Abstract
NAD(P)H:quinone oxidoreductase 1 (NQO1) regulates the stability of the tumor suppressor WT p53. NQO1 binds and stabilizes WT p53, whereas NQO1 inhibitors including dicoumarol and various other coumarins and flavones induce ubiquitin-independent proteasomal p53 degradation and thus inhibit p53-induced apoptosis. Here, we show that curcumin, a natural phenolic compound found in the spice turmeric, induced ubiquitin-independent degradation of WT p53 and inhibited p53-induced apoptosis in normal thymocytes and myeloid leukemic cells. Like dicoumarol, curcumin inhibited the activity of recombinant NQO1 in vitro, inhibited the activity of endogenous cellular NQO1 in vivo, and dissociated NQO1-WT p53 complexes. Neither dicoumarol nor curcumin dissociated the complexes of NQO1 and the human cancer hot-spot p53 R273H mutant and therefore did not induce degradation of this mutant. NQO1 knockdown by small-interfering RNA induced degradation of both WT p53 and the p53 R273H mutant. The results indicate that curcumin induces p53 degradation and inhibits p53-induced apoptosis by an NQO1-dependent pathway.
Keywords: NQO1-dependent pathway, disruption of NQO1–p53 binding, cancer hot-spot p53 mutant, p53-induced apoptosis
Wild-type p53 is a labile tumor suppressor protein whose cellular level is mainly regulated by its rate of proteasomal degradation (reviewed in ref. 1). WT p53 protein can induce growth arrest (1–4) or apoptosis (5–8) and can thus prevent accumulation of DNA-damaged cells that could lead to the development of cancer (reviewed in refs. 1, 9, and 10). This tumor-suppressing activity of WT p53 is abolished by mutations in the p53 gene that occur in >50% of human cancers (1, 11, 12), and the mutant proteins often accumulate in the cancer cells (13). Degradation of p53 is mediated by two alternative pathways, ubiquitin-independent (14, 15) or ubiquitin-dependent. Ubiquitin-dependent degradation of p53 is mediated by different ubiquitin E3 ligases, including Mdm2 (16, 17), Pirh2 (18), and Cop1 (19), that bind and ubiquitinate p53, targeting it to degradation via the 26S proteasome. The ubiquitin-independent pathway is regulated by NAD(P)H:quinone oxidoreductase 1 (NQO1) (14, 15, 20, 21) and is mediated via the 20S proteasome (22). We have shown that NQO1 stabilizes p53, so that NQO1 overexpression increases p53 levels (20, 21), whereas NQO1 knockdown by small interfering RNA (siRNA) decreases the level of p53 (14). NQO1 binds to p53 (23, 24) and dicoumarol, an inhibitor of NQO1 activity that competes with NAD(P)H for binding to NQO1, disrupts NQO1–p53 binding (23), and induces ubiquitin-independent proteasomal degradation of p53 (14, 15, 20, 21, 23). Various other coumarins and flavones that also compete with NAD(P)H for binding to NQO1 also induce ubiquitin-independent p53 degradation (23). However, unlike WT p53 and some p53 mutants, the most frequent hot-spot human cancer p53 mutants (11, 12) were resistant to dicoumarol-induced degradation by increased binding to NQO1 (23).
Curcumin, a natural phenolic compound found in the spice turmeric that gives the yellow color and flavor to curry, has been reported to have many biological effects, including antioxidant and antiinflammatory activities, as well as a chemopreventive effect on chemical carcinogen-induced colon tumors in rats (reviewed in ref. 25). The effects reported for curcumin include induced accumulation of WT p53 and induction of apoptosis in the human breast cancer cell lines MCF7 and TR9–7 (26, 27) and human neuroblastoma cell lines (28). However, in etoposide-treated human RKO colorectal cancer cell line, curcumin was reported to inhibit accumulation of Ser-15 phosphorylated WT p53 and to inhibit induction of G1 growth arrest (29). In view of these apparently contradictory reported effects of curcumin on p53 accumulation and function in different human cancer cell lines, we have studied the effect of curcumin on the stability of p53 in different cells. Here, we show that curcumin inhibits NQO1 activity both in vitro and in vivo. Like dicoumarol, curcumin disrupted the binding of NQO1 to WT p53, induced ubiquitin-independent degradation of p53, and inhibited p53-mediated apoptosis in normal thymocytes and myeloid leukemic cells. Curcumin, like dicoumarol, did not disrupt the binding of NQO1 to the human cancer hot-spot p53 R273H mutant. Thus, an NQO1-dependent pathway regulates induction of p53 degradation and inhibition of p53-induced apoptosis by curcumin.
