Abstract
The αβ heterodimeric T cell receptor (TCR) recognizes peptide antigens that are transported to the cell surface as a complex with a protein encoded by the major histocompatibility complex (MHC). T cells thus evolved a strategy to sense these intracellular antigens, and to respond either by eliminating the antigen-presenting cell (e.g. a virus-infected cell) or by secreting factors that recruit the immune system to the site of the antigen. The central role of the TCR in the binding of antigens as peptide-MHC (pepMHC) ligands has now been studied thoroughly. Interestingly, despite their exquisite sensitivity (e.g. T cell activation by as few as 1 to 3 pepMHC complexes on a single target cell), TCRs are known to have relatively low affinities for pepMHC, with KD values in the micromolar range. There has been interest in engineering the affinity of TCRs in order to use this class of molecules in ways similar to now done with antibodies. By doing so, it would be possible to harness the potential of TCRs as therapeutics against a much wider array of antigens that include essentially all intracellular targets. To engineer TCRs, and to analyze their binding features more rapidly, we have used a yeast display system as a platform. Expression and engineering of a single-chain form of the TCR, analogous to scFv fragments from antibodies, allow the TCR to be affinity matured with a variety of possible pepMHC ligands. In addition, the yeast display platform allows one to rapidly generate TCR variants with diverse binding affinities and to analyze specificity and affinity without the need for purification of soluble forms of the TCRs. The present chapter describes the methods for engineering and analyzing single-chain TCRs using yeast display.
Keywords: T cell receptors, peptide antigens, major histocompatibility complex, yeast display, single-chain T cell receptors, affinity maturation, intracellular antigens
1 Introduction
The structural and biochemical properties of αβ T cell receptors have been revealed through the efforts of many labs over the past 20 years [13, 14, 15, 23, 34, 53]. Among the findings, it has been shown that the variable (V) domains of each chain bind to the specific peptide/MHC ligand in a conserved, diagonal orientation. The structural basis of this invariant orientation is still unclear, but it has been suggested to be due to the: 1) geometry required for the entire T cell complex (αβ TCR, CD3 subunits, and co-receptor CD4 or CD8) to productively engage the pepMHC and to signal the T cell during thymic selection [31, 54], 2) evolutionary pressures that yielded “germline” TCR regions with basal affinity for the helices of MHC molecules [22, 34], or 3) perhaps both [29].
Regardless of the mechanistic or evolutionary basis, the consequence of the conserved diagonal orientation is that it positions the complementarity determining regions (CDRs) of each V region (Vα and Vβ) in ideal proximity to the antigenic components of the ligand. That is, the most hypervariable regions of the TCR (CDR3α and CDR3β) are positioned directly over the most diverse component of the ligand, the bound antigenic peptide. In contrast CDR2 loops are positioned almost exclusively over the MHC helices, whereas CDR1 loops can contact either peptide or MHC. Because TCRs are oriented in this manner, engineering CDR3 loops for affinity maturation may provide the optimal opportunity to maintain the highest level of peptide specificity possible [25, 26, 30, 32, 56]. While there have been several antibodies engineered against individual pepMHC complexes [6, 11, 16, 55], it is not clear whether the antibodies will maintain any type of consistent geometry [33]. In this case, while one might see some peptide selectivity, it remains to be seen if a high level of specificity for the selecting peptide is achieved.
Recent reviews have described the possible applications for TCRs that have been engineered with higher affinity for their specific pepMHC ligands [35, 38, 47]. The applications include use of soluble forms of high-affinity TCRs (picomolar to nanomolar) as targeting components, coupled with other specificities such as anti-CD3 scFv fragments for bispecific agents [32], or cytokines such as IL-10 [51] or IL-15 [49] for use as immunomodulators. Another application involves the introduction of the TCRs into T cells for adoptive therapies [45, 50], but the optimal affinities of these TCRs in order to avoid or minimize cross-reactivities with self-peptides will likely be in the low micromolar to high nanomolar range [8, 25, 57].
The present report focuses on specific methods that our lab has used to affinity mature TCRs, and to more rapidly analyze the binding properties and specificity of these TCRs. By analogy to the development of single-chain antibody fragments (scFv), we have sought to develop strategies that allow expression of only the antigen-binding domains of the TCR as single-chain molecules (Vα-linker-Vβ or Vβ-linker-Vα), called either scTv, or scTCR [1, 28, 37, 43, 47]. The use of stabilized versions of the scTv fragments allows for high levels of expression in the yeast display format, and expression as soluble versions of the stabilized scTv fragments. While the first studies were performed with mouse TCRs, our more recent efforts have applied these same methods to human TCRs. One of the human Vα regions, called Vα2 (IMGT, TRAV12 family) has been shown to be exceptionally stable in the single-chain format, and serves as a platform for engineering human TCRs with different specificities and affinities [1, 43, 47]. Engineering is facilitated by the availability of various forms of the ligands, peptide/HLA class I complexes, some of which are commercially available. The methods involving these different forms, combined with the yeast display system, are also described here. Finally, we describe methods to rapidly discover and to analyze TCR variants with a wide range of affinities.
2 Materials
2.1 Yeast Display Strain, Plasmid, and Primers
Saccharomyces cerevisiae yeast display strain EBY100 (a GAL1-AGA1::URA3 ura3-52 trp1 leu2Δ1 his3Ø200 pep4::HIS2 prb1Δ1.6R can1 GAL)
pCT302 Yeast Display Vector
-
pCT302 Standard Primers
Splice 4L (Forward): 5′ GGCAGCCCCATAAACACACAGTAT
YRS (Reverse): Rev 3′ CGAGCTAAAAGTACAGTGGG
T7 (Reverse): Rev 3′ TAATACGACTCACTATAG
2.2 DNA Purification
Zymoprep Kit II (Zymo Research)
QIAprep Spin Miniprep Kit (Qiagen).
QIAquick Gel Extraction Kit (Qiagen)
QIAquick PCR Purification Kit (Qiagen)
Pellet Paint Co-Precipitant (Novagen)
Agencourt AMPure XP PCR Purification Beads (Beckman Coulter)
2.3 Restriction Enzymes and Ligation
XhoI
NheI
BglII
DpnI
T4 DNA Ligase
Calf Intestinal Alkaline Phosphatase (CIP)
2.4 PCR
FastStart High Fidelity PCR System (Roche)
dATP Solution
dCTP Solution
dGTP Solution
dTTP Solution
Deoxynucleotide (dNTP) solution mix
PfuTurbo DNA Polymerase (Agilent)
Taq DNA Polymerase (Invitrogen)
2.5 LiOAc Yeast Heat Shock Transformation
50% PEG 3350: dissolve 5 g PEG 3350 to a final volume of 10 mL ddH2O, sterile filter and store at room temperature for up to 6 months.
1 M LiOAc: dissolve 16.5 g LiOAc in 250 mL ddH2O, sterile filter and store at room temperature for up to 6 months
10X TE: dissolve 121 mg Tris (10 mM) and 29 mg EDTA (1 mM) in 100 mL ddH2O, sterile filter, and store at room temperature for up to 6 month
Single-stranded carrier: Dissolve 200 mg Salmon Sperm DNA (Sigma) in 100 mL 1X TE buffer, aliquot into 1 mL stocks, and store at −20°C.
2.6 Electrocompetent E. coli Strains
For DNA amplification: Subcloning Efficiency DH5α Competent Cells (Invitrogen)
For Protein expression: BL21(DE3) Competent E. coli (New England Biolabs)
2.7 Yeast Media
YPD media: Dissolve 10 g yeast extract, 20 g bacto-peptone, and 20 g dextrose, bring volume to 1 L with ddH2O, autoclave, and store at room temperature for up to 1 month.
YPD plates: Dissolve 10 g yeast extract, 20 g bacto-peptone, 15 g agar, and 20 g dextrose, bring volume to 1 L dH2O, and autoclave. Cool to ~55°C and pour ~25 mL into 100 mm X 15 mm plates. Cool and store at +4°C for up to 1 month.
SD-CAA media: Dissolve 14.8 g sodium citrate, 4.2 g citric acid monohydrate, 5 g casamino acids, 6.7 g yeast nitrogen base (without amino acids), 20 g dextrose, and 10 mL penicillin-streptomycin (10,000 U/mL), bring volume to 1 L with ddH2O, sterile filter, and store at 4°C for up to 6 months.
SD-CAA plates: Dissolve 91.1 g sorbitol, 7.5 g agar, 7.4 g sodium citrate, and 2.1 g citric acid monohydrate in 400 mL of ddH2O, autoclave, and cool to ~55°C. In a separate container combine 2.5 g casamino acids, 10 g dextrose and 3.35 g yeast nitrogen base (without amino acids) to 100 mL of ddH2O, sterile filter, and add to cooled autoclaved solution. Mix and pour ~25 mL into 100 mm X 15 mm plates. Cool and store at +4°C for up to 6 months.
SG-CAA media: Dissolve 14.8 g sodium citrate, 4.2 g citric acid monohydrate, 5 g casamino acids, 6.7 g yeast nitrogen base (without amino acids), 20 g galactose, and 10 mL penicillin-streptomycin (10,000 U/mL), bring volume to 1 L with ddH2O, sterile filter, and store at 4°C for up to 6 months.
2.8 Yeast Library
1 M Sorbitol: dissolve 45.6 g sorbitol in 250 mL ddH2O, sterile filter, and store at 4°C for up to 6 months.
1M Sorbitol/1 mM CaCl2: dissolve 45.5 g sorbitol and 27 mg of CaCl2 in 250 mL ddH2O, sterile filter, and store at 4°C for up to 6 months.
0.1 M LiAc/10 mM DTT: dissolve 1.65 g lithium acetate (LiAc) and 0.386 g dithiothreitol (DTT) in 250 mL ddH2O, sterile filter, and cool to 4°C for immediate use.
0.2 cm electroporation cuvettes
2.9 Yeast Staining
Phosphate-buffered saline (PBS): Dissolve 8 g NaCl, 0.2 g KCl, 1.15 g Na2HPO4•7H2O, and 0.2 g KH2PO4 (anhydrous); bring volume up to 1 L with ddH2O, adjust pH to 7.4, autoclave, and store at room temperature.
PBS/1% BSA: Dissolve 10 g bovine serum albumin (BSA) in 1 L PBS, sterile filter, and chill to 4°C.
Anti-c-myc, chicken IgY fraction (Invitrogen cat. no. A12181)
HA.11 Clone 16B12 Monoclonal Antibody (anti-HA) (Covance cat. no. MMS-101P)
Alex Fluor 647 Goat anti-Chicken IgG (H+L) (Molecular Probes cat. no. A-21449)
Streptavidin-Phycoerythrin (BD Pharmingen cat. no. 554061)
Alexa Fluor 647 F(ab′)2 Fragment of Goat anti-Mouse IgG (H+L) (Molecular Probes cat. no. A-21237)
BD DimerX HLA-A2:Ig Recombinant Fusion Protein, Human (BD Pharmingen) cat. no. 551263)
2.10 HLA-A2 Expression Plasmids
HLA-A2 Heavy Chain (HLA-A2bsp in pHN1); obtained from the from University of Massachusetts Medical School Tetramer Facility
HLA-A2 Light Chain (β2 microglobulin in pHN1); obtained from the NIH Tetramer Facility
2.11 Bacterial Expression Media
Luria Broth (LB): Dissolve 5 g yeast extract, 10 g tryptone, and 10 g NaCl in 1 L ddH2O, autoclave, and store at room temperature.
Ampicillin (100 mg/mL): Stock solution can be made by dissolving 1 g into 10 mL ddH2O, sterile filtering, and freezing individual aliquots at −20°C.
LB + ampicillin (100 μg/mL) plates: Dissolve 5 g yeast extract, 10 g tryptone, 10 g NaCl, and 15 g agar in 1 L dH2O and autoclave. Cool to ~55°C, then add 1 mL ampicillin stock at 100 mg/mL, swirl, then pour ~25 mL into 100 mm X 15 mm plates. Cool and store at +4°C.
2.12 Inclusion Body Isolation
Lysis Buffer: 50 mM Tris base, 100 nM NaCl, 0.1% NaN3, 1% Triton X-100, 10 mM dithiothreitol (DTT), and 1 mM phenylmethylsulfonyl fluoride (PMSF, dissolved in 1 mL isopropanol), bring volume to 1 L with ddH2O and adjust pH to 8.0 with hydrochloric acid. Buffer can be stored without DTT and PMSF for up to 6 months.
Osmotic Shock Buffer: 20 mM Tris base and 2.5 mM ethylenediaminetetraacetic acid (EDTA); bring volume up to 1 L with ddH2O, adjust pH to 8.0, and chill to 4°C.
Osmotic Shock Buffer with Triton: Add 0.5% Triton X-100 to 1 L osmotic shock buffer and chill to 4°C.
Urea Extraction Buffer: 8 M urea, 25 mM MES, 10 mM EDTA, and 0.1 mM dithiothreitol (DTT) in 10 mL at pH 6.0.
Guanidine Extraction Buffer: 8 M guanidine-HCl, 50 mM Tris, 5 mM EDTA, 5 mM dithiothreitol (DTT) in 10 mL at pH 8.0.
2.13 MHC Refold
Refold Buffer: 400 mM Tris, 400 mM L-Arg, 2 mM EDTA; bring volume up to 200 mL with ddH2O, adjust pH to 8.0, and chill to 4°C.
Injection Buffer: 3 M guanidine-HCl, 10 mM sodium acetate, and 10 mM EDTA; bring volume up to 250 mL with ddH2O and adjust pH to 4.2. Aliquots can be frozen and stored at −20°C.
Dialysis Buffer: 20 mM Tris in 3 L ddH2O, adjust pH to 8.0, and chill to 4°C.
Oxidized glutathione
Reduced glutathione
Phenylmethylsulfonyl fluoride (PMSF)
Regenerated Cellulose 6–8 MWCO Dialysis Tubing
Amicon Ultra-4 Centrifugal Filter Units with Ultracel-10 membrane (Amicon)
2.14 MHC Biotinylation, Purification, and Quantification
In vitro Biotinylation Kit (Avidity)
HPLC Buffer: 20 mM Tris and 50 mM NaCl in 1 L ddH2O, adjust pH to 8.0, and degas.
EDTA Stock: 0.5 M EDTA in 50 mL ddH2O, adjust pH to 8.0, and sterile filter.
BCA Protein Assay kit (Pierce)
2.15 MHC-Restricted Peptides
UV-cleavable peptide, KILGFVFJV, where J is the photolabile amino acid residue, prepared by standard Fmoc-peptide solid phase synthesis using commercially available Fmoc-3-amino-3-(2-nitro)phenyl propionic acid as a building block; store in the dark at −20 °C.
HLA-A2-restricted peptide(s) (20 mg/mL): Dissolve 20 mg of lyophilized peptide into 1 mL DMSO; store at −20°C.
HLA-A2-restricted peptide(s) (2 mg/mL): Perform a 1:10 dilution of 20 mg/mL peptide stock in DMSO in PBS; store at −20°C.
2.16 Magnetic Selection
PBSM: 8 g NaCl, 0.2 g KCl, 1.44 g Na2PO4, 0.24 g KH2PO4, 5 g bovine serum albumin, and 744 mg EDTA in 1 L dH2O, adjust to pH 7.4, sterile filter and store at 4°C.
Microbeads conjugated to anti-mouse IgG (Miltenyi Biotec cat. no. 130-048-401)
Microbeads conjugated to strepavidin (Miltenyi Biotec cat. no. 130-048-101)
Microbeads conjugated to anti-biotin (Miltenyi Biotec cat. no. 130-090-485)
LS Columns (Miltenyi Biotec cat. no. 130-042-401)
2.17 DNA Quantification, Analysis, Site-Directed Mutagenesis, and 454 Sequencing
Qubit dsDNA HS Assay Kit (Life Technologies)
Agilent DNA 7500 Kit (Agilent Technologies)
QuikChange II or QuikChange Lightening Site-Directed Mutagenesis Kit (Agilent)
GS FLX Titanium Sequencing Kit XL+ (Roche)
GS FLX Titanium PicoTiter Plate Kit 70x75 (Roche)
2.18 Equipment
Thermocycler
Two temperature controlled incubator shakers
Electroporator (BioRad Gene Pulser II Electroporation System)
Flow Cytometer and FACS apparatus
Amicon 8400 Stirred Ultrafiltration Cell (Millipore) with regenerated cellulose Ultrafiltration Discs, YM-10, 10 kDa NMWL, 76mm (Millipore cat. no. 13642)
Superdex 200 10/300 GL gel filtration column (GE Biosciences cat. no. 17517501)
CL-1000 Ultraviolet Crosslinker (UVP, LLC)
MidiMACS Separator (Miltenyi Biotec)
MACS MultiStand (Miltenyi Biotec)
Qubit 2.0 Fluorimeter (Life Technologies)
Agilent 2100 Bioanalyzer (Agilent Technologies)
Roche/454 Genome Sequencer FLX+ (Roche)
3 Methods
3.1 Yeast Surface Display of T Cell Receptors
Yeast display allows for surface expression, detection and selection of recombinant proteins via an N- or C-terminal fusion to the yeast agglutinin factor Aga-2 [5]. By forming a disulfide linkage to Aga-1 on the yeast cell surface, 10,000 to 100,000 fusion copies of the protein of interest can be expressed on the yeast cell surface (Fig. 1). Yeast display offers certain advantages over other protein display methods. First, yeast provide eukaryotic expression and processing of the protein of interest as well as the quality control mechanisms that result from the yeast secretory pathway [28, 41, 42]. Second, libraries of yeast can be quantitatively selected for precise affinities or off-rates by fluorescence activated cell sorting (FACS) [4]. Mutants selected by yeast display can also be analyzed directly on the surface of yeast without the need for sub-cloning and expression of large amounts of protein (discussed in section 3.2) [18, 19, 28]. Finally, current library generation protocols have been optimized for the routine generation of large libraries on the order 109–1010 [3, 7]. Here we describe the design and cloning of T cell receptors as yeast-displayed single-chains, and the use of yeast-displayed TCR libraries to engineer TCRs with improved stability.
Fig. 1. Diagram of scTv fragment displayed on surface of yeast.
A scTv fragment with a N-terminus HA tag and a C-terminus c-myc is depicted. The scTv construct is expressed as a fusion as an N-terminal fusion to the AGA-2 yeast mating protein, which forms covalent linkages to AGA-1 on the yeast cell surface. A total of 10,000–100,000 of copies of the scTv are expressed on the surface of each yeast cell and can bind to soluble pepMHC molecules. Expression of the construct can be monitored through antibodies against either tags (HA or c-myc) or antibodies specific for the scTv variable domains.
3.1.1 Design and Cloning of T Cell Receptors for Yeast Display as Single-Chain Variable Fragments
For expression and engineering of T cell receptors on the surface of yeast, we have used scTv fragments consisting of the variable α and β regions linked by a polypeptide. Although other linkers such as Gly/Ser-rich repeats can be used, we have used the charged linker first introduced into a soluble form of the TCR, GSADDAKKDAAKKDGKS [44]. The orientation of the scTv can be constructed in either the Vβ-L-Vα or the Vα-L-Vβ orientation. Expression of scTv fragments has shown higher expression levels than full-length constructs [1, 37], but they require introduction of mutations in order to stabilize the V regions in the absence of C regions. In this section we describe the method for cloning a TCR gene into the pCT302 yeast display vector as a scTv construct. The pCT302 vector contains a galactose promoter and an upstream HA tag (sequence: YPYDVPDYA), which can be used as a probe for expression. We suggest including a C-terminal c-myc epitope tag (sequence: EQKLISEEDL) for monitoring expression of the full-length fusion (Fig. 1). Given that the pCT302 vector does not contain a stop codon, a stop codon should be included at the 3′ end of the scTv sequence, or following the c-myc sequence if used. The following three sections describe cloning the scTV into the yeast display vector pCT302 (section 3.1.2), LiOAc transformation into yeast (section 3.1.3), and a standard yeast induction protocol (section 3.1.4).
3.1.2 Cloning the scTv into the pCT302 Vector
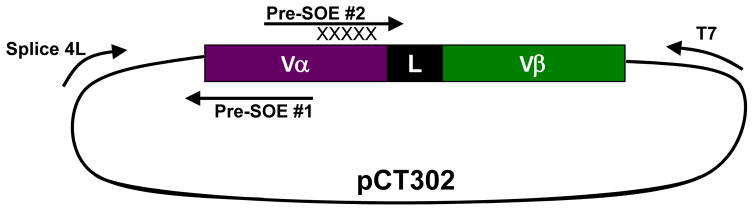
The sequences of the TCR genes to be engineered typically originate from the cloning and sequencing of T cell clones isolated for reactivity against an antigen of interest. As described, we have shown that the human Vα2 region is amenable to expression as a single-chain with different Vβ regions, and thus it is advantageous to identify clones that use this Vα region [1]. Although the scTv construct can be cloned directly from the genes of the isolated T cell clone, we suggest having the scTv gene synthesized for optimized yeast expression. Gene synthesis and codon optimization are available commercially from a variety of companies (e.g., Genscript, DNA2.0, Genewiz). Flanking DNA sequences containing the 5′ NheI and 3′ XhoI and/or BglII restriction sites should be included in original gene synthesis to allow for sub-cloning into the yeast display vector pCT302. We recommend the addition of a C-terminal c-myc epitope tag (sequence: EQKLISEEDL) immediately following the sequence of the scTv in order to assess the population for truncations in future libraries derived from the scTv. Following the c-myc tag, 1–2 stop codons should be introduced prior to the flanking XhoI and/or BglII restriction sites. Once synthesized, the scTv construct can be cloned into the pCT302 vector by ligation into the upstream NheI restriction site and either a downstream XhoI or BglII restriction site (Fig. 2). Traditional cloning methods utilizing competent E. coli, such as DH5α, can then be used to amplify the ligated scTv in the pCT302 vector. pCT302 contains an ampicillin resistant gene for selection in E. coli, which allows for growth in ampicillin concentrations up to 100 μg/ml. Splice4L, T7 and YRS are suggested primers to confirm proper cloning and for mutagenesis described in future sections (Fig. 2).
Fig. 2. scTv in the pCT302 yeast display vector.
Depicted is the pCT302 vector with described cloning sites. The pCT302 vector contains the TrpI gene that allows for the auxotroph EBY100 yeast strand to grow on minimal media, and an ampicillin resistant gene that allows for growth in E. coli. The scTv ORF contains a galactose inducible promoter such that expression of the scTv can be induced by growth in galactose. The 5′ region of the ORF contains the sequence for the yeast mating protein AGA-2 fused to the scTv construct. The scTv, depicted here as a Vα-Linker-Vβ, is flanked by an N-terminal HA epitope tag (HA sequence: YPYDVPDYA), which can be used as a probe for expression, and an added C-terminal c-myc tag (c-myc sequence: EQKLISEEDL) which we recommend adding to the scTv construct to probe for full length variants. For cloning purposes, the standard Splice 4L, YRS or T7 primers can be used for PCR-based mutagenesis and sequencing.
Codon optimize and synthesize the scTv gene of interest in the Vβ-L-Vα or Vα-L-Vβ orientation with a C-terminal c-myc tag (EQKLISEEDL) if desired and two stop codons. Include an N-terminal NheI restriction site (e.g., GCGGCCGCCACC) and a C-terminal XhoI and/or BglII restriction sites (e.g., CTCGAG) on flanking ends for cloning into pCT302 (see Note 1). Typically the manufacturer of the gene will deliver the synthesized gene in a carrier plasmid.
If necessary, amplify codon-optimized gene provided by manufacturer by transforming competent E. coli, such as DH5α, expanding colonies in LB media with the specified antibiotic, and harvesting plasmid using Qiagen Mini-prep kit.
Digest the amplified scTv plasmid with both NheI and XhoI or BglII according to the manufacturer’s protocol. Run entire samples on a 1% agarose gel and gel purify the band containing the scTv (typically ~750 bp for scTvs) with a Qiagen gel extraction kit (see Note 2).
Meanwhile, the pCT302 plasmid should also be digested with the same restriction enzymes used to digest the scTv insert according to the manufacturer’s protocol.
Purify digested pCT302 plasmid using the Qiagen PCR purification kit according to the manufacturer’s protocol.
Dephosphorylate the linearized pCT302 vector using Calf Intestine Phosphatase (CIP) for 1 hour at 37°C according to the manufacturer’s protocol. Alternatively, entire digested vector can be run on a 1% agarose gel and the band containing the digested pCT302 plasmid (typically ~6 Kb) can be gel purified using the Qiagen gel extraction kit according to the manufacturer’s protocol.
Perform ligation of digested scTv insert into the digested pCT302 vector using T4 ligase according to the manufacturers protocol (see Note 3).
Transform 1–2 μL of ligation products into competent E. coli, such as DH5α, and plate entire volumes of transformed E. coli on an LB/Amp plates. Incubate plates for 8–12 hours at 37°C until colonies are visible.
Grow 4–6 colonies in 3 mL of LB media with Amp (100 μg/mL) for 8–12 hours at 37°C.
Purify DNA using Qiagen mini-prep, according to manufacturer’s protocol.
Determine if ligations were successful by performing small (10 μL) test digests with both NheI and XhoI or BglII and running products on a 1% agarose gel. Digestion of the correct product will yield two bands (one for digested pCT302 at ~6 Kb and one for the digested scTv at ~750 bp).
Confirm the sequence of the construct by sequencing using T7, Splice 4L and/or YRS primers.
3.1.3 LiOAc Mediated Transformation of EBY100 Yeast
Once the proper scTv is cloned into the pCT302 vector, the vector is transformed into EBY100 yeast by LiOAc mediated transformation or electroporation. The pCT302 yeast display plasmid contains the trp1 gene, which allows for tryptophan synthesis in EBY100 yeast (Trp− Leu−). Here we describe how to perform a LiOAc mediated heat shock transformation. This protocol has been adapted from Gietz [24].
Streak a YPD plate with EBY100 yeast, preferably from a frozen stock. Grow the yeast for 36–48 hours at 30°C until colonies are visible.
Select an individual colony and transfer to 3 mL of YPD media to start a liquid culture. Grow liquid cultures until dense (density > OD600 8.0; approximately 24–48 hours) shaking at 220 RPM at 30°C.
For each yeast transformation, spin 1 mL of dense yeast culture at 1800 xg for 3 minutes in sterile 1.7 mL microfuge tubes (see Note 4).
Aspirate supernatants and discard.
Wash cells by suspending in a sterile mixture of 800 μL ddH2O, 100 μL 10X TE, and 100 μL 1 M LiAc at room temperature.
Aspirate supernatant leaving ~50 μL of wash mixture and suspend cells in remaining volume.
-
Add the following heat shock mixture to cells in the order listed:
264 μL 50% PEG 3350
36 μL 1M LiOAc
50 μL single-stranded DNA carrier, boiled for 8 minutes on a heat block prior to addition
1–5 μg pCT302 vector containing scTv insert in a volume of 50 μL
Incubate cells at 42°C for 1.5–2.5 hours, gently vortexing occasionally to suspend pelleted cells.
After incubation, remove cells and spin at 16,000 xg for 1 minute.
Aspirate supernatant and discard.
Spin an additional time at 16,000 xg for 1 minute to remove any residual heat shock mixture and discard.
Suspend cells in 1 mL sterile ddH2O to wash.
Transfer 500 μL into a separate tube.
Pellet both tubes at 16,000 xg for 1 minute.
Suspend one aliquot of yeast in 100 μL sterile ddH2O and plate entire volume on an SD-CAA plate. Grow at 30°C for 36–48 hours (until colonies visible).
Suspend the second aliquot in 3 mL SD-CAA media and grow at 30°C for 24–48 hours with shaking. Store at 4°C once dense (see Note 5).
Once colonies are present on plates, expand 1–2 individual colonies each in 3 mL SD-CAA media in glass test tubes. Grow at 30°C for approximately 48 hours until dense (OD600 >8.0).
To confirm the correct sequence, 500 μL of the dense yeast culture should be lysed and DNA harvested using the Zymoprep II kit according to the manufacturer’s protocol (see Note 6).
Transform DH5α cells with 2.5 μL of DNA isolated from Zymoprep kit, and plate on LB/Amp plates. Grow overnight at 37°C
Grow a single colony from the LB/Amp plate in 3 mL liquid cultures with Amp (100 μg/mL) for 8–12 hours.
