our current understanding of antigenic variation in T. brucei. In T. brucei, expression of the variable surface glycoproteins are monoallelic. Variable surface glycoprotein is transcribed by RNA polymerase I rather than the expected RNA polymerase II that normally produces messenger RNA (36). Molecular and cell biology experiments have now shown that the active locus is associated with an extranucleolar focus of RNA polymerase I, whereas nonactive loci are excluded from this area, which the authors call an “expression-site body” (15). Whether the Trypanosoma expression-site body is a discrete structure or merely a localized concentration of polymerase, it may be a useful analogy for var gene transcription, with actively transcribed genes dynamically associating with this transcription-competent area.
Our EM of serial sections demonstrates that, like other eukaryotes, P. falciparum has defined areas of euchromatin and heterochromatin-like material (Fig. 4). Although the nuclear periphery is largely bounded by electron-dense zones, most prominently in schizonts and merozoites, the condensed material does not insulate the entire perinuclear area. At least one small gap is present at the periphery of the nucleus that is free of heterochromatin (Fig. 4). It is striking that such an area is found in all nuclei that were serially sectioned. This area of perinuclear euchromatin may correspond to a transcriptionally competent zone. We have no evidence for the localization of genes in this zone, but we hypothesize that active var genes may preferentially enter such euchromatic regions at the nuclear periphery (Fig. 5). In such a model, localization could provide the context for the decondensed chromatin state of the locus during S phase, allowing epigenetic inheritance of var activity, and may be a region of stronger transcription in ring stages. The existence of transcriptional foci at the nuclear periphery is supported by an elegant study by Duraisingh and colleagues (37), who show preferential colocalization at the nuclear periphery of two actively transcribed reporter genes located at Plasmodium subtelomeres. The authors also show that a similar reporter gene can colocalize with active var genes. This colocalization suggests that any privileged perinuclear position enjoyed by active var genes does not necessarily exclude other active non-var genes.
Fig. 5.
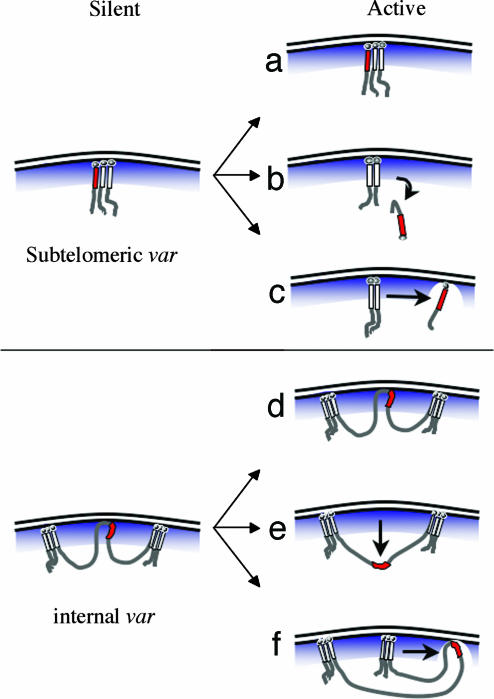
A model for var gene silencing and activation. Inactive subtelomeric var gene loci colocalize with telomeric clusters, reminiscent of cluster-associated silencing in yeast. Several possibilities exist for the mechanism of activation. (a) No positional movement takes place, and other molecular factors determine activation. (b) A var gene is activated after moving away from the silencing effects of the nuclear periphery. (c) The active var gene stays in the nuclear periphery but moves away from telomeric clusters. Our data are inconsistent with possibilities a and b and support option c. The dissociation of the active var gene suggests either a relaxation of the intervening DNA/chromatin structure or exclusion of a single chromosome end from the silencing cluster. This dissociation may allow the movement of the gene from a zone of restricted transcription to a transcriptionally permissive territory. Like subtelomeric var genes, those encoded on internal chromosome positions appear to be silenced by factors positioned at the nuclear periphery. (d–f) Actively transcribed internal var genes could remain in the same position (d), loop out of the periphery to the nuclear core (e), or move to a distinct location within the nuclear periphery (f). Our data are not consistent with e but do not allow us to discern whether internally encoded var genes move within the nuclear periphery. This model does not explain how adjacent var genes on the same chromosome are differentially activated, but an attractive possibility is that chromatin barrier elements (located either between var genes or in var introns) interact with nuclear pores to maintain distinct chromatin states for neighboring genes.
It is unclear whether the delocalization of active var genes from telomere clusters seen by FISH corresponds to a looping of the specific locus or movement of the whole subtelomere to another region. The latter explanation is difficult to exclude, as the weak FISH signal of a single telomere end might be undetectable, although this option would not explain how multiple var genes at the same subtelomere are differentially controlled. Either way, it would be interesting to relate localization of telomeres and var loci to areas of nascent RNA production in P. falciparum, although the necessary incorporation methods are yet to be established.