Materials and Methods
Cells and Cell Culture. Normal thymocytes were obtained from the thymus of 2- to 3-mo-old CD1 mice. The following cell lines were used: p53-null HCT116 human colon carcinoma cells (30), mouse M1 myeloid leukemic cells stably transfected with the mouse p53 mutant A135V (5), and A31N-ts20 cells that have a temperature-sensitive E1 ubiquitin-activating enzyme, which is inactivated at 39°C (31). The p53 mutant A135V is a temperature-sensitive protein that behaves like a tumor-suppressing WT p53 at 32°C and like a mutant p53 at 37°C (32). When cultured at 32°C, the M1 cells carrying A135V p53 (M1-t-p53 cells) undergo apoptosis (5, 10). Normal thymocytes and M1-t-p53 cells were cultured in DMEM containing 100 units/ml penicillin and 100 mg/ml streptomycin supplemented with 10% heat-inactivated (56°C for 30 min) horse serum and cultured at 37°C in a humidified incubator with 10% CO2. HCT116 and A31N-ts20 cells were cultured in DMEM and 10% FBS at 37°C and 32°C, respectively, in a humidified incubator with 5.6% CO2.
Compounds. Dicoumarol (Sigma) was dissolved in 0.13 M NaOH, curcumin (Sigma) in ethanol, NADH and BSA (Sigma) in water, and geldanamycin (Calbiochem) in DMSO.
Plasmids and Transfection. The following plasmids were used: pRc/CMV human WT p53, pRc/CMV human p53 mutant R273H, and pSUPER-NQO1 siRNA (14). Transfection of HCT116 p53-null cells was carried out by the calcium phosphate method, followed by a 10% glycerol shock for 30 sec, at 7 h after transfection. Transfection of A31N-ts20 cells was performed at 32°C by using the lipofectamine reagent (Life Technologies, Grand Island, NY) according to manufacturer's protocol. Cells were transferred to 39°C at 4 h after transfection, and cell extracts were prepared 24 after transfection.
Apoptosis and Cell Viability Assays. Apoptosis in normal thymocytes was induced by γ irradiation at 4 Gy, followed by incubation in culture at 37°C for 5 h. M1-t-p53 myeloid leukemic cells were induced to undergo apoptosis by incubation at 32°C for 23 h. The percentages of apoptosis and cell viability were determined as described (15).
Immunoblot Analysis. Cell extracts and immunoblot analysis were carried out as described (20). The following Abs were used: monoclonal anti-p53 (Pab 240 and Pab 1801), goat anti-NQO1, rabbit anti-IκB (all from Santa Cruz Biotechnology), and monoclonal anti-Actin (Sigma).
Binding of NQO1 to p53. In vitro coimmunoprecipitation was carried out with in vitro reticulocyte lysate-translated [35S]methionine-labeled WT p53 or R273H mutant p53 and recombinant purified NQO1 incubated in Nonidet P-40 buffer (100 mM Tris·HCl, pH 7.5/150 mM NaCl/2 mM EDTA/1 mM NADH/1% Nonidet P-40) for 16 h at 4°C. Protein A/G beads (Santa Cruz Biotechnology) that were preincubated for 1 h at 4°C with mouse anti-p53 mAb (Pab 1801) were added to the mixture, and samples were incubated for an additional 2 h at 4°C. The beads were collected by centrifugation, washed twice with Nonidet P-40 buffer, and washed twice more with either Nonidet P-40 buffer or Nonidet P-40 buffer containing 50 μM dicoumarol or 40 μM curcumin. The beads were then mixed with Laemmli sample buffer and heated at 95°C for 5 min, and samples were loaded on a 12.5% SDS/PAGE gel.
Purification of Recombinant NQO1. Recombinant NQO1 was purified by the Israel Structural Proteomics Center at the Weizmann Institute as described below. The pET28-His-tobacco etch virus (TEV)-NQO1 expression vector (Novagene) was expressed in bacteria (BL 21 Escherichia coli). The bacteria were sonicated in 50 mM Tris·HCl (pH 7.5), 150 mM NaCl, and 1 mM PMSF, and soluble His-TEV-NQO1 was purified by using Ni-NTA column (HiTrap chelating HP, Amersham Pharmacia) followed by gel-filtration chromatography (HiLoad 16/60 Superdex 200, Amersham Pharmacia). Purified His-TEV-NQO1 was cleaved by TEV protease (Molecular Cell Biology, Cardiff University, Cardiff, U.K.), and His-TEV was removed by binding to Ni-NTA column.
NQO1 Activity Assay. NQO1 activity was measured as described in ref. 33 by following the decrease in NADH absorbance at 340 nm by using Sunrise remote control spectrophotometer (Tecan). The reaction mixture in a final volume of 200 μl contained 25 mM Tris·HCl (pH 7.5), 0.01% Tween 20, 0.7 mg/ml BSA (pH 7.4), 40 μM menadione, 5 μM FAD, 200 μM NADH, and 50 ng of recombinant NQO1 or cell extract. Multiple reactions were carried out by using a 96-well plate, and measurements were made at 5-sec intervals over a time period of 10 min. To determine NQO1 activity in cell extracts (untreated cells or cells that were incubated for 5 h with dicoumarol or curcumin), cells were washed with PBS and extracted by sonication in lysis buffer (25 mM Tris, pH 7.5/1 mM EDTA/0.1 mM DTT), and the decrease in NADH absorbance was measured in the absence and presence of 10 μM dicoumarol to measure specific NQO1 activity.
Results
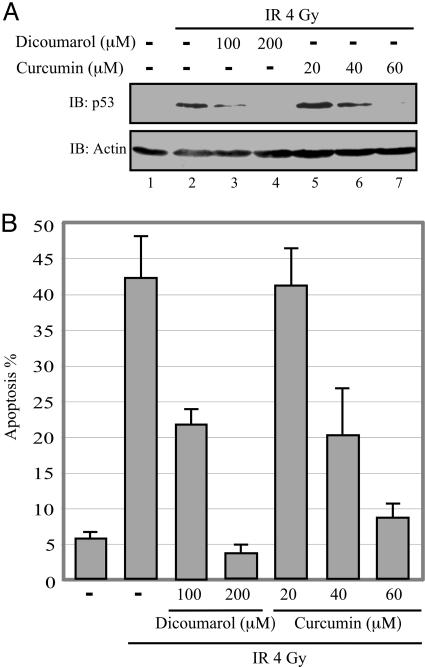
Curcumin Inhibits WT p53 Accumulation and p53-Induced Apoptosis. It has been reported that curcumin may either promote (26–28) or inhibit (29) p53 accumulation in different cancer cell lines. In view of these apparent contradictory effects of curcumin on accumulation of p53 in various cancer cell lines, we examined the effect of curcumin on accumulation of WT p53 and induction of p53-dependent apoptosis in γ-irradiated normal mouse thymocytes. The results show that curcumin inhibited p53 accumulation (Fig. 1A) and induction of p53-dependent apoptosis (Fig. 1B) in a dose-dependent manner, with the strongest effect at 60 μM. Higher concentrations of curcumin were toxic to the thymocytes. The degree of inhibition of p53 accumulation with 60 μM curcumin was similar to that with 200 μM dicoumarol (Fig. 1 A).
Fig. 1.
Curcumin inhibits p53 accumulation and p53-dependent apoptosis in γ-irradiated normal thymocytes. (A) Nonirradiated or 4-Gy γ-irradiated (IR) thymocytes were cultured for 5 h without (–) or with the indicated concentrations of dicoumarol or curcumin. p53 level was determined by Western blotting by using Pab 240 anti-p53 mAb (IB: p53). The blots were then stripped and reprobed with anti-actin mAb (IB: Actin). (B) The percentage of apoptotic cells was analyzed on May-Grünwald Giemsa-stained cytospin preparations by counting 400 cells as described in ref. 20.
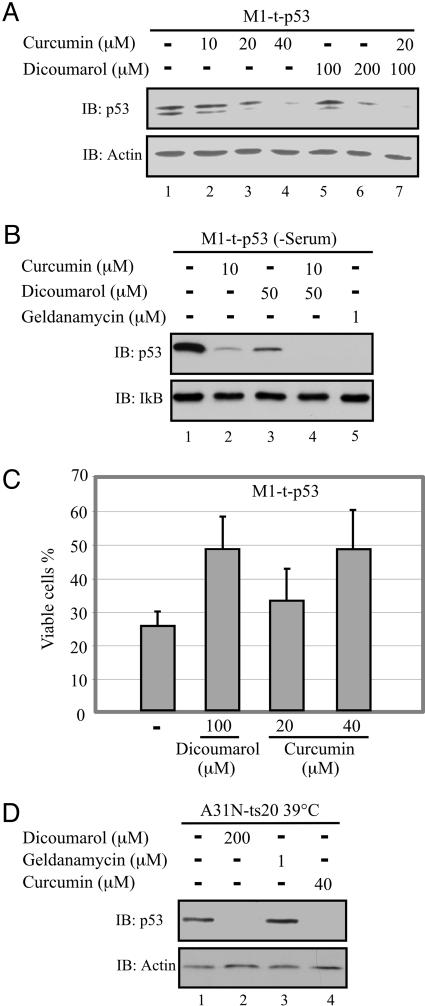
M1-t-p53 myeloid leukemic cells undergo apoptosis when cultured at 32°C (5, 10), when the A135V p53 protein behaves like WT p53. Analysis of the p53 level in M1-t-p53 cells cultured at 32°C for 5 h showed that both curcumin and dicoumarol decreased the level of p53 in a dose-dependent manner (Fig. 2A). Addition of suboptimal concentrations of curcumin (20 μM) and dicoumarol (100 μM) together showed an additive effect on the decrease in p53 level. Like dicoumarol (20, 23), curcumin also induced degradation of the V135A p53 protein in its mutant form when the cells were cultured at 37°C. Both dicoumarol (34) and curcumin (35, 36) bind to serum albumin, so that only a fraction of the added compounds might be available to the cells. When cells were cultured in DMEM without serum, 4-fold lower concentrations of curcumin (10 μM) and dicoumarol (50 μM) were required to induce p53 degradation compared with cultures with serum (Fig. 2B). When both compounds were added together, p53 was further degraded to the same degree as with geldanamycin (Fig. 2B), an inhibitor of the heat shock protein 90 (hsp90) that stabilizes p53. Analysis of p53-induced apoptosis in M1-t-p53 cells has shown that, like dicoumarol (15, 20, 23), curcumin-induced p53 degradation resulted in decreased apoptosis, so that the percentage of viable cells after 23 h at 32°C increased from ≈25% without curcumin to ≈50% with 40 μM curcumin (1Fig. 2C).
Fig. 2.
Curcumin induces ubiquitin-independent degradation of p53 and inhibits p53-induced apoptosis in leukemic cells. (A) M1-t-p53 myeloid leukemic cells were cultured for 5 h at 32°C without (–) or with the indicated concentrations of curcumin or dicoumarol and p53 level was determined. (B) M1-t-p53 cells were cultured for 5 h at 32°C in serum-free medium without (–) or with the indicated concentrations of curcumin, dicoumarol, or geldanamycin, and the p53 level was determined. (C) M1-t-p53 cells were cultured at 32°C without (–) or with the indicated concentrations of dicoumarol or curcumin, and the percentage of viable cells was determined after 23 h as described in ref. 20. (D) A31N-ts20 cells were cultured at 39°C for 24 h and then cultured for an additional 5 h at 39°C in the absence (–) or presence of the indicated concentrations of dicoumarol, geldanamycin, or curcumin, and p53 level was determined.
Curcumin Induces Ubiquitin-Independent Degradation of WT p53. The A31N-ts20 cell line has a temperature-sensitive E1 ubiquitin-activating enzyme that is inactivated at 39°C and causes accumulation of p53 (31). We have confirmed the lack of p53 ubiquitination in these cells at 39°C by showing that overexpression at 39°C of Mdm2, the E3 ubiquitin ligase of p53, did not promote ubiquitin-dependent p53 degradation (14). As shown in ref. 14, the accumulated p53 in A31N-ts20 cells at 39°C was efficiently degraded by dicoumarol (Fig. 2D). Curcumin also induced p53 degradation at 39°C, whereas the hsp90 inhibitor geldanamycin did not (Fig. 2D). Because hsp90 protects its client proteins, including p53, against ubiquitin-dependent degradation and this protection is prevented by geldanamycin (37), our results further confirm the lack of ubiquitination in these cells at 39°C and indicate that curcumin like dicoumarol induced ubiquitin-independent p53 degradation.
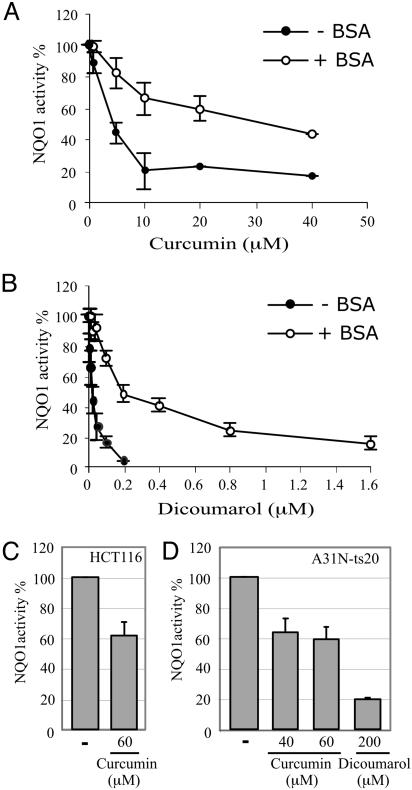
Curcumin Is an Inhibitor of NQO1 Activity. The results described above show that curcumin, like dicoumarol, promotes p53 degradation by an ubiquitin-independent mechanism and protects cells against p53-induced apoptosis. This similarity raised the possibility that curcumin, like dicoumarol, inhibits NQO1 activity. We measured inhibition of NQO1 activity in an in vitro assay using purified recombinant NQO1. Curcumin inhibited NQO1 activity in the in vitro assay, with 50% inhibition at ≈5 μM (Fig. 3A, –BSA), compared with 50% inhibition with dicoumarol at ≈20 nM (Fig. 3B, –BSA). Both curcumin and dicoumarol can bind to serum albumin (34–36). Addition of BSA reduced 8- to 10-fold the NQO1 inhibitory effect of both compounds, and 50% inhibition of NQO1 activity with curcumin and dicoumarol was obtained at ≈40 μM and 200 nM, respectively (Fig. 3 A and B). Thus, curcumin inhibited NQO1 activity, and in this in vitro assay, curcumin was less efficient than dicoumarol as an inhibitor of NQO1 activity (Fig. 3).
Fig. 3.
Curcumin inhibits NQO1 enzymatic activity in vitro and in cells. Enzymatic activity of recombinant NQO1 was determined in the absence (–) or presence (+) of BSA without (–) or with the indicated concentrations of curcumin (A) or dicoumarol (B). NQO1 activity was determined in cell extracts prepared from HCT116 (C) and A31N-ts20 (D) cells cultured for 5 h at 37°C and 32°C, respectively, without (–) or with the indicated concentrations of curcumin or dicoumarol.
We also assayed endogenous cellular NQO1 activity in vivo in extracts prepared from cells cultured in the absence or presence of curcumin or dicoumarol. Using HCT116 and A31N-ts20 cells, we found that 40–60 μM curcumin inhibited endogenous cellular NQO1 activity by 40% (Fig. 3 C and D), and 200 μM dicoumarol inhibited endogenous cellular NQO1 activity by 80% (Fig. 3D). Similar concentrations of both compounds were also required to induce p53 degradation and to inhibit p53-mediated apoptosis (Figs. 1 and 2). These results indicate that the ability of the natural phenolic compound curcumin to induce p53 degradation and inhibit p53-induced apoptosis is related to its ability to inhibit NQO1 activity.
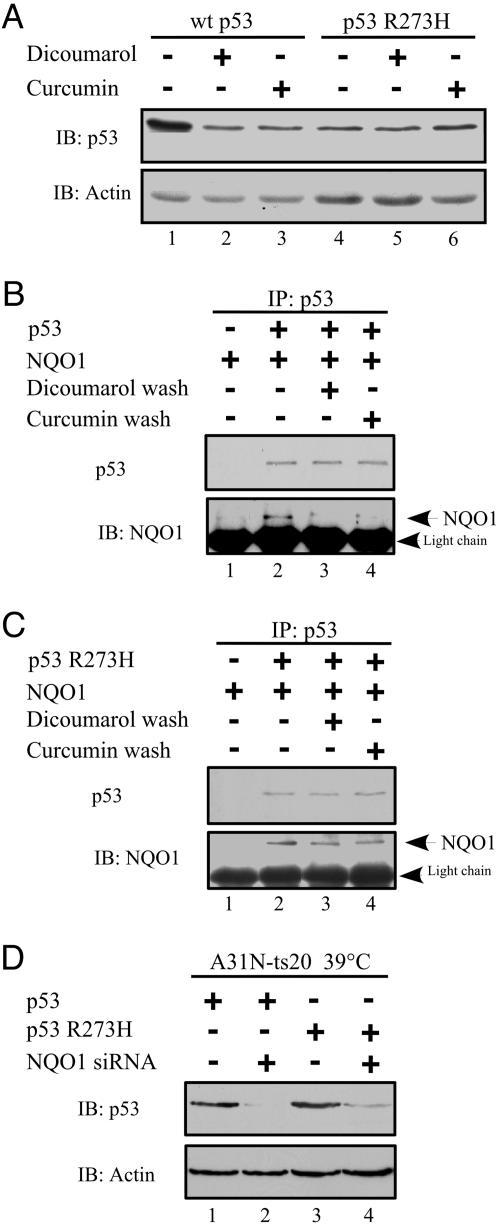
Resistance of the Human Cancer Hot-Spot p53 R273H Mutant to Degradation by Curcumin. Dicoumarol induces degradation of WT p53 and some mutant forms of p53 (14, 15, 20, 21). However, several hot-spot p53 mutants that accumulate in human cancer cells showed a stronger binding to NQO1 and were resistant to degradation by dicoumarol (23). We determined whether a dicoumarol-resistant hot-spot p53 mutant is also resistant to curcumin-mediated degradation. The p53-null HCT116 human colon carcinoma cells were transfected with human WT p53 or the hot-spot p53 R273H mutant. Although WT p53 was degraded to a similar degree by dicoumarol and curcumin, neither curcumin nor dicoumarol induced degradation of the p53 R273H mutant protein (Fig. 4A).
Fig. 4.
Regulation of degradation of WT p53 and the hot-spot p53 R273H mutant. (A) p53-null HCT116 cells were transfected with pRc/CMV human WT p53 or p53 R273H mutant. After transfection (24 h), cells were cultured for 5 h without (–) or with 300 μM dicoumarol or 60 μM curcumin. Cell extracts were prepared, and the p53 level was determined. In vitro [35S]-labeled WT p53 (B) or p53 R273H mutant (C) was incubated alone (–) or together with (+) recombinant NQO1 in the presence of 1 mM NADH. p53 was immunoprecipitated with mouse anti-human p53 Ab (Pab 1801) (IP: p53). The beads were washed without (–) or with (+)50 μM dicoumarol or 40 μM curcumin before elution and separation on SDS/PAGE. p53 was detected by autoradiography, and NQO1 was detected by immunoblotting with goat anti-NQO1 Ab (IB: NQO1). (D) A31N-ts20 cells were transfected at 32°C with pRc/CMV human WT p53 or p53 R273H mutant without or with pSUPER-NQO1, and 4 h later cells were transferred to 39°C. The level of WT p53 and p53 R273H mutant proteins was determined 24 h after transfection by immunoblotting with anti-human p53 (Pab 1801) Ab.
It has been shown that NQO1 binds to WT p53 (23, 24) and that this binding is increased in the presence of NADH (22) and is inhibited by dicoumarol (22, 23), which competes with NADH for NQO1 binding. Binding of NQO1 to the human cancer hot-spot p53 R273H mutant was ≈3-fold higher compared to its binding to WT p53 (23). Analysis of the effect of dicoumarol and curcumin on p53-NQO1 complexes showed that both compounds effectively dissociated NQO1-WT p53 complexes (Fig. 4B) but had little effect on the NQO1–p53 R273H mutant complexes (Fig. 4C). These results again show the similarity between the effects of curcumin and dicoumarol and suggest that curcumin, like dicoumarol, may compete with NADH for binding to NQO1. When NQO1 binding to p53 was reduced by knocking down NQO1 levels with siRNA, both WT p53 and the p53 R273H mutant were degraded under conditions of defective ubiquitination (Fig. 4D). Thus, the failure of dicoumarol and curcumin to effectively dissociate NQO1–p53 R273H complexes can explain the resistance of the p53 R273H mutant to degradation by both of these compounds.
Discussion
Curcumin, a natural phenolic compound found in the spice turmeric, has been reported to have many biological effects (reviewed in ref. 25). Such multiplicity of activities could result from its highly reactive α,β-unsaturated ketone group that can bind covalently to many proteins including the transcription factor NFκB and inhibit their function (38). Here, we report that curcumin induces p53 degradation and, thus, inhibits p53-induced apoptosis. Curcumin induced degradation of p53 under conditions of defective ubiquitination, like several NQO1 inhibitors including dicoumarol (14, 15, 20, 21, 23). Curcumin was also shown to induce degradation of different proteins, including cyclin D1 (39), p185ErbB2 (40, 41), c-Jun (42), C/EBPα, and C/EBPβ (43). It will be interesting to determine whether these proteins are degraded by curcumin also by an ubiquitin-independent mechanism.
NQO1 is a flavoenzyme that catalyzes two electron reduction of quinones and some other electrophiles by using NAD(P)H as an electron donor. We found that curcumin inhibits NQO1 activity. Like dicoumarol that competes with NAD(P)H for binding to NQO1, curcumin also disrupted WT p53-NQO1 complexes and induced p53 degradation. Curcumin inhibits the activity of thioredoxin reductase (29), another flavoprotein that uses NADPH as an electron donor (44). These findings suggest that curcumin may displace NAD(P)H from NQO1 and possibly also from other NAD(P)H using enzymes and, thus, inhibit their activity.
Our studies have shown that curcumin and dicoumarol destabilize WT p53 by inhibiting NQO1 activity and promoting the dissociation of p53–NQO1 complexes. Curcumin and dicoumarol might also affect p53–NQO1 interaction by causing changes in the p53 molecule itself. Many of the cellular functions of p53 are regulated by various redox regulating p53-interacting proteins including thioredoxin (reviewed in ref. 15). Curcumin inhibits the activity of thioredoxin reductase, resulting in reduced binding of p53 to DNA, possibly due to changes in p53 folding (29). In addition to inhibiting NQO1 activity, curcumin might, therefore, also change the folding of p53 in a manner that disrupts NQO1–p53 complex formation. It was recently reported that NAD+ and NADH bind to p53 tetramers and change their conformation (45). NADH also induces a conformational change in NQO1 (46) and increases p53–NQO1 interaction (22). Thus, NAD(P)H could affect both p53 and NQO1 to promote their interaction and p53 stabilization. We have shown that NQO1 is physically associated with the 20S proteasomes and this association prevents the degradation of p53 that is bound to NQO1 (22). Thus, by displacing NAD(P)H and dissociating NQO1–p53 complexes, curcumin and dicoumarol promote the ubiquitin-independent 20S proteasome-mediated degradation of p53. We have previously shown that binding of NQO1 to the human cancer hot-spot p53 R273H mutant was ≈3-fold higher compared to its binding to WT p53 (23). The failure of dicoumarol and curcumin to effectively dissociate NQO1–p53 R273H complexes can explain the resistance of the p53 R273H mutant to degradation by both of these compounds. However, R273H can still be degraded by the ubiquitin-dependent pathway by overexpression of Mdm2 (23) or by the ubiquitin-independent pathway by knocking down NQO1 by siRNA.
Curcumin is a natural compound that is a major constituent of the spice turmeric. Its reported chemopreventive effect on chemical carcinogen-induced colon tumors in rats raised the possibility that it might be a candidate for clinical use as a cancer chemopreventive agent in humans (reviewed in ref. 25). However, caution should be exercised in using curcumin as a cancer chemopreventive agent. It has been shown that NQO1 inactivation by a polymorphic mutation in the NQO1 gene in humans (reviewed in ref. 15) that does not stabilize WT p53 (21), or by knocking out the NQO1 gene in mice (47), is associated with an increased risk of developing cancer. Thus, curcumin-induced WT p53 degradation by inhibiting the activity of NQO1 and dissociating its interaction with p53 could also increase the risk of developing cancer. Studies with curcumin, given to rats and mice and to humans in phase I trials, have shown an increased risk of developing hyperplasia in the colon in rats, thyroid hyperplasia and liver adenoma in mice, and progression of premalignant disease in some patients (reviewed in ref. 29). Our finding that curcumin induces degradation of the tumor suppressor WT p53 also indicates that curcumin treatment in healthy people might lead to accumulation of DNA-damaged cells by inhibiting their p53-induced apoptosis.
Acknowledgments
We thank S. Budilovsky for technical assistance and Dr. M. Oren for the plasmids encoding p53. This work was supported by the Benoziyo Institute of Molecular Medicine, the Dolfi and Lola Ebner Center for Biomedical Research, Dr. and Mrs. Leslie Bernstein, Mrs. Bernice Gershenson, the M. D. Moross Institute for Cancer Research at the Weizmann Institute of Science, and the Israel Academy of Sciences and Humanities.
Abbreviations: NQO1, NAD(P)H:quinone oxidoreductase 1; siRNA, small-interfering RNA; TEV, tobacco etch virus.
References
- 1.Vogelstein, B., Lane, D. & Levine, A. J. (2000) Nature 408, 307–310. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein, B. & Kinzler, K. W. (1992) Cell 70, 523–536. [DOI] [PubMed] [Google Scholar]
- 3.Oren, M. (1992) FASEB J. 6, 3169–3176. [DOI] [PubMed] [Google Scholar]
- 4.Levine, A. J. (1997) Cell 88, 323–331. [DOI] [PubMed] [Google Scholar]
- 5.Yonish-Rouach, E., Resnitzky, D., Lotem, J., Sachs, L., Kimchi, A. & Oren, M. (1991) Nature 352, 345–347. [DOI] [PubMed] [Google Scholar]
- 6.Lotem, J. & Sachs, L. (1993) Blood 82, 1092–1096. [PubMed] [Google Scholar]
- 7.Lowe, S. W., Schmitt, E. M., Smith, S. W., Osborne, B. A. & Jacks, T. (1993) Nature 362, 847–849. [DOI] [PubMed] [Google Scholar]
- 8.Clarke, A. R., Purdie, C. A., Harrison, D. J., Morris, R. G., Bird, C. C., Hooper, M. L. & Wyllie, A. H. (1993) Nature 362, 849–852. [DOI] [PubMed] [Google Scholar]
- 9.Lotem, J. & Sachs, L. (1999) Apoptosis 4, 187–196. [DOI] [PubMed] [Google Scholar]
- 10.Lotem, J. & Sachs, L. (2002) Oncogene 21, 3284–3294. [DOI] [PubMed] [Google Scholar]
- 11.Hollstein, M., Rice, K., Greenblatt, M. S., Soussi, T., Fuchs, R., Sorlie, T., Hovig, E., Smith-Sorensen, B., Montesano, R. & Harris, C. C. (1994) Nucleic Acids Res. 22, 3551–3555. [PMC free article] [PubMed] [Google Scholar]
- 12.Prives, C. & Hall, P. A. (1999) J. Pathol. 187, 112–126. [DOI] [PubMed] [Google Scholar]
- 13.Soussi, T. (2000) Ann. N.Y. Acad. Sci. 910, 121–139. [DOI] [PubMed] [Google Scholar]
- 14.Asher, G., Lotem, J., Sachs, L., Kahana, C. & Shaul, Y. (2002) Proc. Natl. Acad. Sci. USA 99, 13125–13130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Asher, G., Lotem, J., Sachs, L. & Shaul, Y. (2004) Methods Enzymol. 382, 275–293. [DOI] [PubMed] [Google Scholar]
- 16.Haupt, Y., Maya, R., Kazaz, A. & Oren, M. (1997) Nature 387, 296–299. [DOI] [PubMed] [Google Scholar]
- 17.Kubbutat, M. H., Jones, S. N. & Vousden, K. H. (1997) Nature 387, 299–303. [DOI] [PubMed] [Google Scholar]
- 18.Leng, R. P., Lin, Y., Ma, W., Wu, H., Lemmers, B., Chung, S., Parant, J. M., Lozano, G., Hakem, R. & Benchimol, S. (2003) Cell 112, 779–791. [DOI] [PubMed] [Google Scholar]
- 19.Dornan, D., Wertz, I., Shimizu, H., Arnott, D., Frantz, G. D., Dowd, P., O'Rourke, K., Koeppen, H. & Dixit, V. M. (2004) Nature 429, 86–92. [DOI] [PubMed] [Google Scholar]
- 20.Asher, G., Lotem, J., Cohen, B., Sachs, L. & Shaul, Y. (2001) Proc. Natl. Acad. Sci. USA 98, 1188–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asher, G., Lotem, J., Kama, R., Sachs, L. & Shaul, Y. (2002) Proc. Natl. Acad. Sci. USA 99, 3099–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Asher, G., Tsvetkov, P., Kahana, C. & Shaul, Y. (2005) Genes Dev. 19, 316–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Asher, G., Lotem, J., Tsvetkov, P., Reiss, V., Sachs, L. & Shaul, Y. (2003) Proc. Natl. Acad. Sci. USA 100, 15065–15070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anwar, A., Dehn, D., Siegel, D., Kepa, J. K., Tang, L. J., Pietenpol, J. A. & Ross, D. (2003) J. Biol. Chem. 278, 10368–10373. [DOI] [PubMed] [Google Scholar]
- 25.Joe, B., Vijaykumar, M. & Lokesh, B. R. (2004) Crit. Rev. Food Sci. Nutr. 44, 97–111. [DOI] [PubMed] [Google Scholar]
- 26.Bech-Otschir, D., Kraft, R., Huang, X., Henklein, P., Kapelari, B., Pollmann, C. & Dubiel, W. (2001) EMBO J. 20, 1630–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choudhuri, T., Pal, S., Agwarwal, M. L., Das, T. & Sa, G. (2002) FEBS Lett. 512, 334–340. [DOI] [PubMed] [Google Scholar]
- 28.Liontas, A. & Yeger, H. (2004) Anticancer Res. 24, 987–998. [PubMed] [Google Scholar]
- 29.Moos, P. J., Edes, K., Mullally, J. E. & Fitzpatrick, F. A. (2004) Carcinogenesis 25, 1611–1617. [DOI] [PubMed] [Google Scholar]
- 30.Bunz, F., Dutriaux, A., Lengauer, C., Waldman, T., Zhou, S., Brown, J. P., Sedivy, J. M., Kinzler, K. W. & Vogelstein, B. (1998) Science 282, 1497–1501. [DOI] [PubMed] [Google Scholar]
- 31.Chowdary, D. R., Dermody, J. J., Jha, K. K. & Ozer, H. L. (1994) Mol. Cell. Biol. 14, 1997–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michalovitz, D., Halevy, O. & Oren, M. (1990) Cell 62, 671–680. [DOI] [PubMed] [Google Scholar]
- 33.Lind, C., Cadenas, E., Hochstein, P. & Ernster, L. (1990) Methods Enzymol. 186, 287–301. [DOI] [PubMed] [Google Scholar]
- 34.Garten, S. & Wosilait, W. D. (1971) Biochem. Pharmacol. 20, 1661–1668. [DOI] [PubMed] [Google Scholar]
- 35.Pulla Reddy, A. C., Sudharshan, E., Appu Rao, A. G. & Lokesh, B. R. (1999) Lipids 34, 1025–1029. [DOI] [PubMed] [Google Scholar]
- 36.Barik, A., Priyadarsini, K. I. & Mohan, H. (2003) Photochem. Photobiol. 77, 597–603. [DOI] [PubMed] [Google Scholar]
- 37.Whitesell, L., Sutphin, P. D., Pulcini, E. J., Martinez, J. D. & Cook, P. H. (1998) Mol. Cell. Biol. 18, 1517–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brennan, P. & O'Neill, L. A. (1998) Biochem. Pharmacol. 55, 965–973. [DOI] [PubMed] [Google Scholar]
- 39.Mukhopadhyay, A., Banerjee, S., Stafford, L. J., Xia, C., Liu, M. & Aggarwal, B. B. (2002) Oncogene 21, 8852–8861. [DOI] [PubMed] [Google Scholar]
- 40.Hong, R. L., Spohn, W. H. & Hung, M. C. (1999) Clin. Cancer Res. 5, 1884–1891. [PubMed] [Google Scholar]
- 41.Tikhomirov, O. & Carpenter, G. (2003) Cancer Res. 63, 39–43. [PubMed] [Google Scholar]
- 42.Uhle, S., Medalia, O., Waldron, R., Dumdey, R., Henklein, P., Bech-Otschir, D., Huang, X., Berse, M., Sperling, J., Schade, R. & Dubiel, W. (2003) EMBO J. 22, 1302–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Balasubramanian, S. & Eckert, R. L. (2004) J. Biol. Chem. 279, 24007–24014. [DOI] [PubMed] [Google Scholar]
- 44.Mustacich, D. & Powis, G. (2000) Biochem. J. 346, 1–8. [PMC free article] [PubMed] [Google Scholar]
- 45.McLure, K. G., Takagi, M. & Kastan, M. B. (2004) Mol. Cell. Biol. 24, 9958–9967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winski, S. L., Faig, M., Bianchet, M. A., Siegel, D., Swann, E., Fung, K., