Abstract
The extracellular N-terminal domain (NTD) is the largest region of NMDA receptors; however, biological roles for this ectodomain remain unknown. Here, we determined that the F-box protein, Fbx2, bound to high-mannose glycans of the NR1 ectodomain. F-box proteins specify ubiquitination by linking protein substrates to the terminal E3 ligase. Indeed, ubiquitination of NR1 was increased by Fbx2 and diminished by an Fbx2 dominant-negative mutant. When expressed in hippocampal neurons, this Fbx2 dominant-negative mutant augmented NR1 subunit levels and NMDA receptor-mediated currents in an activity-dependent fashion. These results suggest that homeostatic control of synaptic NR1 involves receptor retrotranslocation and degradation by the ubiquitin-proteasome pathway.
Keywords: endoplasmic reticulum degradation pathway, neuronal activity, NFB42, proteasome, ubiquitylation
N-methyl-d-aspartate (NMDA)-type glutamate receptors play central roles in neuronal development and information storage in the mammalian brain. These neurotransmitter receptors are glutamate-gated cation channels whose permeability to Ca2+ underlies aspects of synaptic plasticity. On the other hand, excess Ca2+ influx through NMDA receptors mediates cell death in certain neurodegenerative processes. Therefore, neurons must precisely control NMDA-receptor levels. Indeed, global neuronal activity negatively regulates the density of synaptic NMDA receptors (1–4).
NMDA receptors all contain the NR1 subunit, one or more subunits of NR2A-D, and NR3 subunits that form a heterotetramer (5). NMDA-receptor subunits have a large extracellular domain, three transmembrane domains, and a cytosolic C-terminal tail (5). Proteins interacting with the NMDA-receptor tail help to anchor NMDA receptors and traffic the NMDA receptors to synapses (6). However, identified protein interactions with the extracellular domains of NMDA receptor are limited (7, 8). The extracellular domain of NMDA receptors comprises a large N-terminal domain (NTD) and a ligand-binding domain. Although the NTD is the largest part of NMDA receptors (≈400 aa), the functions of the NTD remain mysterious.
NMDA-receptor proteins concentrate at the postsynaptic density (PSD), a specialized apparatus beneath synapses that comprises receptors, scaffolding molecules, and signal-transduction enzymes. Synaptic transmission modulates composition of the PSD, in part, by activity-dependent ubiquitination and degradation of PSD components (3). Several scaffolding molecules including PSD95, Shank, GKAP, and AKAP79/109 are identified as substrates for ubiquitination (3, 9).
The ubiquitin-proteasome system (UPS) targets numerous cellular proteins for degradation. Several reports demonstrate that the UPS is also involved in synaptic transmission and cognitive function. Pharmacological inhibition of the UPS increases evoked synaptic transmission at Drosophila neuromuscular junctions (10) and leads to an increase in glutamate-evoked responses in Aplysia (11). Moreover, abnormalities of the UPS in the brain contribute to Parkinson's disease, Alzheimer's disease, prion disease, amyotrophic lateral sclerosis, and polyglutamine disease (12). Furthermore, patients with schizophrenia show decreased expression of genes that mediate the UPS (13).
Ubiquitin conjugation involves the sequential activities of three enzymes: ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin-ligase enzyme (E3) (14, 15). E3 determines selectivity for ubiquitination by bridging target proteins to E2 and ubiquitin. The SCF-E3 complex contains Skp1, Cul1, Rbx1, and an F-box protein. Whereas Skp1, Cul1, and Rbx1 are common subunits, specific E3s contain distinct F-box proteins, which determine substrate specificity. Modular F-box proteins contain the signature F-box domain, which binds to Skp1, and specialized F-box-associated regions that recognize specific substrates (14, 15).
Fbx2/NFB42/Fbs1 is a neuron-specific F-box protein that recognizes N-linked high-mannose oligosaccharides on substrate proteins (16, 17). Because mannose modifications occur in the lumen of the endoplasmic reticulum (ER), Fbx2 must recognize substrates after their retrotranslocation from the ER as part of the ER degradation (ERAD) pathway (18). The ERAD pathway mediates protein quality control to ensure that aberrant polypeptides do not transit through the secretory pathway. Misfolded or unwanted proteins in the ER are dislocated to the cytosol for degradation by the proteasome (18). This retrotranslocation/degradation by the ERAD system can also dispose of proteins internalized by endocytosis/phagocytosis (19–22).
Here, we used expression cloning and identified Fbx2 protein as a binding partner for the high-mannose glycans from the NTD of NR1. Fbx2 augmented NR1 ubiquitination, and a dominant-negative Fbx2 (dnFbx2) mutant lacking the F-box domain blocked NR1 ubiquitination. We found that overexpression of this dnFbx2 mutant increased the density of extrasynaptic NMDA receptors in hippocampal neurons in an activity-dependent manner. We propose that interaction between the NR1 ectodomain and the Fbx2 ubiquitin ligase E3 participates in the homeostatic control of NMDA receptors.
Methods
Production of Alkaline Phosphatase (AP)-Fusion Proteins. Nucleotides encoding the NTD of NR1a (amino acids 21–397), NR1b (amino acids 21–418), or GluR2 (amino acids 22–415) were cloned into the pAP5-tag vector (GenHunter, Nashville, TN). Plasmids were transfected into COS-7 cells, and the media containing the secreted AP-tagged fusions were harvested after 72 h. AP activity in the media was quantitated as described in ref. 23 by using p-nitrophenylphosphate as substrate.
Staining of Cultured Cells with AP Fusions. Hippocampal neurons or COS-7 cells were stained with AP-tagged fusion proteins as described in ref. 23. Briefly, cells were washed with TBS, fixed with 4.5% formalin for 15 min, permeabilized with 0.1% Triton X-100 in Hank's balanced salt solution (HBAH), 0.5 mg/ml BSA, 0.1% NaN3, and 20 mM Hepes (pH 7.4), and incubated for 90 min at 22°C with the AP-fusion protein. The cells were then washed with HBAH, fixed with acetone-formalin [65% acetone and 8% formalin in 20 mM Hepes (pH 7.4)], and incubated for 100 min at 65°C to inactivate endogenous phosphatase activity. The cells were stained in buffer [100 mM NaCl/5 mM MgCl2/100 mM Tris·Cl (pH 9.5)] containing 0.17 mg/ml 5-bromo-4-chloro-3-indolyl phosphate (BCIP) and 0.33 mg/ml nitroblue tetrazolium (NBT) solution for several hours to visualize AP activity. The reaction was stopped by the addition of PBS.
Quantitative Binding Assay. COS-7 cells expressing Fbx2 were incubated with AP-fused NTD and fixed with acetone-formalin, and endogenous phosphatase activity was inactivated by heating. The cells were rinsed with AP-substrate buffer (1 mM MgCl2 in 2 M diethanolamine, pH 9.8) and incubated with 3.3 mg/ml p-nitrophenylphosphate. Monitoring absorbance at 420 nm quantitated bound AP.
Expression Cloning. Twenty-four pools of 5,000 arrayed cDNA clones from a mouse adult-brain library (OriGene Technologies, Rockville, MD) were transfected into COS-7 cells, and NR1-NTD-AP binding was detected by staining with BCIP-NBT. Six positive clones were isolated from 120,000 cDNA clones. All six clones independently encoded Fbx2.
Antibody Production. Bacterially expressed full-length Fbx2 protein was used as antigen. Antibodies were raised in guinea pigs and rabbits, and the antisera were purified on a column containing the immunizing antigen.
Coimmunoprecipitation Analyses. Human embryonic kidney cells (293 T cells) transfected with Fbx2 and NR1 cDNA were solubilized in RIPA buffer [150 mM NaCl/1% Nonidet P-40/0.5% deoxycholate/0.1% SDS/50 mM Tris·Cl (pH 7.4)], and insoluble materials were removed by centrifugation at 100,000 × g. The supernatant was precleared with protein G Sepharose (Amersham Pharmacia) and incubated with anti-NR1 antibody (BD Biosciences, San Diego). The immunocomplexes were precipitated with protein G Sepharose, separated by SDS/PAGE, and immunoblotted. Whole rat brain was homogenized in buffer containing 20 mM Tris·Cl (pH 9.0), 1 mM PMSF, 10 μM MG132, and 10 μM leupeptin. Deoxycholate (1%) was added, and the mixture was incubated at 37°C for 1 h. After centrifugation (100,000 × g) for 1 h at 4°C, the supernatant was dialyzed overnight against 0.1% Triton X-100 and 50 mM Tris·Cl (pH 7.4). The lysate was centrifuged (100,000 × g) for 1 h at 4°C, and the supernatant was cleared with protein A Sepharose (Sigma). The cleared lysate was incubated with guinea pig anti-Fbx2 antibody, and the immune complexes were precipitated with protein A Sepharose and eluted with SDS sample buffer.
Overlay Assay. Extracts from COS7 cells transfected with NTD-AP were treated with PNGase F or Endo H (NEB, Beverly, MA) at 37°C for 1 h according to the manufacturer's instruction, separated by SDS/PAGE, and transferred to poly(vinylidene difluoride) membrane. The membrane was washed with buffer A [20 mM Hepes (pH 7.5)/50 mM KCl/10 mM MgCl2/1mMDTT/0.1% Nonidet P-40], and was soaked in 6 M guanidine hydrochloride in buffer A for 10 min at 4°C. Proteins on the blot were refolded by diluting the guanidine hydrochloride in half and incubated for 10 min at 4°C. This dilution series was repeated four times until the concentration of guanidine hydrochloride was 0.18 M. The membranes were then blocked with 5% skim milk in buffer A followed by incubation overnight at 4°C with 300 ng/ml His-tagged recombinant Fbx2 protein purified from Escherichia coli. The membrane was washed with 100 mM KCl in PBS and 0.1% Triton X-100 and incubated with rabbit anti-Fbx2 antibody in 5% milk in 0.1% Tween-20 in Tris-buffered saline followed by horseradish-peroxidase-conjugated anti-rabbit IgG. Immunoreactivity was visualized with SuperSignal West Pico chemiluminescent substrate (Pierce).
Immunocytochemistry. Hippocampal cultures were prepared as described in ref. 24. The cells were seeded (28,000 cm–2) on poly(d-lysine)-coated, 12-mm coverslips and maintained in Neurobasal/B-27 medium (Invitrogen) containing 0.5 mM glutamine. Half of the medium was replaced every week. The cells were used for immunocytochemistry at days 25–30 in vitro. Semliki-forest viral (SFV) vectors encoding GFP-fused Fbx2 were constructed, and infectious SFV particles were generated as described in ref. 25. For staining, neurons were fixed with 4% paraformaldehyde in PBS for 1 min and then with 100% methanol for 10 min at –20°C, washed with 0.02% Triton X-100 in PBS for 15 min, and, finally, washed with PBS for 15 min. After blocking with BSA (8%) in PBS for 1 h, the cells were incubated with mouse anti-NR1 antibody (1:500) (BD Biosciences), mouse anti-PSD95 (1:200) (Affinity BioReagents, Golden, CO), mouse anti-synaptophysin (1:200) (Sigma) or rabbit anti-Sec61β (1:1,000; Upstate Biotechnology, Lake Placid, NY), and guinea pig anti-Fbx2 (1:1,000) followed by Alexa Fluor 488 and Cy3- or Cy5-conjugated secondary antibodies. Images were captured with a laser scanning fluorescent microscope LSM510 (Zeiss) equipped with ×40 and ×63 objectives, and fluorescence was quantified by using nih image (National Institutes of Health, Bethesda).
Electrophysiology. Recordings were made from infected neurons 48 h after infection by using 3- to 5-MΩ glass electrodes filled with an internal solution [115 mM CsMeSO3/20 mM CsCl/10 mM Hepes/2.5 mM MgCl2/4 mM Na2-ATP/0.4 mM Na-GTP/10 mM Na-phosphocreatine/0.6 mM EGTA/0.1 mM spermine/5 mM QX314 (pH 7.2)]. The external perfusion medium consisted of 119 mM NaCl, 2.5 mM KCl, 1.3 mM MgSO4, 2.5 mM CaCl2, 1 mM NaH2PO4, 26 mM NaHCO3, and 11 mM glucose equilibrated with 95% O2 and 5% CO2 and included 20 μM glycine. Infected pyramidal cells were identified by using fluorescence microscopy. Somatic outside-out patches were clamped at +40 mV. Currents were evoked by local application of 1 mM NMDA for 2 s in the presence of 20 μM glycine. Responses were collected with an Axopatch-1D amplifier (Axon Instruments, Foster City, CA), filtered at 2 kHz, digitized at 5 kHz, and analyzed online by using igor pro software (WaveMetrics, Lake Oswego, OR). All data are expressed as mean ± SEM. The statistical significance for within-group comparisons was determined by one-way repeated-measures ANOVA followed by Tukey's posttest.
Ubiquitination Assay. Transfected human embryonic kidney 293T cells were treated with MG132 for 10 h before harvesting extracts. NR1 was immunoprecipitated as described above and blotted with antiubiquitin (Research Diagnostics, Flanders, NJ), anti-NR1 (BD Biosciences). Cortical cultures prepared as described above (16 days in vitro; 10 × 106 cells per 10-cm dish) were treated with 40 μM bicuculline (BIC) or 2 μM tetrodotoxin (TTX) in the presence of 10 μM MG132 for 8 h. The cells were solubilized in 20 mM Tris·Cl (pH 7.4), 1% SDS, 1 mM PMSF, and 10 μM MG132, and after centrifugation at 100,000 × g, supernatants were neutralized with 1% Triton X-100. NR1 immunoprecipitates from these supernatants were separated by SDS/PAGE and blotted with antiubiquitin antibody.
Results
Identification of Fbx2 as a NR1-NTD-Binding Partner. To determine whether molecules interact with the NTD of NR1 (Fig. 1a), we constructed a mammalian expression plasmid that drives secretion of the NTD of NR1 as a fusion with AP. The binding of such probes can be detected sensitively by using BCIP and NBT as substrates (23). The NTD (NR1a)-AP probe strongly bound to cultured hippocampal neurons (Fig. 1b) but did not stain a variety of nonneuronal cell lines (data not shown). To identify the neuronal NR1 interacting protein, COS7 cells were transfected with pools of clones from a mouse-brain cDNA library probed with the NTD (NR1)-AP fusion (Fig. 1c). We isolated six positives from 120,000 clones, and all six independently encoded Fbx2, a component of E3 ubiquitin ligase (16).
Fig. 1.
Expression cloning Fbx2 by interaction with the NTD of NR1. (a) Schematic representation of NR1. (Left) The extracellular region of NMDA receptor subunits comprises a large NTD and a ligand-binding domain (S1 and S2). (Right) Schematic of the secreted AP fusion. (b) Specific binding of the NTD of NR1 to hippocampal neurons. The AP-fused NTD of NR1 was applied to fixed and permeabilized neurons. The probe was detected by staining with BCIP-NBT. The NTD(NR1)-AP probe gave robust labeling in the somatodendritic regions. No labeling is detected in the cells treated with AP alone. (c) Expression cloning of cDNAs encoding proteins that interact with the NTD of NR1. A rare positively stained cell is observed in the screening of 5,000 pooled clones (Left). A positive pool from the second round of screening (50 cDNA clones) shows many more labeled cells (Center). All transfected cells stain when transfected with an isolated clone encoding Fbx2.
Fbx2 Binds to High-Mannose Glycans on NR1. Fbx2 protein was initially identified as a neuron-specific F-box that associates with high-mannose-type glycans (17) (Fig. 2a). Because Fbx2 is cytosolic, interaction with the glycans from NR1 requires receptor retrotranslocation. To determine whether this interaction occurs in a cellular environment, coimmunoprecipitation analysis was performed by using cotransfected 293T cells. We found that NR1 coimmunoprecipitated with wild-type Fbx2. NR1 associated with an F-box-deleted mutant, Fbx2(ΔF), with slightly weaker affinity than wild type but did not associate with a C terminus-deleted mutant, Fbx2(ΔC) (Fig. 2b). This result indicates that NR1 interacts with the FBA domain. Furthermore, we found that NR1 but not GluR2 immunoprecipitated with Fbx2 from brain extracts (Fig. 2c).
Fig. 2.
Fbx2-dependent ubiquitination of NR1 mediated interaction with the glycosylated ectodomain of NR1. (a) Structure of Fbx2 and deletion mutants. The F-box and F-box-associated (FBA) domains are indicated. (b) The FBA region mediates NR1 binding. FLAG-tagged Fbx2 variants were transfected into COS7 cells. The cell lysates were immunoprecipitated with anti-FLAG antibody and then immunoblotted with anti-NR1. Wild-type Fbx2 and Fbx2(ΔF) but not Fbx(ΔC) were coimmunoprecipitated with Fbx2. (c) Fbx2 protein interacts with NR1 in vivo. Rat brain lysates were immunoprecipitated with two independent anti-Fbx2 antibodies or control IgG and immunoblotted for NR1, GluR2, and Fbx2. (d) Fbx2 interacts with high-mannose glycans on NR1. Proteins from COS7 cells expressing Myc-tagged NTD of NR1 were treated with EndoH or peptide N-glycosidase F (PNGaseF). Immunoblotting demonstrates deglycosylation (Left). Far Western blotting shows that EndoH or PNGaseF treatment abolished interaction of NTD(NR1)-AP with Fbx2. (e) Both NR1 alternatively spliced variants (NR1a and NR1b), but not GluR2, bind to Fbx2. COS7 cells expressing Fbx2 were fixed, permeabilized, and probed with NTD of NR1a, NR1b, or GluR2 (GR2). Bound AP was quantitated enzymatically by using p-nitrophenylphosphate as substrate. Error bar, SEM. (f) Fbx2 constructs or GFP were cotransfected with full-length NR1 and ubiquitin into COS7 cells. MG132 (10 μM) was applied 10 h before harvesting. Cell lysates were immunoprecipitated with anti-NR and blotted with antiubiquitin or anti-NR1 antibodies.
Far Western blotting revealed that interaction of NR1 and Fbx2 required high-mannose glycosylation of NR1, because the interaction was disrupted by treatment of NR1 with EndoH, which trims only high-mannose glycans, or with PNGaseF, which prunes all N-linked sugars (Fig. 2d). Although a substantial fraction of GluR2 harbors high-mannose glycans (26, 27), GluR2 did not bind Fbx2 (Fig. 2 c and e). These data show that protein determinates help specify Fbx2-binding properties. Because two splice variants, NR1a and NR1b, occur in the NTD region of NR1 (5), we examined the binding of these NR1 variants to Fbx2. Fig. 2e shows that both variants bound Fbx2 with similar efficiency.
Fbx2 Ubiquitinates NR1. To determine whether Fbx2 induces NR1 ubiquitination, 293T cells were cotransfected with NR1, Fbx2, and HA-tagged ubiquitin. NR1 was immunopurified and probed for ubiquitin or anti-HA. Strong NR1 polyubiquitin was detected in the Fbx2(WT)-transfected cells treated with the proteasome inhibitor MG132 (Fig. 2f). On the other hand, an F-box-deleted mutant Fbx2(ΔF) blocked the minimal polyubiquitination found in a control transfection with GFP. These results suggest that retrotranslocated NR1 is ubiquitinated by Fbx2 and then degraded by the proteasome pathway. They also indicate that the Fbx2(ΔF), which binds NR1, can inhibit receptor ubiquitination.
Fbx2 Is Localized in Somatodendritic Regions and Spines. Immunofluorescent labeling showed that Fbx2 localizes to somatodendritic regions and spines. NR1 and Fbx2 partly colocalize in somatodendritic regions (Fig. 3). Furthermore, some punctate Fbx2 staining colocalized with NR1 and/or PSD95 in synaptic spines. Synaptophysin puncta were often juxtaposed to Fbx2 puncta but were rarely overlapping, consistent with a postsynaptic function for Fbx2. Sec61β, which can serve as a channel for retrotranslocation, was localized to neuronal soma and dendrites but was rarely detected in spines. Sec61 is a well described candidate for the protein-conducting channel of retrotranslocation (28). Sec61β, a component of the Sec61 channel, localized to neuronal soma and dendrites but was rarely detected in spines. Therefore, the channels for retrotranslocation serving for dendritic Fbx2 are likely to be different from Sec61.
Fig. 3.
Fbx2 localizes to the somatodendritic region and to synaptic spines. Cultured hippocampal neurons were stained with antibodies as indicated. Green arrowheads show Fbx2 immunoreactivity in the spines. Red arrowheads show punctate staining juxtaposed to Fbx2 in the spines. Yellow arrowheads show colocalized puncta. Rectangles show the regions magnified. The enlarged image of Sec61β staining is the result of prolonged exposure to enhance details along the dendrite.
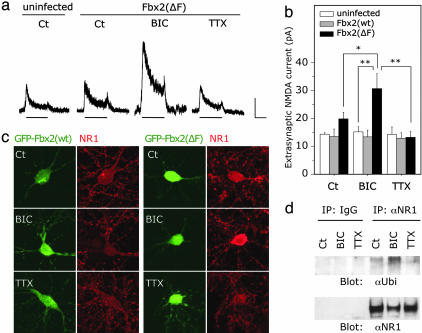
Fbx2 Mediates Activity-Dependent Degradation of NR1. Previous studies showed that neuronal activity homeostatically reduces synaptic NMDA-receptor activity (1) and that proteasome inhibitors block this NR1 removal from the PSD (3). This activity-dependent decrease in synaptic NR1 may reflect the lateral migration of synaptic NMDAR to extrasynaptic sites (29). To evaluate possible roles for Fbx2 in this process, we overexpressed GFP-tagged Fbx2(WT) or Fbx2(ΔF) in hippocampal cultured neurons and recorded NMDA-receptor-mediated currents in outside-out somatic membrane patches (Fig. 4 a and b). These patches contain extrasynaptic NMDA receptors. We found a 2-fold increase in NMDA-receptor-mediated responses in neurons expressing Fbx2(ΔF) that were treated with the GABAA-receptor antagonist BIC (30.6 ± 5.3 pA, n = 16 vs. 14.3 ± 0.6 pA, n = 7 in uninfected cells). This effect was activity-dependent, because Fbx2(ΔF) overexpression did not affect NMDA currents under control conditions or in cultures treated with TTX (control, 19.8 ± 2.9 pA, n = 15; TTX, 13.2 ± 2.1 pA, n = 14). Overexpression of Fbx2(WT) did not affect NMDA currents (control, 13.5 ± 2.6 pA, n = 5; BIC, 13.4 ± 2.4 pA, n = 7; TTX, 12.8 ± 2.0 pA, n = 7). Paralleling these electrophysiological changes, we found that Fbx2(ΔF) dramatically increased NR1 immunoreactivity in somatic regions of neurons treated with BIC (256 ± 24%) (Fig. 4c). We also detected activity-dependent ubiquitination of NR1 (Fig. 4d).
Fig. 4.
Activity-dependent degradation of NR1 by Fbx2. (a and b) NMDAR-mediated responses in extrasynaptic outside-out patches are selectively increased in BIC-treated neurons infected with Fbx2(ΔF). (a) Sample traces of NMDA-evoked currents (2 s, 1 mM NMDA plus 20 μM glycine); bar indicates NMDA application. Ct, control (no drug treatment); BIC, 40 μM for 48 h; TTX, 2 μM for 48 h. Calibration, 10 pA, 1 s.(b) Quantification of NMDA peak current. Uninfected (Ct, n = 7; BIC, n = 11; TTX, n = 10); Fbx2(WT) (Ct, n = 5; BIC, n = 7; TTX, n = 7); Fbx2(ΔF) (Ct, n = 15; BIC, n = 16; TTX, n = 14). *, P < 0.05; **, P < 0.01. (c) Immunostaining of cultured hippocampal neurons expressing GFP-tagged Fbx2 constructs. NR1 immunoreactivity (red) in the somatic region was greatly increased in BIC-treated neurons expressing Fbx2(ΔF). (d) Activity-dependent ubiquitination of NR1. Cortical cultures were treated as control (Ct) or with BIC or TTX in the presence of MG132 for 8 h. Lysates were purified with NR1 antibodies or nonimmune IgG, and these immunprecipitates were blotted with antiubiquitin antibody.
Discussion
This study demonstrates that activity-dependent changes in neuronal NMDA receptors involve retrotranslocation and degradation by an Fbx-2 ubiquitination-proteosome pathway. Whereas neuronal activity down-regulates synaptic NMDA-receptor subunit density and channel function, biochemical studies show that total neuronal NR1 levels are unchanged (3, 4). These reports (3, 4) imply that activity disperses NMDA receptors away from synapses. We find that augmenting neuronal activity in the presence of dominant-negative Fbx2 increases extrasynaptic NMDA-receptor density. This finding suggests a model whereby activity releases synaptic NR1, which relocates to the dendrites and soma for retrotranslocation and degradation (Fig. 5). Fbx2 localized in the cytosolic region recognizes the retrotranslocated ectodomain of NR1 and ubiquitinates it. The ubiquitinated NR1 is degraded by cytosolic proteasomes.
Fig. 5.
Model for activity-dependent degradation of NR1. MDA receptors are released from synaptic regions by activity. Fbx2 recognizes high-mannose glycans on retrotranslocated NR1 to promote ubiquitination and degradation.
The cellular sites for NR1 retrotranslocation and ubiquitination require elucidation. Membrane protein dislocation to the cytosol can occur by retrograde trafficking through the classical Sec61 translocation (30). Recent studies also suggest that a novel protein Derlin can also support protein translocation (31, 32) to the cytosolic compartment. We find that Fbx2 localizes to some synaptic spines and that Fbx2 and Sec61 both occur broadly within the somatodendritic domain. The cellular distribution of Derlin in neurons and its possible roles in ERAD in the brain remains unknown.
Although ERAD was first discovered as a mechanism for eliminating unfolded proteins in the ER, recent studies indicate that this pathway can also degrade mature proteins after endocytosis (19–22). We propose a similar mechanism for Fbx2 regulation of NMDA receptors. The sugar moieties of cell-surface NR1 are high-mannose (33). This unusual feature helps explain the sensitivity of NMDA receptors to the Fbx2 pathway. That Fbx2 inhibition causes NMDA-receptor accumulation at the plasma membrane suggests that internalized receptors can recycle to the plasma membrane and that ubiquitination controls both receptor retrotranslocation and degradation. This finding is consistent with previous studies showing that polyubiquitin serves as a ratcheting mechanism, because p97, an ATPase associated to a variety of cellular activities binds to ubiquitinated proteins at the surface of the ER and “pulls” the ubiquitinated proteins into the cytosol (34).
Because NMDA receptors play central roles in synaptic plasticity but can also mediate neurotoxicity when overstimulated, neurons must precisely regulate these receptors. This work demonstrates a central role for retrotranslocation and Fbx2-mediated ubiquitination in this homeostatic process. These experiments also provide the first evidence that ERAD participates in the regulation of neurotransmitter receptors in the brain.
Acknowledgments
This work was supported by grants from the National Institutes of Health (to D.S.B. and R.A.N.), the Christopher Reeves Paralysis Foundation (to D.S.B.), the International Human Frontier Science Program (to D.S.B. and N.R.), Bristol-Myers Squibb (to R.A.N.), the Uehara Memorial Foundation (to A.K.), and the American Heart Association (to A.K.). D.S.B. is an established investigator of the American Heart Association. R.A.N. is a member of the Keck Center for Integrative Neuroscience and the Silvio O. Conte Center for Neuroscience Research.
Author contributions: R.A.N. designed research; A.K. and N.R. performed research; and D.S.B. wrote the paper.
Abbreviations: AP, alkaline phosphatase; BCIP, 5-bromo-4-chloro-3-indolyl phosphate; BIC, bicuculline; ER, endoplasmic reticulum; ERAD, ER degradation; NBT, nitroblue tetrazolium; NTD, N-terminal domain; PSD, postsynaptic density; TTX, tetrodotoxin.
References
- 1.Watt, A. J., van Rossum, M. C., MacLeod, K. M., Nelson, S. B. & Turrigiano, G. G. (2000) Neuron 26, 659–670. [DOI] [PubMed] [Google Scholar]
- 2.Crump, F. T., Dillman, K. S. & Craig, A. M. (2001) J. Neurosci. 21, 5079–5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ehlers, M. D. (2003) Nat. Neurosci. 6, 231–242. [DOI] [PubMed] [Google Scholar]
- 4.Mu, Y., Otsuka, T., Horton, A. C., Scott, D. B. & Ehlers, M. D. (2003) Neuron 40, 581–594. [DOI] [PubMed] [Google Scholar]
- 5.Mori, H. & Mishina, M. (1995) Neuropharmacology 34, 1219–1237. [DOI] [PubMed] [Google Scholar]
- 6.Wenthold, R. J., Prybylowski, K., Standley, S., Sans, N. & Petralia, R. S. (2003) Annu. Rev. Pharmacol. Toxicol. 43, 335–358. [DOI] [PubMed] [Google Scholar]
- 7.Takasu, M. A., Dalva, M. B., Zigmond, R. E. & Greenberg, M. E. (2002) Science 295, 491–495. [DOI] [PubMed] [Google Scholar]
- 8.Dalva, M. B., Takasu, M. A., Lin, M. Z., Shamah, S. M., Hu, L., Gale, N. W. & Greenberg, M. E. (2000) Cell 103, 945–956. [DOI] [PubMed] [Google Scholar]
- 9.Colledge, M., Snyder, E. M., Crozier, R. A., Soderling, J. A., Jin, Y., Langeberg, L. K., Lu, H., Bear, M. F. & Scott, J. D. (2003) Neuron 40, 595–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Speese, S. D., Trotta, N., Rodesch, C. K., Aravamudan, B. & Broadie, K. (2003) Curr. Biol. 13, 899–910. [DOI] [PubMed] [Google Scholar]
- 11.Zhao, Y., Hegde, A. N. & Martin, K. C. (2003) Curr. Biol. 13, 887–898. [DOI] [PubMed] [Google Scholar]
- 12.Ciechanover, A. & Brundin, P. (2003) Neuron 40, 427–446. [DOI] [PubMed] [Google Scholar]
- 13.Middleton, F. A., Mirnics, K., Pierri, J. N., Lewis, D. A. & Levitt, P. (2002) J. Neurosci. 22, 2718–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weissman, A. M. (2001) Nat. Rev. Mol. Cell Biol. 2, 169–178. [DOI] [PubMed] [Google Scholar]
- 15.Nakayama, K. I., Hatakeyama, S. & Nakayama, K. (2001) Biochem. Biophys. Res. Commun. 282, 853–860. [DOI] [PubMed] [Google Scholar]
- 16.Erhardt, J. A., Hynicka, W., DiBenedetto, A., Shen, N., Stone, N., Paulson, H. & Pittman, R. N. (1998) J. Biol. Chem. 273, 35222–35227. [DOI] [PubMed] [Google Scholar]
- 17.Yoshida, Y., Chiba, T., Tokunaga, F., Kawasaki, H., Iwai, K., Suzuki, T., Ito, Y., Matsuoka, K., Yoshida, M., Tanaka, K. & Tai, T. (2002) Nature 418, 438–442. [DOI] [PubMed] [Google Scholar]
- 18.McCracken, A. A. & Brodsky, J. L. (2003) BioEssays 25, 868–877. [DOI] [PubMed] [Google Scholar]
- 19.Lencer, W. I. & Tsai, B. (2003) Trends Biochem. Sci. 28, 639–645. [DOI] [PubMed] [Google Scholar]
- 20.Sandvig, K. & van Deurs, B. (2002) FEBS Lett. 529, 49–53. [DOI] [PubMed] [Google Scholar]
- 21.Guermonprez, P., Saveanu, L., Kleijmeer, M., Davoust, J., Van Endert, P. & Amigorena, S. (2003) Nature 425, 397–402. [DOI] [PubMed] [Google Scholar]
- 22.Houde, M., Bertholet, S., Gagnon, E., Brunet, S., Goyette, G., Laplante, A., Princiotta, M. F., Thibault, P., Sacks, D. & Desjardins, M. (2003) Nature 425, 402–406. [DOI] [PubMed] [Google Scholar]
- 23.Flanagan, J. G., Cheng, H. J., Feldheim, D. A., Hattori, M., Lu, Q. & Vanderhaeghen, P. (2000) Methods Enzymol. 327, 19–35. [DOI] [PubMed] [Google Scholar]
- 24.Kato, A., Fukuda, T., Fukazawa, Y., Isojima, Y., Fujitani, K., Inokuchi, K. & Sugiyama, H. (2001) Eur. J. Neurosci. 13, 1292–1302. [DOI] [PubMed] [Google Scholar]
- 25.DiCiommo, D. P. & Bremner, R. (1998) J. Biol. Chem. 273, 18060–18066. [DOI] [PubMed] [Google Scholar]
- 26.Greger, I. H., Khatri, L., Kong, X. & Ziff, E. B. (2003) Neuron 40, 763–774. [DOI] [PubMed] [Google Scholar]
- 27.Greger, I. H., Khatri, L. & Ziff, E. B. (2002) Neuron 34, 759–772. [DOI] [PubMed] [Google Scholar]
- 28.Romisch, K. (1999) J. Cell Sci. 112, 4185–4191. [DOI] [PubMed] [Google Scholar]
- 29.Tovar, K. R. & Westbrook, G. L. (2002) Neuron 34, 255–264. [DOI] [PubMed] [Google Scholar]
- 30.Tsai, G. & Coyle, J. T. (2002) Annu. Rev. Pharmacol. Toxicol. 42, 165–179. [DOI] [PubMed] [Google Scholar]
- 31.Ye, Y., Shibata, Y., Yun, C., Ron, D. & Rapoport, T. A. (2004) Nature 429, 841–847. [DOI] [PubMed] [Google Scholar]
- 32.Lilley, B. N. & Ploegh, H. L. (2004) Nature 429, 834–840. [DOI] [PubMed] [Google Scholar]
- 33.Huh, K. H. & Wenthold, R. J. (1999) J. Biol. Chem. 274, 151–157. [DOI] [PubMed] [Google Scholar]
- 34.Flierman, D., Ye, Y., Dai, M., Chau, V. & Rapoport, T. A. (2003) J. Biol. Chem. 278, 34774–34782. [DOI] [PubMed] [Google Scholar]