The lack of colocalization between central var genes and telomeric clusters (data not shown) indicates that additional factors are responsible for silencing these loci. The close association between internal var loci and the nuclear periphery implies that protein factors attach these areas to the inside of the nuclear envelope. Sequence elements common to the 5′ UTRs of internal var genes but absent in subtelomeric var genes (38) may mediate such regulation, and indeed, proteins that bind differentially to subtelomeric and internal var promoters have been described (39). Peripheral silencing mechanisms for internal chromosome genes also have been induced (albeit artificially) in yeast (9). The activation of internal var genes, like subtelomeric var genes, could be associated with nuclear peripheral movements, but we currently lack markers to test this hypothesis.
The only other studies to specifically address var gene silencing have focused on the conserved introns of var genes, which are similarly sized and contain an A+T-rich repeat structure, suggesting that the intron may play some role of its own. Indeed, a reporter gene driven by a var 5′ UTR could be silenced when flanked by a matching var intron (40). This silencing was accomplished only when parasites bearing the plasmid had passed through S phase (40). S-phase processes have been proposed to play a role in some histone modifications and transcriptional silencing, but these functions remain controversial (41). A region within var introns also appears to possess promoter activity, possible generating smaller RNA molecules that may be involved in laying down the chromatin code (42). The same var introns are predicted to bind AT-hook domain proteins (42), which have been implicated in modulation of chromatin structure and intranuclear attachments (43), including tethering to the nuclear periphery (44). Such an attachment may be responsible for the common peripheral location that we find for internal and subtelomeric var genes (Fig. 2). It is noteworthy that one var gene (var1CSA) with an anomalous intron is apparently unable to be silenced (18), supporting a role for the var intron in silencing.
One of the major questions unanswered by our model (Fig. 5) for var activation is how adjacent var genes only 10 kb apart in a subtelomeric group can be differentially activated. An answer might be found in the var introns, which are predicted to contain chromatin-spreading boundaries (42), elements that can buffer adjacent genes, maintaining one region in a condensed conformation while the neighboring area is decondensed. Insulators of heterochromatin spread (also called barrier activity) are poorly understood, but an important recent report indicates that some barrier activity requires physical interactions with the nuclear pore complex at the nuclear periphery (45). Perhaps adjacent var genes may be kept differentially active by a similar mechanism. Interactions with nuclear pores might explain the localization of active var genes, which may remain at the nuclear periphery to maintain the chromatin boundary that keeps the adjacent var genes silenced.
Conclusion
Of the subtelomeric and internal var genes studied, all were preferentially found at the nuclear periphery, irrespective of activity. This localization points to distinct silencing and activation domains at the nuclear periphery. For subtelomeric var genes, a mechanism akin to TPE likely creates a default transcriptional suppression that can be modified by additional factors. We show that subtelomeric genes move out of physical telomere clusters to be transcribed, whereas additional markers are needed to determine whether comparable movements are associated with activation of internal var genes. The nuclear periphery often is characterized as a transcriptionally silent territory; our data indicate that it also contains areas of active transcription. It is not possible to determine whether the movements we observed are a consequence of their derepression or are, in fact, the cause of activation. Either way, such movements are likely to be accompanied by chromatin modifications in the surrounding chromosomal areas. Future studies should address the protein complexes that tether subtelomeric and internal var clusters to the nuclear periphery as well as the enzymes and possible RNA species that control the modification of chromatin. In addition, the genetic elements that are responsible for peripheral tethering and silencing (probably var promoters and/or var introns) deserve further attention. The regions flanking the var2csa gene, for example, are distinct from those of other subtelomeric var genes and are different again from those of central var genes. It remains to be shown which factors are common to all var genes and which factors are specific to subclasses. The possible existence of a physical transcription factory at the nuclear periphery also needs closer investigation, possibly through localization of elements of the transcription machinery in living cells.
Acknowledgments
We thank Freddy Frischknecht and Frank Feuerbach for critical reading of the manuscript, Sebastian Ulbert for helpful discussion, Michael Lanzer (University of Heidelberg) for the C1 parasites, and Tom Wellems (National Institutes of Health) for Dd2 parasites. This work was supported by Australian National Health and Medical Research Council C. J. Martin Fellowship 251775 (to S.A.R.) and partially supported by European Union Sixth Framework Programme BioMalPar Grant LSHPCT-2004-503578.
Author contributions: S.A.R. and A.S. designed research; S.A.R. and C.S.-B. performed research; S.A.R., C.S.-B., and A.S. analyzed data; and S.A.R. and A.S. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TPE, telomere position effect; CSA, chondroitin sulfate A.
References
- 1.De Las Penas, A., Pan, S. J., Castano, I., Alder, J., Cregg, R. & Cormack, B. P. (2003) Genes Dev. 17, 2245–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freitas-Junior, L. H., Bottius, E., Pirrit, L. A., Deitsch, K. W., Scheidig, C., Guinet, F., Nehrbass, U., Wellems, T. E. & Scherf, A. (2000) Nature 407, 1018–1022. [DOI] [PubMed] [Google Scholar]
- 3.Scherf, A., Hernandez-Rivas, R., Buffet, P., Bottius, E., Benatar, C., Pouvelle, B., Gysin, J. & Lanzer, M. (1998) EMBO J. 17, 5418–5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, Q., Fernandez, V., Sundstrom, A., Schlichtherle, M., Datta, S., Hagblom, P. & Wahlgren, M. (1998) Nature 394, 392–395. [DOI] [PubMed] [Google Scholar]
- 5.Fernandez, V., Chen, Q., Sundstrom, A., Scherf, A., Hagblom, P. & Wahlgren, M. (2002) Mol. Biochem. Parasitol. 121, 195–203. [DOI] [PubMed] [Google Scholar]
- 6.Deitsch, K. W., del Pinal, A. & Wellems, T. E. (1999) Mol. Biochem. Parasitol. 101, 107–116. [DOI] [PubMed] [Google Scholar]
- 7.Sandell, L. L. & Zakian, V. A. (1992) Trends Cell Biol. 2, 10–14. [DOI] [PubMed] [Google Scholar]
- 8.Gottschling, D. E., Aparicio, O. M., Billington, B. L. & Zakian, V. A. (1990) Cell 63, 751–762. [DOI] [PubMed] [Google Scholar]
- 9.Andrulis, E. D., Neiman, A. M., Zappulla, D. C. & Sternglanz, R. (1998) Nature 394, 592–595. [DOI] [PubMed] [Google Scholar]
- 10.Tham, W. H., Wyithe, J. S., Ko Ferrigno, P., Silver, P. A. & Zakian, V. A. (2001) Mol. Cell 8, 189–199. [DOI] [PubMed] [Google Scholar]
- 11.Scherf, A., Figueiredo, L. M. & Freitas-Junior, L. H. (2001) Curr. Opin. Microbiol. 4, 409–414. [DOI] [PubMed] [Google Scholar]
- 12.Freitas-Junior, L. H., Hernandez-Rivas, R., Ralph, S. A., Montiel-Condado, D., Ruvalcaba-Salazar, O. K., Rojas-Meza, A. P., Māncio-Silva, L., Leal-Silvestre, R. S. & Scherf, A. (2005) Cell, in press. [DOI] [PubMed]
- 13.Figueiredo, L. M., Freitas-Junior, L. H., Bottius, E., Olivo-Marin, J. C. & Scherf, A. (2002) EMBO J. 21, 815–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horrocks, P., Kyes, S., Pinches, R., Christodoulou, Z. & Newbold, C. (2004) Mol. Biochem. Parasitol. 134, 193–199. [DOI] [PubMed] [Google Scholar]
- 15.Navarro, M. & Gull, K. (2001) Nature 414, 759–763. [DOI] [PubMed] [Google Scholar]
- 16.Trager, W. & Jensen, J. (1976) Science 193, 673–675. [DOI] [PubMed] [Google Scholar]
- 17.Andrews, K. T., Pirrit, L. A., Przyborski, J. M., Sanchez, C. P., Sterkers, Y., Ricken, S., Wickert, H., Lepolard, C., Avril, M., Scherf, A., et al. (2003) Mol. Microbiol. 49, 655–669. [DOI] [PubMed] [Google Scholar]
- 18.Kyes, S. A., Christodoulou, Z., Raza, A., Horrocks, P., Pinches, R., Rowe, J. A. & Newbold, C. I. (2003) Mol. Microbiol. 48, 1339–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kraemer, S. M. & Smith, J. D. (2003) Mol. Microbiol. 50, 1527–1538. [DOI] [PubMed] [Google Scholar]
- 20.Guinet, F. & Wellems, T. E. (1997) Mol. Biochem. Parasitol. 90, 343–346. [DOI] [PubMed] [Google Scholar]
- 21.Salanti, A., Staalsoe, T., Lavstsen, T., Jensen, A. T., Sowa, M. P., Arnot, D. E., Hviid, L. & Theander, T. G. (2003) Mol. Microbiol. 49, 179–191. [DOI] [PubMed] [Google Scholar]
- 22.Su, X. Z., Heatwole, V. M., Wertheimer, S. P., Guinet, F., Herrfeldt, J. A., Peterson, D. S., Ravetch, J. A. & Wellems, T. E. (1995) Cell 82, 89–100. [DOI] [PubMed] [Google Scholar]
- 23.Weiler, K. S. & Wakimoto, B. T. (1995) Annu. Rev. Genet. 29, 577–605. [DOI] [PubMed] [Google Scholar]
- 24.Baur, J. A., Zou, Y., Shay, J. W. & Wright, W. E. (2001) Science 292, 2075–2077. [DOI] [PubMed] [Google Scholar]
- 25.Upcroft, P., Chen, N. & Upcroft, J. A. (1997) Genome Res. 7, 37–46. [DOI] [PubMed] [Google Scholar]
- 26.Maillet, L., Boscheron, C., Gotta, M., Marcand, S., Gilson, E. & Gasser, S. M. (1996) Genes Dev. 10, 1796–1811. [DOI] [PubMed] [Google Scholar]
- 27.Gotta, M., Laroche, T., Formenton, A., Maillet, L., Scherthan, H. & Gasser, S. M. (1996) J. Cell Biol. 134, 1349–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barry, J. D., Ginger, M. L., Burton, P. & McCulloch, R. (2003) Int. J. Parasitol. 33, 29–45. [DOI] [PubMed] [Google Scholar]
- 29.Feuerbach, F., Galy, V., Trelles-Sticken, E., Fromont-Racine, M., Jacquier, A., Gilson, E., Olivo-Marin, J. C., Scherthan, H. & Nehrbass, U. (2002) Nat. Cell Biol. 4, 214–221. [DOI] [PubMed] [Google Scholar]
- 30.Hediger, F., Neumann, F. R., Van Houwe, G., Dubrana, K. & Gasser, S. M. (2002) Curr. Biol. 12, 2076–2089. [DOI] [PubMed] [Google Scholar]
- 31.Csink, A. K. & Henikoff, S. (1996) Nature 381, 529–531. [DOI] [PubMed] [Google Scholar]
- 32.Brown, K. E., Baxter, J., Graf, D., Merkenschlager, M. & Fisher, A. G. (1999) Mol. Cell 3, 207–217. [DOI] [PubMed] [Google Scholar]
- 33.Maniatis, T. & Reed, R. (2002) Nature 416, 499–506. [DOI] [PubMed] [Google Scholar]
- 34.Hozak, P., Hassan, A. B., Jackson, D. A. & Cook, P. R. (1993) Cell 73, 361–373. [DOI] [PubMed] [Google Scholar]
- 35.Cook, P. R. (1999) Science 284, 1790–1795. [DOI] [PubMed] [Google Scholar]
- 36.Kooter, J. M. & Borst, P. (1984) Nucleic Acids Res. 12, 9457–9472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duraisingh, M. T., Voss, T. S., Marty, A. J., Duffy, M. F., Good, R. T., Thompson, J. K., Freitas-Junior, L. H., Scherf, A., Crabb, B. S. & Cowman, A. F. (2005) Cell, in press. [DOI] [PubMed]
- 38.Voss, T. S., Thompson, J. K., Waterkeyn, J., Felger, I., Weiss, N., Cowman, A. F. & Beck, H. P. (2000) Mol. Biochem. Parasitol. 107, 103–115. [DOI] [PubMed] [Google Scholar]
- 39.Voss, T. S., Kaestli, M., Vogel, D., Bopp, S. & Beck, H. P. (2003) Mol. Microbiol. 48, 1593–1607. [DOI] [PubMed] [Google Scholar]
- 40.Deitsch, K. W., Calderwood, M. S. & Wellems, T. E. (2001) Nature 412, 875–876. [DOI] [PubMed] [Google Scholar]
- 41.Smith, J. S. & Boeke, J. D. (2001) Science 291, 608–609. [DOI] [PubMed] [Google Scholar]
- 42.Calderwood, M. S., Gannoun-Zaki, L., Wellems, T. E. & Deitsch, K. W. (2003) J. Biol. Chem. 278, 34125–34132. [DOI] [PubMed] [Google Scholar]
- 43.Girard, F., Bello, B., Laemmli, U. K. & Gehring, W. J. (1998) EMBO J. 17, 2079–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caslini, C., Alarcon, A. S., Hess, J. L., Tanaka, R., Murti, K. G. & Biondi, A. (2000) Leukemia 14, 1898–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ishii, K., Arib, G., Lin, C., Van Houwe, G. & Laemmli, U. K. (2002) Cell 109, 551–562. [DOI] [PubMed] [Google Scholar]