Isolate and purify DNA using a mini-prep kit and sequence DNA using either Splice 4L, T7 or YRS primers to confirm the correct identity of the plasmid.
Yeast cultures in SD-CAA media can be maintained in liquid culture at 4°C for several months; however, it is recommended that they be freshly expanded within 1 week of use (e.g., by expanding 10–50 μL of dense yeast in 3 mL fresh SD-CAA media for 12–24 hours). Frozen stocks of yeast cultures can also be snap frozen on dry ice and stored at −80°C in 10% DMSO indefinitely.
3.1.4 Standard Yeast Induction with SG media
EBY100 yeast containing the pCT302 plasmid are induced to express the scTv when transferred into galactose-minimal media. Here we describe a yeast induction procedure.
Determine the OD600 of the dense SD-CAA culture. An OD600 = 1 is equivalent to a concentration of approximately 1 X 107 cells/mL.
Yeast cells will be induced at a final concentration of 1 X 107 cells/mL. Determine the number of cells required for the volume of yeast being induced, typically 2 X 107, and transfer into sterile microfuge tubes (see Note 7).
Spin cells down at 1,800 xg for 3 minutes at 4°C and suspend cells in 1 mL of cold SG-CAA media.
Repeat wash two additional times.
After last wash, suspend yeast to a final concentration of 1 X 107 cells/mL and transfer to a glass test tube.
Induce cultures for 48 hours at 20°C while shaking at 220 rpm.
Induced yeast can be stored at 4°C for approximately 2 weeks.
3.1.5 Design and Selection of Yeast Libraries Generated by Error-Prone PCR
Because scTv fragments lack TCR constant regions that stabilize the TCR on the surface of the T cell, scTv constructs typically require mutations to generate a stabilized form to express on the surface of yeast. Typically the resultant mutations occur at the interface of the variable α and β domains, or now exposed region of the variable region that is normally buried by the constant region [1, 28, 37, 41, 56]. Recently, we showed that the use of the highly stable human Vα2 region, enhances stabilization of the scTv on the surface of yeast and that the introduction of a serine substitution at the polymorphic residue 49 in Vα2 confers additionally stability [1]. We routinely introduce the F49S mutation into the Vα of our Vα2-containing scTv fragments during gene synthesis.
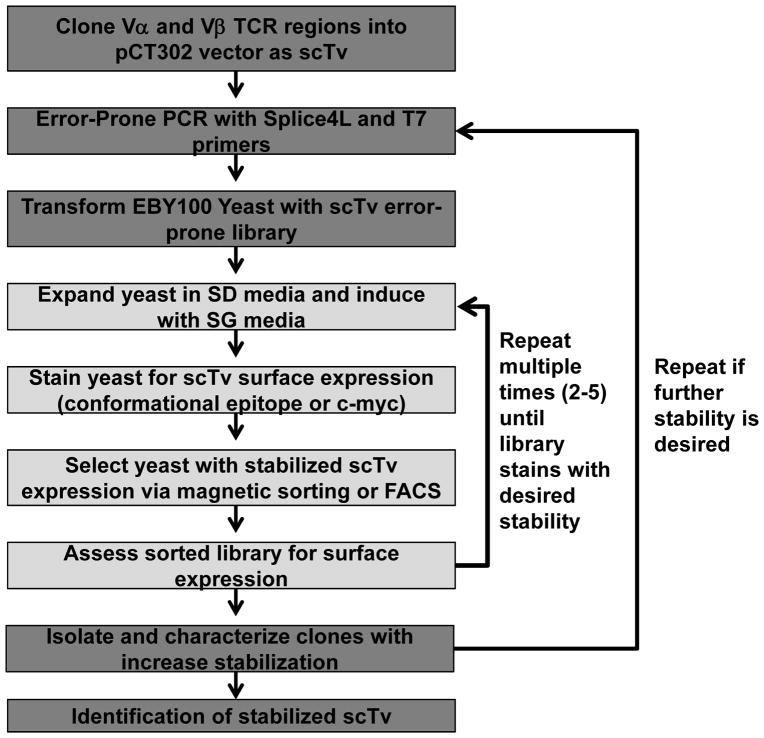
Despite the increased stability conferred by the use of Vα2, further mutagenesis is often desirable in order to achieve higher surface expression. In this section we describe a method for creating a mutated scTv library by error-prone PCR, from which stabilized clones can be selected. Induced yeast are stained with a conformational-specific antibody that recognizes epitopes on the Vβ and/or the Vα chain, if available (see Note 8). Yeast cells that express the most stable scTv mutants are isolated by magnetic bead sorting and/or high-speed FACs. In order to eliminate TCR library variants that result from truncations through introduction of pre-mature stop codons, c-myc specific antibodies can be used to select for constructs expressing the full-length scTv if a C-terminal c-myc tag is added to the scTv construct. A flow chart for the selection of surface-stabilized scTvs is shown in Fig. 3. The following six sections describe error prone library generation (section 3.1.6), preparation of electrocompetent yeast (section 3.1.7), staining yeast cells (section 3.1.8), and both magnetic and FACS-based selections (sections 3.1.9, 3.1.10. and 3.1.11).
Fig. 3. Sample selection scheme for stabilized scTv fragment.
This is a selection scheme that can yield surface stabilized scTvs. Stabilized scTv constructs can be selected for either by antibodies specific for conformational epitopes or by antibodies specific for the c-myc tag.
3.1.6 Preparation of Error-Prone scTv PCR Insert and pCT302 Vector
Random mutagenesis of the scTv can be conducted by error-prone PCR, using Splice4L and T7 primers, which flank the scTv region of the gene. This method produces a 0.5% error rate as described previously [39]. Error-prone scTv constructs, along with NheI and XhoI digested pCT302 vector are simultaneously electroporated into EBY100 yeast. Overlap between digested pCT302 vector and error-prone PCR inserts allow the PCR products to be introduced into pCT302 by a process of homologous recombination. This method can generate yeast libraries in the range of 108–1010 independent mutants, using multiple electroporations.
-
For error-prone PCR, assemble 100 uL reactions in a PCR tubes (see Note 9):
220 μM dATP
200 μM dCTP
340 μM dGTP
2.4 mM dTTP
0.3 ng/uL template (scTv in pCT302 vector)
250 nM Splice 4L primer (forward primer)
250 nM T7 primer (reverse primer)
5 ng/μL bovine serum albumin
3.325 mM MgCl2
0.5 mM MnCl2
1:10 dilution of 10X Taq Polymerase Reaction Buffer
1 μL Taq Polymerase
-
Place PCR tubes in thermocycler and run the following protocol:
95°C for 1 minute
-
25 cycles of:
95°C for 1 minute
50°C for 1 minute
72°C for 3 minutes
72°C for 5 minutes
4°C forever
Confirm amplification of the gene by running 10 μL of PCR product on a 1% agarose gel to determine if correct scTv fragment amplified. For typical scTvs, the region between Splice4L and T7 is approximately 1.2 Kb.
Purify and concentrate PCR combined product with the Qiagen PCR Purification kit according to the manufacturer’s protocol. For Qiagen kits, spin columns hold a maximum of 10 μg of plasmid. Calculate the number of columns to use by approximating DNA concentration from the agarose gel ran in step #3 and elute each in >30 μL ddH2O.
In order to reduce background from the remaining template, digest the PCR reaction with DpnI according to the manufacturer’s protocol.
Perform an additional Qiagen PCR Purification as before and determine concentration. Because the scTv library will be introduced into pCT302 via homologous recombination, no further digest is required.
Meanwhile, digest pCT302 vector with NheI and XhoI restriction enzymes according to manufacturer’s protocol.
Perform Qiagen PCR Purification according to the manufacturer’s protocol. For Qiagen kits, spin columns hold a maximum of 10 μg of plasmid. Calculate the number of columns to use by approximating DNA concentration from the agarose gel ran in step #3 and elute each in >30 μL ddH2O.
Run the entire volume of digested vector on a 1% agarose gel and gel purify the band containing the digested pCT302 plasmid (typically ~5.7 Kb) using the Qiagen gel extraction kit according to the manufacturer’s protocol. It is recommended that the undigested pCT302 be ran on the same gel to distinguish digested from undigested DNA.
Mix 4 μg of error prone mutagenized DNA insert and 1 μg of double digested/gel-purified pCT302 in a 1.7 mL Eppendorf tube for each electroporation. Controls consisting of insert only and vector only should also be included to determine background (see Note 10).
Pellet DNA in 1.7 mL Eppendorf tube using Novagen Pellet Paint Co-Precipitant, according to manufacturer’s protocol (see Note 11). Store pelleted DNA at −20°C until ready for transformation into yeast.
3.1.7 Preparation of Electrocompetent Yeast
Once error-prone DNA and digested pCT302 have been obtained, yeast libraries are generated by electroporation. This protocol has been adapted and optimized from previous reported methods [3, 7, 47]:
Streak a YPD plate with EBY100 yeast, preferably from a frozen stock. Grow the yeast for 36–48 hours at 30°C until colonies are visible.
Select an individual colony and transfer to 3 mL of YPD media to start a liquid culture. Grow liquid cultures for 6–12 hours shaking at 220 RPM at 30°C.
Expand liquid culture of EBY100 cells into 50 mL YPD culture in a 250 mL Erlenmeyer flask. Grow cultures overnight (12–16 hours) at 30°C with shaking.
The next morning, innoculate a pre-warmed YPD liquid culture to an OD600 of 0.2 from overnight cultures. 50 mL of culture are required for each electroporation. Shake culture at 220 RPM at 30°C (see Note 12).
Monitor OD600 hourly, until an OD600 of 1.5 is reached (approximately 6 hours) (see Note 13).
Transfer 50 mL of culture into 50 mL conical tubes (e.g.,10 total for a 500 mL culture). Pellet by spinning at 2000 xg for 5 minutes and discard supernatants. 50 mL of culture (or one conical tube) will be used for each electroporation. From this step on, it is critical that all reagents and cells remain on ice for the duration of the procedure, except where indicated.
Wash cells in each conical tube in 25 mL cold sterile ddH2O, pellet cells by spinning at 2000 xg for 5 minutes. Discard supernatant.
Wash cells in 25 mL of cold 1M sorbitol/1mM CaCl2, pellet cells by spinning at 2000 xg for 5 minutes. Discard supernatant.
Suspend cells in 25 mL 0.1M LiAc/10 mM DTT solution. Loosen caps to allow for aeration and secure lids with tape to maintain a sterile environment. Incubate cells at 30°C while shaking at 220 RPM for 30 minutes.
Remove cells and place on ice.
Pellet cells at 2000 xg for 5 minutes at 4°C.
Remove supernatant and wash cells in 25 mL cold 1M sorbitol/1 mM CaCl2 per tube. Pellet cells at 2000 xg for 5 minutes. Discard supernatant.
Wash pelleted cells one time in 25 mL cold 1M Sorbitol (No CaCl2). Pellet cells at 2000 xg for 5 minutes at 4°C. Discard supernatant.
Suspend cells in each conical tube in 1M sorbitol (No CaCl2) to a final volume of 250 μL using repeat pipetting.
Suspend precipitated DNA from pellet paint (section 3.1.6) in 10 μL of ddH2O.
Add 250 μL of EBY100 yeast cells to DNA. Mix well by pipetting and then transfer to a pre-chilled 2 mm electroporation cuvette. Let cuvettes incubate on ice for 5 min (see Note 14).
Using a BioRad Gene Pulser II Electroporation System, electroporate cells at 2.5 kV, 25 μF capacitance with a 0.2 cm gap cuvette. Typical time constant ranges are from 3.0 to 4.5 ms.
Following pulse remove the cuvette and immediately add 1 mL of a 1:1 mixture of YPD media and 1M sorbitol mixture to cells. Mix cells/YPD/Sorbitol mixture completely, and transfer contents of the cuvette into a sterile glass test tube.
Add an additional 1 mL of 1:1 mixture of YPD media and 1M sorbitol to the cuvette and transfer to the glass tube to wash any residual cells in the cuvette.
Incubate electroporated yeast cells in glass test tubes at 30°C for 1 hour without shaking.
After 1 hour incubation, gently vortex cells using the lowest setting on a benchtop vortex to suspend.
Transfer cells into conical tubes, combining electroporations from the same libraries into a single tube.
Pellet cells by spinning at 900 xg for 5 minutes and suspend in 10 mL SD-CAA.
Determine the library size by generating 1:100, 1:1,000, 1:10,000, 1:100,000, 1:1 X 106 dilutions from the 10 mL of cells in SD-CAA. For control electroporations, only plate undiluted cells.
Streak 10 μL of each dilution on SD-CAA plates and grow at 30°C for 2–3 days until colonies are visible. The number of colonies on each plate corresponds to a total library size of 1 X 105, 1 X 106, 1 X 107, 1 X 108, and 1 X 109, respectively. To calculate diversity, average the library sizes calculated from the number of colonies for plates where colonies could be reliably counted. Background on controls should be minimal (i.e. less than 10 colonies), and the values should be subtracted from the diversity.
Transfer the remaining library from the 10 mL combined electroporations into 100 mL SD-CAA media for each electroporation (e.g., if 8 electroporations are done, expand in 800 mL). Control electroporations can be discarded after plating.
Expand culture for 36–48 hours at 30°C, 220 RPM until saturation is reached (OD600 > 8.0). Pellet cultures in conical tubes in order to remove cellular debris by centrifuging at 2000 xg for 5 minutes.
Decant supernatants and suspend sufficient cells to oversample the anticipated diversity in a SD-CAA to an OD600 of 1.0 and expand until dense (OD600 > 8.0)(see Note 15).
Yeast libraries in SD-CAA media can be maintained in liquid culture at 4°C for several months; however, it is recommended that they be freshly expanded within 1 week of use. Frozen stocks of yeast cultures in 10% DMSO can be snap frozen on dry ice and stored at −80°C indefinitely. Sufficient aliquots should be made to oversample the library diversity calculated in step #24.
The diversity of each library should be further assessed by isolating plasmids from at least 10 individual colonies for each library and sequencing (protocol in steps #18–21 in section 3.1.3; see Note 16).
3.1.8 Staining Induced Yeast Cultures
Once an error-prone library has been made, it will be necessary to determine the initial staining profile with antibodies specific for conformational epitopes and c-myc. Here we describe the protocol to assess the staining profile of the library.
Induce the yeast library as described in section 3.1.4.
Aliquot 50 μL of induced yeast to flow tubes and add 1 mL PBS/1% BSA
Spin tubes down at 1800 xg and aspirate supernatant.
Suspend cells in 50 μL of primary antibody staining mixture (see Note 17).
Incubate cells on ice for 30–60 minutes
Add 1 mL of PBS/1% BSA, spin cells at 1800 xg and aspirate supernatant.
Suspend cells in 50 μL of secondary staining reagent staining mixture (see Note 18).
Incubate cells on ice for 30–45 minutes.
Add 1 mL of PBS/1% BSA and spin cells at 1800 xg for 3 minutes, aspirate supernatant.
Wash cells one additional time with 1 mL PBS/1% BSA.
Suspend cells in 500 μL of PBS/1% BSA.
Read cells using flow cytometer.
3.1.9 Selection of Stabilized scTvs from Error-Prone Library
In order to isolate scTv clones with enhanced stability on the surface of yeast, selections can be conducted using either magnetic cell sorting or fluorescence activated cell sorting (FACS). Whereas FACS allows for more precise selection, it can be time-intensive and costly, especially when large libraries are selected. For this reason, our lab has found that magnetic cell sorting is both cost and time efficient for initial selections of stabilized scTv clones. Following 1–2 magnetic sorts, the diversity of the library is decreased and more precise sorts can be performed with FACS if desired. To isolate stabilized scTv clones, it is necessary to use an antibody specific for a conformational epitope. If truncations are noted from assessment of the staining profile of the library population as described in section 3.1.8 at any step of the sort progression, selection can be conducted based on c-myc staining, which allows for the population to contain only full-length scTv fragments. The following two sections describe selection via magnetic beads (section 3.1.10) and FACS (section 3.1.11).
3.1.10 Selection of Stabilized scTvs from Error-Prone Library Using Magnetic Cell Sorting
Here we describe a general magnetic sorting protocol that can be applied to use of either antibodies against Vα or Vβ conformational epitopes or the C-myc epitope tag. This magnetic protocol is adapted from Chao [7].
Induce sufficient yeast cells in order to oversample the library diversity as described in section 3.1.4 (see Note 19).
Determine the concentration of the cells post-induction by taking the OD600. An OD600=1 is equivalent to a concentration of approximately 1 X 107 cells/mL.
Determine the volume of cells needed to sample tenfold the library size for staining and selection. Pellet the required volume of yeast at 1800 xg for 5 minutes in a 50 mL conical tube.
Aspirate supernatant and wash cells one time with 25 mL PBSM.
Spin cells down again at 1800 xg for 5 minutes and aspirate supernatant.
Suspend cells in PBSM containing the appropriate amount of primary staining reagent (see Note 20).
Incubate at 4°C with inversion or on ice with mild agitation every 10–15 minutes to keep cells in suspension for 1 hour.
After incubation, suspend cells to a volume of 25 mL PBSM and spin at 1800 xg for 5 minutes to wash.
Aspirate supernatant and suspend cells in 5 mL of PBSM buffer. Add 200 μL of Ig-linked microbeads to suspension and mix by inversion.
Incubate for 30–45 minutes with inversion or on ice with mild agitation every 10–15 minutes to keep cells in suspension.
Pellet cells at 1800 xg for 5 minutes, aspirate supernatant, and suspend cells in 7 mL of PBSM buffer.
Meanwhile, place an LS column in the magnetic stand assembly and equilibrate with 3 mL cold PBSM buffer. Discard flow through. Apply the 7 mL of cells in PBSM media to the column. Cells that are bound to antibody remain stuck to the column.
Once cells have run through column, remove the LS column from the magnetic assembly and quickly reinsert column into magnetic assembly to allow beads to re-orientate the column.
Wash column in the magnetic assembly with 10–20 mL of PBSM buffer to allow unbound cells trapped in the column to flow through. Discard flow through.
Remove column from magnetic stand assembly and elute cells by adding 7 mL of SD-CAA media and pushing cells into a collection tube using the provided plunger.
Bring volume up to 10 mL and plate serial dilutions as done in step 24–25 of section 3.1.7 to determine the cells captured (see Note 21).
Expand the collected cells in 250 mL of SD-citrate-CAA media
Expand culture for 24–48 hours at 30°C, shaking at 220 rpm until dense (OD600 > 8.0).
Induce cells and stain to assess surface expression of c-myc tag or conformational epitope according to protocols described in sections 3.1.4 and 3.1.8.
If the desired surface expression level has not been achieved, another round of selection can be conducted using the same or different conformational epitope or c-myc antibody, either by another magnetic bead selection or by a FACS selection as described in the next section.
Once the library has been substantially enriched for stabilized scTv fragments, plate an aliquot on SD-CAA plates to isolate single colonies.
Pick approximately ten colonies, expand in SD-CAA media, and induce as described in section 3.1.4.
Stain cultures as described in section 3.1.8 and characterize individual colonies using flow cytometry to select for a highly stable variant(s). Sequences can be determined by harvesting plasmid and sequencing as described in steps #18–21 in section 3.1.3.
Following expansion of yeast, frozen stocks of SD-CAA cultures can be snap frozen on dry ice and stored at −80°C in 10% DMSO indefinitely.
3.1.11 Selection of Stabilized scTvs from Error-Prone Library Using FACS
Sorting via FACS can provide selections of more precise cell populations. To sort error-prone libraries for stabilized scTv clones we describe the following protocol:
Induce sufficient yeast cells in order to oversample the current library diversity as described in section 3.1.4.
Determine the concentration of the cells post-induction by taking the OD600. An OD600=1 is equivalent to approximately 1 X 107 cells/mL.
Determine the volume of cells needed to sample tenfold the current library size for staining and selection. Pellet the required volume of yeast at 1800 xg for 3 minutes in a microfuge tube (see Note 22).
Aspirate supernatant and wash cells one time with 1 mL PBS/1%BSA.
Spin cells down again at 1800 xg for 3 minutes and aspirate supernatant.
Suspend cells in PBS/1% BSA containing the appropriate amount of primary staining reagent (see Note 23).
Incubate on ice for 1 hour.
After incubation, suspend cells to a volume of 1 mL PBS/1% BSA and spin at 1800 xg for 3 minutes to wash.
Suspend cells in a sufficient volume of fluorescently labeled secondary reagent and incubate for 30–45 minutes on ice
Wash cells in 1 mL PBS/1% BSA for a total of 2–3 washes.
Suspend cells to a concentration of 1 x 107 cells in PBS/1% BSA.
Sort cells via FACS, sampling 10-fold the current library diversity and collect in a tube containing SD-CAA media (see Note 24).
Following sort, expand collected cells in 50 mL SD-CAA media and grow until dense (OD600 > 8.0). Depending on the number of cells collected, this can take anywhere from 1–3 days.
Induce and stain to assess surface expression of c-myc tag or conformational epitope according to protocols described in sections 3.1.4 and 3.1.8.
If the desired surface expression level has not been achieved, another round of selection can be conducted using the same or different conformational epitope or c-myc antibody by FACS selection.
Once the library has been substantially enriched for stabilized scTv fragments, plate an aliquot on SD-CAA plates to isolate single colonies.
Pick approximately ten colonies, expand in SD-CAA media, and induce as described in section 3.1.4.
Stain cultures as described in section 3.1.8 and characterize individual colonies using flow cytometry to select for a highly stable variant(s). Sequences are determined by isolating plasmid and sequencing as described in steps #18–21 in section 3.1.3.
Following expansion of yeast, frozen stocks of SD-CAA cultures can be snap frozen on dry ice and stored at −80°C in 10% DMSO indefinitely.
3.2 Peptide-MHC Ligands for Selection and Analysis of T Cell Receptors
A variety of soluble pepMHC ligands with different valencies can be utilized for selections or binding analyses of TCR clones and libraries on the surface of yeast. These reagents include expressed and refolded biotinylated pepMHC monomers, immunoglobulin-linked dimers, and streptavidin-conjugated tetramers (Fig, 4). Multimeric pepMHC reagents, such as immunoglobulin-based dimers [12] and streptavidin-based tetramers [2], are able to increase the detection of weak TCR:pepMHC interactions through avidity effects (see Note 25). For TCRs with higher binding affinities, the use of monomeric reagents is preferred, as multivalent MHC reagents often do not distinguish between several fold changes in mean fluorescence intensity with affinities in the mid-low nanamolar range [46]. Here we describe the generation of pepMHC reagents, and the use of such reagents to stain TCRs on the surface of yeast. Yeast library selection using these reagents is discussed later in section 3.4.1.
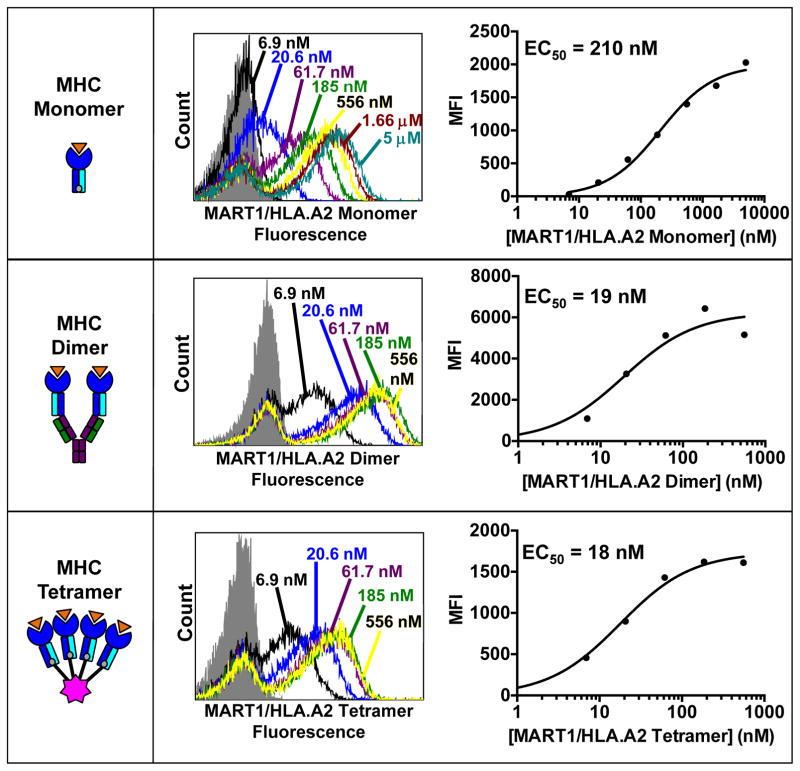
Fig. 4. Peptide-MHC ligands for the selection and analysis of T cell receptors.
(Left panels) Schematics of pepMHC monomers, dimers, and tetramers are shown, where orange represents peptide, blue represents MHC heavy chain, and cyan represents MHC light chain. MHC monomers (top) are biotinylated and are used with a PE-conjugated SA secondary reagent to stain yeast-displayed TCRs. MHC dimers (middle) are fused to an mouse immunoglobulin and are used with a fluorophore-conjugated Goat anti-Mouse IgG secondary antibody to stain yeast-displayed TCRs. MHC tetramers (bottom) are directly bound to a PE-conjugated streptavidin molecule and can be used to stain yeast-displayed TCRs directly. (Center panels) Flow cytometry histograms showing the yeast staining profiles of the T1-S18.45 scTv, which binds to MART-1/HLA-A2, with the three pepMHC reagents at the indicated concentrations. The gray shaded curve represents yeast stained with secondary reagent only. (Right panels) Plots of the mean fluorescent intensity (MFI) obtained from the flow histograms at each concentration. EC50 values obtained from nonlinear regression analysis of the plots are shown in the inset.
3.2.1 Expression of Biotinylated Peptide-MHC Ligands
In this section we describe the expression and purification of soluble HLA-A2 monomers. HLA-A2 monomers can be used directly to stain TCR on the surface of yeast utilizing fluorophore-conjugated streptavidin as a secondary reagent, or they can be pre-incubated with fluorophore-conjugated streptavidin to form tetramers as described in section 3.2.9. HLA-A2 heavy chain and light chain are expressed separately as inclusion bodies, and refolded together in a 3:1 mass ratio along with a peptide of choice (Fig. 5A). The HLA-A2 heavy chain contains a biotinylation substrate peptide (bsp) sequence that allows for in vitro biotinylation. Refolded pepHLA-A2 monomers can be stored in aliquots at −80 °C for up to 12 months. The following four sections describe E. coli growth and induction (section 3.2.2), isolation of inclusion bodies (section 3.2.3), MHC refolds (section 3.2.4), and MHC biotinylation and purification (section 3.2.5). This protocol is adapted from [20, 21].
Fig. 5. Generation of peptide-MHC monomers and tetramers.
(A) Flowchart for the preparation monomers and tetramers. (B) UV-cleavable HLA-A2-binding peptide containing an unnatural amino acid (orange) at position 8 which is cleaved on exposure of UV radiation, allowing for loading of other peptides.
3.2.2 Growth and Induction of E. coli
Transform HLA-A2 heavy chain (with bsp tag) and HLA-A2 light chain (β2 microglobulin) into an E. coli strain optimized for protein expression, such as BL21(DE3), according to the manufacturers protocol and plate on LB plates containing 100 μg/mL ampicillin (see Note 26).
Innoculate single colonies of HLA-A2 heavy and light chain into 3 mL LB cultures with 100 μg/mL ampicillin and grow for 4–8 hours at 37°C with rapid shaking to ensure adequate aeration.
Expand 3 mL cultures into 250 mL LB cultures with 100 μg/mL ampicillin and grow 8–12 hours at 37°C with rapid shaking to ensure adequate aeration.
Subculture into 6 X 6 L flasks each containing 1.5 L pre-warmed LB media each (total of 9 L) with 100 μg/mL ampicillin to an OD600 of 0.1.
Grow cells to an OD600 of approximately 1.0. It is recommended to take a 1 mL sample of the culture at this stage for characterization pre-induction on an SDS-PAGE gel.
To induce expression, add IPTG to a final concentration of 0.7–1.0 mM.
Induce for 2–4 hours at 37°C.
Following induction, it is recommended to take a second 1 mL sample of the induced culture. Cells from both samples, pre-induction and post induction, can be pelleted in a microfuge for 5 min, suspended in SDS sample loading buffer, and ran on an SDS-PAGE gel to verify protein expression (i.e. a prominent band at about 36 kDa for HLA-A2 heavy chain or 12 kDa for HLA-A2 light chain in the post-induction samples compared to the pre-induction sample)
Harvest E. coli by spinning in a centrifuge at 4,200 xg at 4°C, keeping cell pellets on ice from this point onward. Growth can be slowed by transfer of flasks to 4°C when multiple centrifuge runs are required.
3.2.3 Isolation of Inclusion Bodies
Suspend pelleted cells from entire 9 L preparation in 100 mL ice-cold lysis buffer. If desired, suspended cells can be left in lysis buffer on ice overnight.
Filter the suspended cells through a course filter, such as a tea strainer prior to microfluidization (see Note 27).
Prime the microfluidizer in osmotic shock buffer with triton prior to use.
Pass cell solution in lysis buffer through the microfluidizer 3–5 times until the consistency is like that of water.
Centrifuge lysed cell suspension at 11,000 xg at 4°C for 30–60 minutes to pellet inclusion bodies.
Discard supernatant and fully suspend pellet in 40 mL osmotic shock buffer with triton to wash (see Note 28).
Centrifuge at 11,000 xg at 4°C for 30 minutes.
Discard supernatant and repeat wash 3 more times, once more with osmotic shock buffer with triton followed by two times with osmotic shock buffer without triton.
Following the fourth wash, pellets should be white. If the pellets still appear brown, perform additional washes.
Suspend inclusion bodies in 10 mL urea extraction buffer and incubate at room temperature with rotation for 1–2 hours. For heavy chain extractions, we find that guanidine extraction buffer has higher yields.
Centrifuge at 11,000 xg at 4°C for 45 minutes to remove any insoluble matter. Retain supernatant.
Perform a BCA assay or other protein quantification assay to determine protein concentration of the supernatant.
Aliquot 10 mg of protein into microfuge tubes.
Snap freeze tubes on dry ice and store at −80°C.
3.2.4 MHC Refold
Chill 200 mL refold buffer to 4°C. Keep refold mixture at 4°C and stirring at all times.
Dissolve 62 mg oxidized glutathione and 308 mg reduced glutathione in a small volume of ddH2O (<2 mL) and add to the chilled refold buffer
Then, dissolve 7.2 mg PMSF and 12 mg of HLA-A2-restricted peptide of choice in a small volume of DMSO (<2 mL) and add to chilled refold buffer (see Note 29). UV-cleavable peptide (discussed in section 3.2.6) can be used to allow for rapid exchange of peptides after refolding.
Dissolve one 10 mg aliquot of HLA-A2 light chain inclusion bodies and one 10 mg aliquot of HLA-A2 heavy chain inclusion bodies each in 0.5 mL injection buffer, and add inclusion bodies drop by drop into the chilled refold mixture. Inclusion bodies can be incubated at 37°C to assist in dissolution if needed.
In 8–12 hour intervals, add two additional 10 mg aliquots of HLA-A2 heavy chain inclusion bodies dissolved in 0.5 mL injection buffer (total of 30 mg; 3:1 mass ratio heavy to light chain).
Incubate for 8–12 hours following last addition.
Concentrate the ~200 mL of refold mixture down to 30 mL using an Amicon 8400 Stirred Ultrafiltration Cell with a 10 kDa molecular weight cutoff membrane, or similar concentration device.
Collect concentrated refold mixture, and transfer into dialysis tubing (6–8 MWCO) pre-soaked in dialysis buffer. Leave 3–4 cm of dead space to allow for osmotic expansion.
Allow refold mixture to dialyze in 0.5–1 L of cold dialysis buffer while stirring.
Change dialysis buffer 2–3 additional times in 2–4 hours intervals.
Collect dialyzed refold mixture and concentrate entire mixture down to 3–5 mL using an Amicon 10,000 MWCO filter.
Transfer concentrated protein into a chilled tube and centrifuge for 1 minute at maximum speed to pellet any precipitated protein (see Note 30).
3.2.5 MHC Biotinylation and Purification
Using the Avidity Biotinylation Kit, prepare the reaction mixture by combining 7 parts HLA-A2 refold:1 part Biomix A:1 part Biomix B:1 part excess Biotin.
Transfer into 1 mL aliquots, and add 2 uL of provided BirA enzyme to every 1 mL of reaction mixture and incubate for 8–12 hours on ice.
Pool reactions and purify HLA-A2 monomers by size exclusion chromatography on a Superdex 200 10/300 column with HPLC Buffer.
Determine fractions that contain purified HLA-A2 via SDS-PAGE gel. HLA-A2 monomers are ~48 kDa but typically dissociate into individual chains on SDS-PAGE gels (~36 kDa for heavy chain and ~12 kDa for light chain).
Pool fractions and determine protein concentration via BCA assay.
Confirm biotinylation of pepMHC monomers via a gel shift assay. Boil biotinylated pepMHC in SDS-PAGE sample dye for 10 min, cool on ice, and add streptavidin in excess. Incubate for 20 min on ice, then run on an SDS-PAGE gel. Biotinylation is indicated by an upward shift in the molecular weight of streptavidin-conjugated pepMHC monomers. Controls should include unbiotinylated pepMHC and streptavidin only.
Add 1:100 dilution of 0.5 M EDTA solution to a final concentration of 5 μM.
Snap freeze in small 50–100 uL aliquots on dry ice and store stocks at −80°C. Aliquots should be used within 1 week of thawing and should not be refrozen.
3.2.6 UV-Exchanged Peptide-MHC Ligands
To allow for rapid generation of pepMHC detection reagents, we produce soluble HLA-A2 molecules refolded in the presence of a UV-cleavable peptide [52]. In this system, HLA-A2 monomers are refolded in large batch in the presence of the UV-cleavable peptide as described above, which can then be exchanged with individual HLA-A2 restricted peptides of interest by incubating with excess peptide in the presence of UV light (Fig. 5B). In this section, we describe the procedure for the UV-exchange reaction.
Thaw biotinylated HLA-A2 monomers from −80°C storage on ice.
Make 115 uL reaction mixtures of HLA-A2 monomers and 100-fold excess HLA-A2-restricted peptide (from 20 mg/mL DMSO stocks) in PBS in 0.6 mL Eppendorf tubes. Although routinely done at 5 μM, we have successfully performed efficient UV exchange reactions with final HLA-A2 monomer concentrations ranging from 1 to 30 μM.
Put 0.6 mL Eppendorf tubes containing the exchange reaction in an ice box with lids open.
Expose tubes to 30–60 minutes of >350 nm UV-exposure at power level 2000 X 100 μJ/cm2 in 15 minute intervals, vortexing for 1–2 seconds between intervals.
Following UV treatment, allow mixtures to incubate for at least one hour before use. Overnight incubation is recommended.
3.2.7 Monomeric Peptide-MHC Ligands
Monomeric peptide-MHC ligands can be used with a fluorophore-conjugated streptavidin secondary reagent to stain yeast displayed TCRs. Because monomeric reagents lack avidity effects, it is recommended to perform minimal washes following the primary incubation step, especially when fast TCR off-rates are anticipated. Here we describe staining of yeast-displayed TCRs with monomeric pepMHC.
Aliquot 50 μL of induced yeast cells (typically 1–2.5 X 106 cells)
Add 0.5 mL of ice-cold PBS/1% BSA.
Pellet cells at 1,800 Xg at 4°C for 3 minutes.
Aspirate supernatant
Suspend cells in 50 μL primary staining mixture including the monomeric pepHLA-A2 monomer and excess peptide (1 μM) in PBS/1% BSA (see Note 31).
Incubate on ice for 1 hour.
Add 0.5 mL of ice-cold PBS/1% BSA, pellet, and aspirate supernatant to wash.
Suspend cells in 50 uL 1:100 SA-PE, or other desired SA-fluorochrome conjugate, in PBS/1% BSA.
Incubate on ice for 30 minutes.
Add 0.5 mL of ice-cold PBS/1% BSA, pellet, and aspirate supernatant to wash.
Analyze using flow cytometry.
3.2.8 Dimeric Peptide-MHC Ligands
Immunoglobulin-linked pepMHC dimers (commercially available as “DimerX,” BD Biosciences) consist of two pepMHC molecules fused to the antigen binding sites of a mouse IgG. Dimers provide an alternative to performing lengthy MHC expression and refolds, and allow for detection with an anti-mouse IgG secondary antibody rather than a streptavidin conjugate. Here we describe staining of yeast-displayed TCRs with pepMHC dimer.
Load HLA-A2:Ig dimers according to manufacturer’s protocol. Typically we use a 120–160-fold excess of peptide.
Incubate overnight at 37°C.
Aliquot 50 μL of induced yeast cells (typically 1–2.5 X 106 cells)
Add 0.5 mL of ice-cold PBS/1% BSA.
Pellet cells at 1,800 Xg at 4°C for 3 minutes.
Aspirate supernatant.
Suspend cells in 50 μL primary staining mixture including pepHLA-A2:Ig dimers and excess peptide (1 μM) in PBS/1% BSA.
Incubate on ice for 1 hour.
Add 0.5 mL of ice-cold PBS/1% BSA, pellet, and aspirate supernatant to wash.
Suspend cells in 50 uL 1:100 goat anti-mouse F(ab’)2 Alexa 647, or other desired antibody-fluorochrome conjugate, in PBS/1% BSA.
Incubate on ice for 30 minutes.
Add 0.5 mL of ice-cold PBS/1% BSA, pellet, and aspirate supernatant to wash.
Analyze using flow cytometry.
3.2.9 Tetrameric Peptide-MHC Ligands
Biotinylated pepMHC monomers can be used to form multivalent tetramers for the staining of yeast-displayed TCRs. Tetramers are formed by pre-incubation of pepMHC monomers with a streptavidin-fluorochrome, which allows for flow cytometry analysis of yeast in a single staining step. Although we prefer SA-PE, other fluorochrome conjugates, such as SA-APC and SA-FITC, can be used as well. Here we describe tetramer formation and staining of yeast-displayed TCRs. Tetramer protocol is adapted from Altman [2].
Prepare biotinylated pepMHC monomers by refolding with desired peptide or by UV-peptide exchange.
Determine the amount of SA-PE required for a given amount of pepMHC monomers assuming four biotin-binding sites per streptavidin molecule based on the molecular weights of SA-PE (MW ~293 kDa) and pepMHC (MW ~48 kDa).
Add the SA-PE to the pepMHC monomers in 4 equal additions, incubating for 5–10 minutes between additions. Keep tubes on ice.
Allow at least one hour of incubation prior to staining, although overnight incubation is preferred.
Aliquot 50 μL of induced yeast cells (typically 1–2.5 X 106 cells)
Add 0.5 mL of ice-cold PBS/1% BSA.
Pellet cells at 1,800 xg at 4°C for 3 minutes.
Aspirate supernatant.
Suspend cells in 50 μL staining mixture including pepHLA-A2 tetramers and excess peptide (1 μM) in PBS/1% BSA.
Incubate on ice for 1 hour.
Add 0.5 mL of ice-cold PBS/1% BSA, pellet, and aspirate supernatant to wash.
Repeat wash to avoid background.
Analyze using flow cytometry.
3.3 Binding Affinity Analysis of Yeast-Displayed T Cell Receptors
A major advantage of yeast display over other display methods is that it allows for the ability to assess binding affinities of yeast expressing individual TCR clones or mutants directly on the surface of yeast without the need to express large quantities of protein. Here we describe the use of yeast-displayed TCRs to determine TCR binding affinities, and do mutational analyses.
3.3.1 Binding Titrations with Monomeric or Multimeric Peptide-MHC Ligands
Yeast can be titrated with various concentrations of pepMHC ligand, analyzed by flow cytometry, and KD values can be determined as a ligand concentration yielding half-maximum bound. KD values determined by yeast display are typically within 10-fold of SPR determined binding affinities (e.g. Fig. 4 shows titrations and EC50 values of the T1-S18.45 TCR with pepMHC monomers, dimers, and tetramers, whose affinity was measured by SPR as 45 nM) [43]. Values determined by staining with monomeric pepMHC ligands tend to be several-fold higher than affinities determined by SPR, as samples are washed during staining and are read in excess buffer to avoid background. Due to avidity effects, values measured by multimeric pepMHC reagents tend to be several-fold lower. Here we describe how to determine approximate binding affinities of yeast displayed TCRs.
Stain yeast according to the protocol in section 3.2.7, 3.2.8, or 3.2.9 at multiple concentrations of pepMHC, ranging from ~10-fold below to ~20-fold above the anticipated KD if monomers are being used, or ~20-fold below to ~10-fold above if multimeric pepMHC ligands are being used.
Controls should include secondary staining alone, as well as null pepMHC ligands to assess peptide specificity.
Analyze samples by flow cytometry and determine the mean fluorescent intensity (MFI) of each sample.
Adjust data for background by subtracting the MFI of a control stained only with a secondary reagent, such as SA-PE or goat anti-mouse F(ab′)2 Alexa 647, or a null pepMHC ligand from the experimental MFI values.
Plot data of adjusted MFI versus ligand concentration, or as percent maximum bound ligand versus ligand concentration, and determine the KD as the ligand concentration yielding half maximum bound using nonlinear regression analysis as in Fig. 4.
3.3.2 Alanine Scanning Mutagenesis of T Cell Receptors
Alanine scanning mutagenesis of yeast-displayed TCRs can be used to determine the binding energy contributions of residues involved in pepMHC binding (e.g. [8, 43]). CDR loop residues are changed to alanine via site directed mutagenesis and binding is assessed with soluble pepMHC molecules on the surface of yeast via flow cytometry. The fold change in binding affinity due to mutation can be determined by taking the ratio of the ligand concentration yielding half-maximum bound of the mutant TCR to the wild-type TCR. Here we describe the procedure for generating yeast-displayed site-directed alanine mutants and determine changes in binding affinity.
Recover pCT302 plasmid containing TCR of interest to serve as a template for PCR mutagenesis using the Yeast Zymoprep II kit according to the manufacturer’s protocol.
Transform 2.5 μL of Zymoprep DNA into Subcloning Efficiency DH5α Chemically Competent E. coli according to the manufacturers protocol, and plate on LB plates containing 100 μg/mL ampicillin. Grow 8–12 hours at 37°C.
Innoculate a single colony from plates into 3 mL LB cultures with 100 μg/mL ampicillin and grow for 4–8 hours at 37°C with rapid shaking to ensure adequate aeration.
Recover plasmid using QIAprep Spin Miniprep Kit and eluting in 50 μL ddH2O
Sequence using the YRS primer.
Design primers for desired alanine mutants via the QuikChange Primer Design tool available on Aglient Technologies’ website, replacing the amino acid at the position of interest with alanine.
Perform PCR reactions, DpnI digests, and transformation into XL1-Blue Supercompetent Cells using a QuikChange II or QuikChange Lightening Site-Directed Mutagenesis Kit according to the manufacturer’s protocol. Plate reactions on LB plates containing 100 μg/mL ampicillin.
Grow a colony from each QuikChange reaction in 3 mL LB with 100 μg/mL ampicillin and grow for 4–8 hours at 37°C with rapid shaking to ensure adequate aeration.
Rescue plasmids from bacteria via QIAprep Spin Miniprep Kit and elute in 50 μL ddH2O.
Sequence plasmids to confirm mutants have been made successfully.
Transform mutagenized plasmid into EBY100 yeast according to the protocol in section 3.1.3.
Perform binding titrations of mutants and wild type TCRs as described in section 3.3.1.
Determine the ligand concentration yielding half maximum bound using nonlinear regression analysis for mutants, and divide by that of the wild type TCR to determine the fold change in binding affinity.
Experiments with independently induced yeast cells should be repeated in triplicate.
3.4 Fine-Tuning the Affinities of Yeast Displayed T Cell Receptors
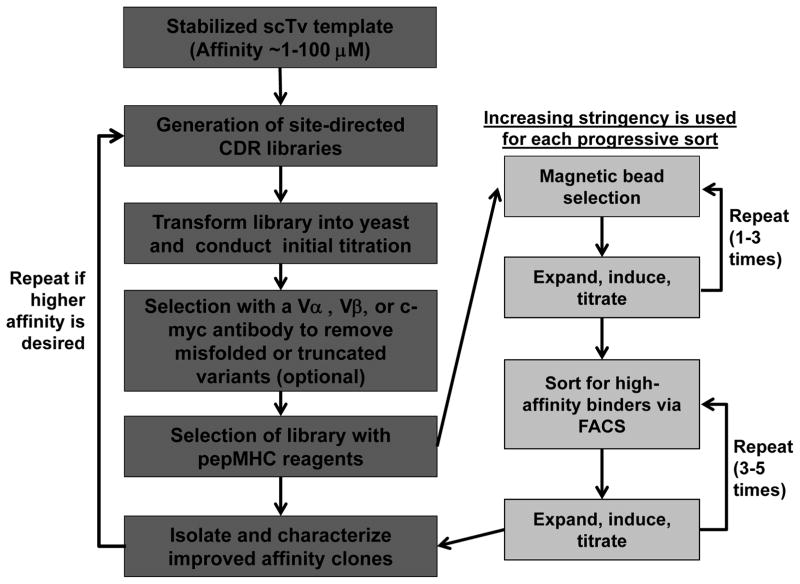
Upon the generation of a stable scTv mutant, site-directed libraries can be generated in the CDR loops of the stabilized TCR in order to select for TCRs with improved binding affinities to pepMHC [25, 26, 43, 56]. Although the affinities of wild type TCRs for pepMHC are low (KD values in the range of 1–100 μM) [48], engineering by yeast display can yield TCRs with over 1,000-fold improvements in binding affinity. Here we describe the affinity maturation of TCRs via site-directed mutagenesis and the modulation of affinities of the resultant high affinity TCRs by using single codon libraries and deep sequencing.
3.4.1 Site-Directed Mutagenesis of T Cell Receptors for Affinity Maturation
In order to increase the affinity of a scTv for its cognate pepMHC ligand, site-directed mutagenesis libraries can be generated in TCR loops which contact the peptide in order to select for variants with improved binding properties, such as affinities or off-rates. As discussed above, the interface of the TCR V regions that contact the pepMHC is comprised of six loops called complementarity determining regions (CDRs), three in each Vα and Vβ. The relative positions of these loops is conserved, such that the regions derived from the germline encoded regions of the TCR (i.e. especially CDR2) are positioned over the MHC helices whereas the diverse regions derived from the junctions of somatically rearranged gene segments (i.e. CDR3) are positioned primarily over the peptide [22, 40]. Because the CDR3 loops are thought to contribute primarily to peptide specificity, we focus our affinity maturation libraries on CDR3 loops, although engineering in other regions have yielded specific high-affinity TCRs [10, 17].
For affinity maturation of stabilized scTvs, site-directed mutagenesis libraries in the CDR loops of a scTv are created by splice by overlap extension (SOE) PCR [27] (Fig. 6). Primers containing degenerate codons in selected positions in CDR loops are used to generate a library of scTv constructs. The use of NNS or NNK nucleotide composition for degenerate codons, where N is any nucleotide (A,T,C, or G), S is either G or C, and K is either G or T, allows for any of the 20 amino acids to be encoded at degenerate positions and limits the potential diversity of the library (see Note 32). The following three sections describe the site-directed mutagenesis of scTvs (section 3.4.2), and the generation and selection of affinity maturation libraries (section 3.4.3 and 3.4.4, respectively).
Fig. 6. A schematic of splicing by overlap extension (SOE) PCR for creating site-directed CDR libraries.
In order to generate site-directed libraries in CDR loops, the N-terminal portion of the gene and C-terminal portion of the gene between standard Splice4L and T7 primers are amplified separately to include a 25–35 nucleotide overlap just prior to the degenerate regions. Pre-SOE #1 is generated via PCR of the Splice4L primer and a reverse primer designed to contain the 25–35 nucleotide overlap and an additional 25–35 nucleotides upstream. Pre-SOE #2 is generated via PCR of a forward primer designed to contain the 25–35 nucleotide overlap followed by the degenerate (NNS or NNK) codons and an addition 25–35 nucleotides downstream. Following separate amplification of PreSOEs #1 and #2, a SOE reaction containing both Pre-SOEs and Splice4L and T7 primers is used to amplify the entire gene product from Splice4L to T7. The resultant PCR product can be used directly to generate libraries via homologous recombination in yeast.
3.4.2 Site-Directed Mutagenesis of scTvs via SOE PCR
Site directed mutagenesis of up to 5 codons in scTv CDR loops is accomplished via SOE PCR. For each library, two primers are designed to be used with standardized Splize4L and T7: The first is a reverse primer used to amplify the N-terminal end of the scTv along with the Splice4L forward primer up to an overlap region just prior to the degenerate codons (i.e. Pre-SOE #1). The second primer is a forward primer used to amplify the C-terminal region of the gene. This primer includes the complement of the overlap sequence used in the first primer followed by the degenerate codons to be used along with the T7 reverse primer (i.e. Pre-SOE #2). Following the amplification of Pre-SOEs #1 and #2, the SOE product can be formed by the amplification of both Pre-SOEs with both Splice4L and T7 primers, The overlap region allows for the Pre-SOEs to prime off one another generating complete full scTv constructs. A diagram of the procedure is depicted in Fig. 6.
Design primers shown in Fig. 6 for SOE PCR using NNS or NNK sequences for sites intended to be degenerate (see Note 33).
-
Assemble 4–8 reactions of each of the following Pre-SOE PCR reactions in PCR tubes. This step generates the N- and C-terminal ends of the scTv that will be extended via the overlap in the net PCR step.
-
5′ Pre-SOE #1:
81.5 μL ddH2O
2.5 μL 10 mM dNTP mix
10 μL Pfu 10X Buffer (provided with polymerase)
2 μL 10 μM Splice 4L 5′ primer
2 μL 10 μM Pre-SOE #1 3′ primer
1 μL 3 ng/uL yeast display plasmid containing template scTv, typically stabilized via error prone mutagenesis as described in section 3.1.5
1 μL Pfu Turbo polymerase
-
3′ Pre-SOE #2:
81.5 μL ddH2O
2.5 μL 10 mM dNTP mix
10 μL PFU 10X Buffer (provided with polymerase)
2 μL 10 μM Pre-SOE #2 5′ primer (which contains degenerate codons)
2 μL 10 μM T7 primer
1 μL 3 ng/uL yeast display plasmid containing template scTv, typically stabilized via error prone mutagenesis as described in section 3.1.5
1 μL PfuTurbo polymerase
-
-
Place the 100 μL reactions in a PCR tubes and run the following PCR program in a thermocycler (see Note 34):
95°C for 1 minute
-
5 cycles of:
95°C for 1 minute
55–65°C for 1 minutes
72°C for 3 minutes
-
25 cycles of:
95°C for 1 minutes
50–60°C for 1 minutes
72°C for 3 minutes
72°C for 5 minutes
4°C forever
Confirm that the PCR products are the correct size by running PCR fragments on a 1% agarose gel (see Note 35).
Combine successful PCR reactions.
Purify and concentrate PCR combined product with the Qiagen PCR Purification kit according to the manufacturer’s protocol. For Qiagen kits, spin columns hold a maximum of 10 μg of plasmid. Calculate the number of columns to use by approximating DNA concentration from the agarose gel ran in step #4 and elute each in >30 μL ddH2O.
-
Set up ten or more identical SOE PCR reactions in PCR tubes as follows:
80.5 μL ddH2O
2.5 μL 10 mM dNTP
10 μL Pfu 10X Buffer
2 μL 10 mM Splice 4L primer
2 μL 10 mM T7 primer
1 μL 5′ Pre-SOE #1 (100 ng/uL)
1 μL 3′ Pre-SOE #2 (100 ng/uL)
1 μL PfuTurbo polymerase
-
Place PCR tubes in thermocycler and run the following program:
95°C for 1 minute
-
5 cycles of:
95°C for 1 minute
55–65°C for 1 minutes
72°C for 3 minutes
-
25 cycles of:
95°C for 1 minutes
50–60°C for 1 minutes
72°C for 3 minutes
72°C for 5 minutes
4°C forever
Confirm that the SOE PCR products are the correct size (~1.2 Kb for standard scTv fragments) by running a small amount of each reaction on a 1% agarose gel.
Combine successful SOE reactions into the same tube.
Purify and concentrate PCR combined product with the Qiagen PCR Purification kit according to the manufacturer’s protocol as in step #6.
Run entire samples on a 1% agarose gel and gel purify the band containing the scTv library with a Qiagen gel extraction kit.
Assess the purity of gel-purified product by running a small volume of eluted SOE on a 1% agarose gel (see Note 36).
Store PCR products at −20°C until ready to pellet DNA with digested pCT302 prior to electroporation.
3.4.3 Generation of Affinity Maturation Libraries
In order to generate scTv site-directed mutagenesis libraries 4 μg of SOE products are precipitated with 1 μg digested pCT302 vector as described in steps #7–11 of section 3.1.6. The same library protocol used to generate error-prone libraries as described in section 3.1.7 is used this time with the SOE product instead of error-prone PCR product. If multiple libraries containing different degenerate regions will be combined for selections, we recommend performing separate electroporations and pooling the libraries according to the diversities determined my limiting dilutions after libraries have been expanded (see Note 37).
3.4.4 Selection of scTvs with Improved Affinities to Peptide-MHC
As performed with error-prone libraries to select for clones with improved stability (as described in section 3.1.5), magnetic bead selections or FACS can be used to select mutants from scTv CDR libraries that bind with improved affinity to pepMHC. In order to assess the initial library population, staining should be performed with antibodies that recognize conformational epitopes of the Vα and/or Vβ chains (section 3.1.8). If significant loss in binding occurs, we recommend performing a single magnetic bead selection using this antibody as was done in section 3.1.10 to eliminate scTv variants which may no longer fold correctly due to destabilizing mutations in CDR loops. In addition selection can be performed with the anti-c-myc antibody to eliminate truncated variants initially or at any stage of the directed evolution process.
Once the library has been excluded of misfolded and/or truncated scTv clones, selections with pepMHC reagents are performed. Typically, in order to assess the library for binding to pepMHC, a small aliquot of the library is induced and titrated with a variety of concentrations of a pepMHC reagent as in section 3.3.1 at each step to determine selection criteria. In initial libraries prior to selection we often see little to no staining and recommend the use of 1–2 magnetic bead selections using higher concentrations of primary pepMHC reagents (see Note 38). Following 1–2 magnetic selections, a positively staining population usually emerges to which more stringent selections can be applied (i.e., staining with low concentrations of pepMHC reagents as described in section 3.2). We recommend the use of a variety of pepMHC and secondary reagents in order to decrease the risk of isolation of non-specific clones (see Note 39). In general we perform a total of 3–7 selections to isolate scTvs with low nanamolar affinities (see Note 40). A general selection workflow is shown in Fig. 7.
Fig. 7. Schematic of an example of the method for selecting higher affinity scTv fragments.
Depicted is a general workflow for isolated high affinity scTv fragments from CDR libraries. Initial searches are conducted via magnetic cell sorting or by using high-avidity reagents such as pepMHC tetramers or dimers. Final searches can be conducted via FACS with more stringent selection conditions, such as pepMHC monomers.
Following isolation of clones that bind with improved affinity to the cognate pepMHC, additional libraries can be generated in other CDR regions using the improved scTv as a template for further library generation and selection. Stabilized and high affinity scTvs can be expressed and refolded at high levels in E. coli and used as soluble reagents. Although not the focus of this review, this protocol has been recently described in detail in [47].
3.4.5 Single-Codon Libraries and Deep Sequencing Approaches to Select T Cell Receptor Mutants with Desired Affinities
Although TCRs engineered by yeast display as described can yield KD values in the low nanamolar range, some applications of TCR engineering only require modest improvements from the wild type TCR binding affinity. In the case of adoptive T cell therapies a strategy is to engineer TCRs just above the threshold where they are independent of the CD8 coreceptor (KD values <1–5 μM), such that they allow for class I-restricted TCRs to mediate the activity of CD4+ T cells [8, 57]. However, it is thought that TCRs with affinities that are much higher (i.e. KD values in low nanamolar range or below) may increase the chance of TCR cross-reactivity and potential adverse effects in adoptive therapies [50]. One strategy to reverse engineer a high affinity TCR for lower affinity to reduce the risk of cross-reactivity involves the generation of single codon libraries at CDR loop positions thought to interact primarily with MHC helices [9, 43]. Libraries are selected by staining yeast populations with a given concentration of pepMHC ligand (typically near the KD of the TCR) followed by FACS. Yeast populations pre- and post-selection are analyzed with deep sequencing approaches to determine which residues at the degenerate position were positively and negatively selected, and at what frequency. The output of such analysis is a range of potential amino acid substitutions from TCRs that span the range from high-affinity of the original affinity-matured TCR to 100-fold lower affinities (Fig. 8).
Fig. 8. Sample data output of deep sequencing of single-codon libraries.
Yeast display libraries are generated at a single position of interest and selected by FACS, and sequence distribution is determined by 454 sequencing. Amino acid residues that were positively and negative selected are indicated on the y-axis as a function of the logarithm of the ratio of the amino acid frequency post-selection divided by the frequency pre-selection. Structurally similar amino acids are grouped together by color.
To guide the selection of CDR positions to vary in degenerate, single-codon libraries, a body of literature identifying evolutionarily conserved amino acids involved in MHC restriction is available [34]. Recently we showed that a key CDR2α position residue, Y51 of the Vα2 region, yielded 10–20 fold reductions in TCR affinity when mutated to alanine. Using the single-codon library approach described here in a high affinity TCR T1-S18.45, which recognizes MART-1/HLA-A2, we were able to determine a range of positively and negatively selected residues spanning the affinity range of the TCR [43]. Similarly, a study of CDR2β residue 46 of the mouse TCR 2C and high-affinity variant m33 of T cells selected both in vitro and in vivo were collectively able to span a 10,000-fold range for binding to SIY/Kb [9]. Here we describe the preparation, selection, and analysis of single codon libraries using this strategy.
Prepare single-codon yeast display libraries via SOE PCR based on NNS or NNK nucleic acid composition as described in section 3.4.2 and 3.4.3.
Perform a single selection with pepMHC at or within 10-fold of the KD of the high affinity template TCR according to the protocol in section 3.4.4. Collect up to the top 10% of all positively staining yeast cells.
Expand pre- and post-selected yeast in SD culture.
Perform plasmid preparations of yeast pre- and post-selection via Yeast Zymoprep II according to the manufacturer’s protocol. We recommend performing 4–6 zymoprep reactions for each library population and combining to obtain adequate diversity.
Design PCR amplicon sets for each library according to the manufacturer’s protocol for the next generation sequencer of choice. As we use Roche/454 Genome Sequencer FLX+, we use the protocol in the “Roche Technical Bulletin: TCB-09013_AmpliconFusionPrimerDesignGuidelines.pdf” available online. Steps 6–15 to follow describe sample preparation, sequencing, and analysis using this system. This technique will tag the DNA from each library with a unique barcode sequence, called a MID sequence. Later, the DNA can be pooled from multiple libraries for sequencing, but the results can be sorted by that MID sequence for analysis.
Synthesize PCR primers designed in step 5 and HPLC purify.
Perform PCR using the FastStart High Fidelity PCR Kit according to the manufacturer’s protocol using 2 ng of template zymoprep DNA and performing 8 amplicons for each reaction. It is crucial not to allow contamination from sample to sample, as this assay is very sensitive.
Following PCR, combine all amplicons from the same library in the same tube and purify PCR product with AMPure XP Beads according to the manufacturer’s protocol (see Note 41).
Quantify DNA with the Qubit dsDNA Quantification Kit according to the manufacturer’s protocol. It is important to have sufficient DNA for eventual sequencing (see step 13). If the DNA amount is low, run more PCR reactions (back to step 7).
Run 1 μL of each amplicon on a DNA 7500 Bioanlyzer chip to assess the quality (i.e. expected fragment size, absence of primer dimers). If multiple bands are seen, amplicons should be run on a 2% agarose gel, gel purified, and analysis should be repeated prior to proceeding.
Pool libraries to be run on the same sequencing chip in equimolar concentration based on the size from bioanalyzer chip and concentration from Qubit quantification. In general we combine up to 6 different library amplicons, each with its own MID, in a single sample to be ran on 1/16th of a chip, and obtain >20,000 total sequences per 1/16th of a chip. Typically, a sample of at least 20 μL with a DNA concentration of at least 5 ng/μL is desired.
Dilute samples to 1 X 106 molecules/μL for sequencing.
In order to determine the sequences of the amplicon pool, emulsion-based clonal amplification and sequencing is performed on a Roche/454 Genome Sequencer FLX+ system for 400 flow cycles according to the manufacturer’s instructions. The library is sequenced on designated regions of a 70x75 PicoTiter Plate with the Roche XL+ sequencing kit, and signal processing and base calling are performed using the bundled 454 Data Analysis Software.
Group the resulting sequences according to MID sequence for each library to be analyzed individually.
Align the gene sequences within each library and focus on the region of interest (i.e., the single degenerate codon region). Translate into amino acid sequences, and count the number of sequences with each amino acid (or stop codon) at the degenerate position. For this analysis we use BioPerl software for alignment and tabulate the raw output with a simple R script that counts the number of sequences for each amino acid at the degenerate position for each library and selection scheme.
Determine the frequency of each amino acid pre- and post-selection by dividing the count for each amino acid (or stop codon) by the total number of sequences in the analysis.
Selection can be represented as the logarithm of the frequency of a particular amino acid or stop codon post-selection divided by the frequency pre-selection and plotted on a bar graph as in Fig. 8.
Acknowledgments
This work was supported by various NIH grants over the years (DMK), including current NIH grants P01 CA097296 (DMK), T32 GM070421 (SNS), and F30 CA180723 (DTH), and a grant from the Melanoma Research Alliance (DMK). We thank Dane Wittrup for helpful discussions, and past and current members of the Kranz lab.
Footnotes
We recommend avoiding EcoRI, NotI, and additional XhoI restriction sites in the codon optimization in order to facilitate cloning into pET expression vectors if expression of scTvs may be desired later on (reviewed previously in [47]).
Alternatively, PCR primers can be designed to amplify the scTv gene. PCR products should be digested with DpnI to remove the E. coli derived template plasmid (which is methylated), then double digested with both NheI and XhoI or BglII according to the manufacturer’s protocol. Following digestion, digest PCR products should be purified via Qiagen PCR Purification Kit prior to ligation into pCT302.
We find that optimal ligation conditions include 25–100 ng of digested pCT302 with a 1:3 or 1:6 molar ratio of digested insert and overnight incubation at 16°C. Controls should include digested scTv and digested pCT302 only to determine background.
Although freshly grown EBY100 yeast are optimal, expanded EBY100 yeast stored at 4°C in YPD liquid culture can be used up to 1 month for transformations.
If yeast grown on plates following transformation do not yield colonies, the expanded SD-CAA liquid cultures can be used to streak additional plates.
Because the purity of the DNA isolated from yeast is often low, the plasmid should be amplified in E. coli prior to sequencing. As an alternative, yeast colony PCR can be used to determine the sequence of the inserted plasmid; however, we typically find that it is beneficial to rescue the pCT302 plasmid in cases where library mutants are being assessed in order to minimize cloning at future steps.
For inductions for general staining of yeast, typically 2 mL cultures are made for induction (containing 2 X 107 cells total) in glass test tubes. When larger libraries are induced, typically enough cells to cover the diversity of the library at least 10-fold are induced in 250–500 mL Erlenmeyer flasks. For smaller cultures, washes are done routinely in 1.7 mL microfuge tubes with 1 mL SG-CAA media. For larger inductions done in 15 mL or 50 mL conical tubes, 5 mL and 25 mL washes, respectively, should be done.
Anitbodies specific for TCR Vα and Vβ are widely commercially available (e.g. Beckman-Coulter, BD Pharmingen, Thermo Scientific). To determine whether an antibody recognizes a conformational epitope, prior to staining, incubate induced yeast at a series of different temperatures (from 4°C to 80°C) for 30 minutes. Conformational epitopes will show a decrease in binding when incubated at elevated temperatures due to irreversible denaturation of the scTv [36]. Staining of yeast with antibodies that bind to linear epitopes will show maintained (or increased) binding even at elevated temperatures.
In order to assure sufficient PCR product is formed, it is recommended to do several 100 μL PCR reactions simultaneously (typically 4–8). Reactions can be combined for future steps.
Typically, each electroporation (requiring 4 μg insert, 1 μg vector, and 50 mL of yeast) will yield 1 X 107 – 1 X 108 transformants. Routinely we generate libraries with 8 electroporations, yielding libraries around 1 X 108 – 1 X 109 in size. If a library of 1 X 109 is required, we recommend doing no fewer than 20 electroporations.
Allow the pellet to dry of 20–30 minutes. To confirm that the pellet is dry the 1.7 Eppendorf tube can be tapped on bench top. If the pellet is fully dried, it will dislodge from the bottom of the tube.
For example, if 8 electroporations are being performed for a library with 2 controls, a 500 mL culture of yeast in a 2 L Erlenmeyer flask should be grown. For cultures greater than 500 mL, multiple flasks should be used to ensure adequate aeration.
The doubling time of EBY100 yeast is 1.5–2 hours.
Be sure to not touch the metal sides of the cuvettes as it decreases the electroporation efficiency. We also recommend wiping the metal sides of the cuvette with kimwipe immediately prior to electroporation.
For example, if the calculated library size is 1 X 108, we recommend expanding no less that 1 X 109 cells.
If the sequences of wild type or the same mutants occur multiple times, it is recommended to repeat error prone PCR and library generation. For future attempts, the amount of template DNA for error prone PCR can be decreased and the DpnI digest of product performed for a longer time.
We recommend diluting anti-HA antibody 1:50 in PBS/1% BSA and anti-c-myc 1:50 in PBS/1% BSA. For antibodies for the specific Vα and Vβ domains, we recommend using a several concentrations initially to determine the optimal staining concentration.
For the anti-HA antibody (Covance), we recommend the use of Alexa Fluor 647 F(ab′)2 Fragment of Goat anti-Mouse IgG (H+L) (Molecular Probes) at a 1:100 dilution. For the c-myc antibody, we recommend the use of Alex Fluor 647 Goat anti-Chicken IgG (H+L) (Molecular Probes) at a 1:100 dilution.
For example, if the diversity of the library is 1 X 108, we recommend inducing no less that 1 X 109 cells (i.e. 100 mL of culture at 1 X 107).
For large amounts of yeast (e.g., 1 X 108 – 1 X 109 cells) it is necessary to stain in a volume to maintain cell suspension (from 0.5–1.0 mL of primary reagent) and ensure the staining reagent is not depleted (approximate 10,000–100,000 of scTv fragments displayed on each yeast cell).
By determining the number of cells captured, the approximate percentage of cells selected from the total library can be used to determine the approximate new diversity. For example, if 10% of total cells are captured in a library with a size of 1 X 108, the new diversity can be estimated to be 1 X 107 following the selection.
Typically we perform FACS selections for libraries that have diversities of 1 X 107 or less. If the diversity is higher, we recommend the use of magnetic bead selections to decrease the library size prior to FACS. If FACS is used for libraries of > 1 X 107, the volume of reagents and washes must be increased to account for the greater cell number.
Typically 100 μL volumes are sufficient for libraries < 1 X 107.
We suggest collecting the top 0.5%–2% of stained cells. If fewer than 2% of cells stain positive, collect the entire positively staining population.
Typically similar FACs titration profiles are seen for dimers and tetramers, although dimers typically show higher staining intensity as they utilize an anti-mouse IgG secondary reagent that amplifies the signal.
It is recommended that test inductions be done prior to large preparations to ensure adequate expression. BL21 stocks can be frozen at −80°C in 10% glycerol and be used to seed future starter cultures.
Other methods besides microfluidization can be used to release inclusion bodies from E. coli, including sonication, French press, and homogenization. Since we use microfluidization, it is described in this protocol.
It is essential that all clumps from pellets be fully suspended. We recommend the use of a small blending tool, such as the Cordless Mini Mixer (NorPro Model # 2273) to assist in suspension.
HLA-A2-restricted peptides, synthesized by solid phase synthesis, can be used in their crude form (>70% purity) without additional purification. We do not see significant binding differences when purified peptides are used.
It is recommended to retain 20 μL of refolded HLA-A2 as a pre-biotinylated sample for SDS-PAGE.
We find it is beneficial to spike in excess peptide into staining mixtures in order to shift the equilibrium of MHC to peptide-bound form, especially in cases where the peptide may be a weak MHC binder.
The theoretical diversity of libraries derived from NNS or NNK libraries can be determined by (4 X 4 X 2)X, where X is the number of degenerate codons. Typically libraries are made degenerate in up to 5 codons, such that the theoretical library size of (4 X 4 X 2)5, or 3.4 X 107, can easily be achieved with the yeast library protocol described in section 3.1.7.
When creating SOE primers it is necessary to have sufficient overlap between primers so Pre-SOE #1 and Pre-SOE #2 can bind to and extend off one another. We usually use SOE primers that contain 25–35 nucleotides of overlap depending on the GC content. Primers ideally will have similar melting temperatures to Splice 4L and T7 primers (approx. 60°C). In addition to having an overlap, we generally design the PreSOE #1 reverse primer to include an additional 25–35 nucleotides upstream to the overlap, and the PreSOE #2 primer to include an addition 25–35 nucleotides downstream to the degenerate residues.
We find that preforming 4 rounds of PCR at a higher annealing temperature allows for better fidelity in future cycles. For our standard protocol, the first four cycles are at an annealing temperature of 60°C and the following 24 cycles are at 55°C; however, one should consider the melting temperature of the primers designed in step #1 when choosing an appropriate annealing temperature.
If the standard PCR protocol has low or no yield, we recommend trying a lower annealing temperature to amplify Pre-SOE DNA. If bands appear as smears, PCR reactions should be repeated at a higher annealing temperature to increase fidelity. If the correct PCR product band is seen but there are extra bands present on the agarose gel (e.g., due to primer dimerization), Pre-SOEs can be alternatively gel purified using the Qiagen Gel Purification kit prior to SOE PCR.
The band containing the scTv gene should not contain a smear. If a smear appears, try repeating SOE reactions at an elevated annealing temperature
Alternatively, SOE PCR products of libraries can be combined in equimolar ratios prior to homologous recombination; however, this does not allow for precise calculation of the representation of each library in the final population.
For example, 5 μM or greater concentrations of monomeric pepMHC (section 3.2.7) used with either SA- or anti-biotin-conjugated beads, or 100 nM or greater concentrations of dimeric pepMHC with anti-Mouse IgG-conjugated beads (section 3.2.8).
Sometimes when consecutive selections are performed with the same secondary reagents, we isolate clones that bind non-specifically to the secondary reagent (e.g., SA-PE, Goat anti-Mouse IgG). We recommend switching secondary reagents every selection in order to minimize the isolation of non-specific binders. For example, if conducting two magnetic sorts with a biotinylated primary reagent, we recommend switching between streptavidin and anti-biotin microbeads.
Generally the number of selections that can be performed relies on the diversity of library. Following each selection, the new diversity of the library can be roughly estimated by the percent capture (i.e. for magnetic selections) or percent sorted (i.e. for FACS selections). Once the estimated diversity is on the order of 1–10, individual clones should be isolated and sequenced to determine whether sufficient diversity is present to warrant further sorting.
It is important to have fairly pure, homogenously-sized PCR product prior to sequencing. Any remaining primer in the mix will significantly impede sequencing results.
References
- 1.Aggen DH, Chervin AS, Insaidoo FK, et al. Identification and engineering of human variable regions that allow expression of stable single-chain T cell receptors. Protein Engineering, Design, & Selection. 2011;24:361–372. doi: 10.1093/protein/gzq113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altman JD, Moss PaH, Goulder PJR, et al. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. [Google Scholar]
- 3.Benatuil L, Perez JM, Belk J, et al. An improved yeast transformation method for the generation of very large human antibody libraries. Protein Eng Des Sel. 2010;23:155–159. doi: 10.1093/protein/gzq002. [DOI] [PubMed] [Google Scholar]
- 4.Boder ET, Wittrup KD. Yeast surface display for directed evolution of protein expression, affinity, and stability. Methods Enzymol. 2000;328:430–444. doi: 10.1016/s0076-6879(00)28410-3. [DOI] [PubMed] [Google Scholar]
- 5.Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotech. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- 6.Chames P, Willemsen RA, Rojas G, et al. TCR-like human antibodies expressed on human CTLs mediate antibody affinity-dependent cytolytic activity. J Immunol. 2002;169:1110–1118. doi: 10.4049/jimmunol.169.2.1110. [DOI] [PubMed] [Google Scholar]
- 7.Chao G, Lau WL, Hackel BJ, et al. Isolating and engineering human antibodies using yeast surface display. Nat Protoc. 2006;1:755–768. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- 8.Chervin AS, Stone JD, Holler PD, et al. The impact of TCR-binding properties and antigen presentation format on T cell responsiveness. J Immunol. 2009;183:1166–1178. doi: 10.4049/jimmunol.0900054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chervin AS, Stone JD, Soto CM, et al. Design of T-cell receptor libraries with diverse binding properties to examine adoptive T-cell responses. Gene Therapy. 2013 doi: 10.1038/gt.2012.80. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chlewicki LK, Holler PD, Monti BC, et al. High-Affinity, Peptide-Specific T Cell Receptors Can Be Generated by Mutations in CDR1, CDR2 or CDR3. J Mol Biol. 2005;346:223–239. doi: 10.1016/j.jmb.2004.11.057. [DOI] [PubMed] [Google Scholar]
- 11.Cohen CJ, Sarig O, Yamano Y, et al. Direct phenotypic analysis of human MHC class I antigen presentation: visualization, quantitation, and in situ detection of human viral epitopes using peptide-specific, MHC-restricted human recombinant antibodies. J Immunol. 2003;170:4349–4361. doi: 10.4049/jimmunol.170.8.4349. [DOI] [PubMed] [Google Scholar]
- 12.Dal Porto J, Johansen TE, Catipovic B, et al. A soluble divalent class I major histocompatibility complex molecule inhibits alloreactive T cells at nanomolar concentrations. Proc Natl Acad Sci U S A. 1993;90:6671–6675. doi: 10.1073/pnas.90.14.6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis MM, Boniface JJ, Reich Z, et al. Ligand recognition by alpha beta T cell receptors. Annu Rev Immunol. 1998;16:523–544. doi: 10.1146/annurev.immunol.16.1.523. [DOI] [PubMed] [Google Scholar]
- 14.Davis MM, Krogsgaard M, Huppa JB, et al. Dynamics of cell surface molecules during T cell recognition. Annu Rev Biochem. 2003;72:717–742. doi: 10.1146/annurev.biochem.72.121801.161625. [DOI] [PubMed] [Google Scholar]
- 15.Davis MM, Krogsgaard M, Huse M, et al. T cells as a self-referential, sensory organ. Annu Rev Immunol. 2007;25:681–695. doi: 10.1146/annurev.immunol.24.021605.090600. [DOI] [PubMed] [Google Scholar]
- 16.Denkberg G, Stronge VS, Zahavi E, et al. Phage display-derived recombinant antibodies with TCR-like specificity against alpha-galactosylceramide and its analogues in complex with human CD1d molecules. Eur J Immunol. 2008;38:829–840. doi: 10.1002/eji.200737518. [DOI] [PubMed] [Google Scholar]
- 17.Dunn SM, Rizkallah PJ, Baston E, et al. Directed evolution of human T cell receptor CDR2 residues by phage display dramatically enhances affinity for cognate peptide-MHC without increasing apparent cross-reactivity. Protein Sci. 2006;15:710–721. doi: 10.1110/ps.051936406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feldhaus M, Siegel R, Opresko L, et al. Flow-cytometric isolation of human antibodies from a nonimmune Saccharomyces cerevisiae surface display library. Nat Biotechnol. 2003;21:163–170. doi: 10.1038/nbt785. [DOI] [PubMed] [Google Scholar]
- 19.Gai SA, Wittrup KD. Yeast surface display for protein engineering and characterization. Curr Opin Struct Biol. 2007;17:467–473. doi: 10.1016/j.sbi.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garboczi DN, Ghosh P, Utz U, et al. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- 21.Garboczi DN, Utz U, Ghosh P, et al. Assembly, specific binding, and crystallization of a human TCR-alphabeta with an antigenic Tax peptide from human T lymphotropic virus type 1 and the class I MHC molecule HLA-A2. J Immunol. 1996;157:5403–5410. [PubMed] [Google Scholar]
- 22.Garcia KC, Adams JJ, Feng D, et al. The molecular basis of TCR germline bias for MHC is surprisingly simple. Nat Immunol. 2009;10:143–147. doi: 10.1038/ni.f.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia KC, Teyton L, Wilson IA. Structural basis of T cell recognition. Annu Rev Immunol. 1999;17:369–397. doi: 10.1146/annurev.immunol.17.1.369. [DOI] [PubMed] [Google Scholar]
- 24.Geitz RD, Schiestl RH, Willems A, et al. Studies on the mechanism of high efficiency transformations of intact yeast cells. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 25.Holler PD, Chlewicki LK, Kranz DM. TCRs with high affinity for foreign pMHC show self-reactivity. Nat Immunol. 2003;4:55–62. doi: 10.1038/ni863. [DOI] [PubMed] [Google Scholar]
- 26.Holler PD, Holman PO, Shusta EV, et al. In vitro evolution of a T cell receptor with high affinity for peptide/MHC. Proc Natl Acad Sci U S A. 2000;97:5387–5392. doi: 10.1073/pnas.080078297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horton RM, Cai ZL, Ho SN, et al. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- 28.Kieke MC, Shusta EV, Boder ET, et al. Selection of functional T cell receptor mutants from a yeast surface-display library. Proc Natl Acad Sci U S A. 1999;96:5651–5656. doi: 10.1073/pnas.96.10.5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kranz DM. Two mechanisms that account for major histocompatibility complex restriction of T cells. F1000 Biology Reports. 2009;1:55–58. doi: 10.3410/B1-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y, Moysey R, Molloy PE, et al. Directed evolution of human T-cell receptors with picomolar affinities by phage display. Nat Biotechnol. 2005;23:349–354. doi: 10.1038/nbt1070. [DOI] [PubMed] [Google Scholar]
- 31.Li Y, Yin Y, Mariuzza RA. Structural and biophysical insights into the role of CD4 and CD8 in T cell activation. Front Immunol. 2013;4:206. doi: 10.3389/fimmu.2013.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liddy N, Bossi G, Adams KJ, et al. Monoclonal TCR-redirected tumor cell killing. Nat Med. 2012;18:980–987. doi: 10.1038/nm.2764. [DOI] [PubMed] [Google Scholar]
- 33.Mareeva T, Martinez-Hackert E, Sykulev Y. How a T cell receptor-like antibody recognizes major histocompatibility complex-bound peptide. J Biol Chem. 2008;283:29053–29059. doi: 10.1074/jbc.M804996200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marrack P, Scott-Browne JP, Dai S, et al. Evolutionarily conserved amino acids that control TCR-MHC interaction. Annu Rev Immunol. 2008;26:171–203. doi: 10.1146/annurev.immunol.26.021607.090421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oates J, Jakobsen BK. ImmTACs: Novel bi-specific agents for targeted cancer therapy. Oncoimmunology. 2013;2:e22891. doi: 10.4161/onci.22891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orr BA, Carr LM, Wittrup KD, et al. Rapid Method for Measuring ScFv Thermal Stability by Yeast Surface Display. Biotechnol Prog. 2003;19:631–638. doi: 10.1021/bp0200797. [DOI] [PubMed] [Google Scholar]
- 37.Richman SA, Aggen DH, Dossett ML, et al. Structural features of T cell receptor variable regions that enhance domain stability and enable expression as single-chain ValphaVbeta fragments. Mol Immunol. 2009;46:902–916. doi: 10.1016/j.molimm.2008.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richman SA, Kranz DM. Display, engineering, and applications of antigen-specific T cell receptors. Biomol Eng. 2007;24:361–373. doi: 10.1016/j.bioeng.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 39.Richman SA, Kranz DM, Stone JD. Biosensor detection systems: engineering stable, high-affinity bioreceptors by yeast surface display. Methods Mol Biol. 2009;504:323–350. doi: 10.1007/978-1-60327-569-9_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 41.Shusta EV, Holler PD, Kieke MC, et al. Directed evolution of a stable scaffold for T-cell receptor engineering. Nat Biotechnol. 2000;18:754–759. doi: 10.1038/77325. [DOI] [PubMed] [Google Scholar]
- 42.Shusta EV, Kieke MC, Parke E, et al. Yeast polypeptide fusion surface display levels predict thermal stability and soluble secretion efficiency. J Mol Biol. 1999;292:949–956. doi: 10.1006/jmbi.1999.3130. [DOI] [PubMed] [Google Scholar]
- 43.Smith SN, Sommermeyer D, Piepenbrink KH, et al. Plasticity in the Contribution of T Cell Receptor Variable Region Residues to Binding of Peptide-HLA-A2 Complexes. J Mol Biol. 2013 doi: 10.1016/j.jmb.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soo Hoo WF, Lacy MJ, Denzin LK, et al. Characterization of a single-chain T cell receptor expressed in E. Coli. Proc Natl Acad Sci. 1992;89:4759–4763. doi: 10.1073/pnas.89.10.4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soto CM, Stone JD, Chervin AS, et al. MHC-class I-restricted CD4 T cells: a nanomolar affinity TCR has improved anti-tumor efficacy in vivo compared to the micromolar wild-type TCR. Cancer Immunol Immunother. 2013;62:359–369. doi: 10.1007/s00262-012-1336-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stone JD, Artyomov MN, Chervin AS, et al. Interaction of streptavidin-based peptide-MHC oligomers (tetramers) with cell-surface TCRs. J Immunol. 2011;187:6281–6290. doi: 10.4049/jimmunol.1101734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stone JD, Chervin AS, Aggen DH, et al. T cell receptor engineering. Methods Enzymol. 2012;503:189–222. doi: 10.1016/B978-0-12-396962-0.00008-2. [DOI] [PubMed] [Google Scholar]
- 48.Stone JD, Chervin AS, Kranz DM. T-cell receptor binding affinities and kinetics: impact on T-cell activity and specificity. Immunology. 2009;126:165–176. doi: 10.1111/j.1365-2567.2008.03015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stone JD, Chervin AS, Schreiber H, et al. Design and characterization of a protein superagonist of IL-15 fused with IL-15Ralpha and a high-affinity T cell receptor. Biotechnol Prog. 2012;28:1588–1597. doi: 10.1002/btpr.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stone JD, Kranz DM. Role of T cell receptor affinity in the efficacy and specificity of adoptive T cell therapies. Front Immunol. 2013;4:244. doi: 10.3389/fimmu.2013.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stone JD, Yin Y, Mo M, et al., editors. Engineering High-Affinity T Cell Receptor/Cytokine Fusions for Therapeutic Targeting. InTech; 2012. [Google Scholar]
- 52.Toebes M, Coccoris M, Bins A, et al. Design and use of conditional MHC class I ligands. Nat Med. 2006;12:246–251. doi: 10.1038/nm1360. [DOI] [PubMed] [Google Scholar]
- 53.Van Der Merwe PA, Davis SJ. Molecular interactions mediating T cell antigen recognition. Annu Rev Immunol. 2003;21:659–684. doi: 10.1146/annurev.immunol.21.120601.141036. [DOI] [PubMed] [Google Scholar]
- 54.Van Laethem F, Tikhonova AN, Pobezinsky LA, et al. Lck availability during thymic selection determines the recognition specificity of the T cell repertoire. Cell. 2013;154:1326–1341. doi: 10.1016/j.cell.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Verma B, Neethling FA, Caseltine S, et al. TCR mimic monoclonal antibody targets a specific peptide/HLA class I complex and significantly impedes tumor growth in vivo using breast cancer models. J Immunol. 2010;184:2156–2165. doi: 10.4049/jimmunol.0902414. [DOI] [PubMed] [Google Scholar]
- 56.Weber KS, Donermeyer DL, Allen PM, et al. Class II-restricted T cell receptor engineered in vitro for higher affinity retains peptide specificity and function. Proc Natl Acad Sci U S A. 2005;102:19033–19038. doi: 10.1073/pnas.0507554102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao Y, Bennett AD, Zheng Z, et al. High-affinity TCRs generated by phage display provide CD4+ T cells with the ability to recognize and kill tumor cell lines. J Immunol. 2007;179:5845–5854. doi: 10.4049/jimmunol.179.9.5845. [DOI] [PMC free article] [PubMed] [Google Scholar]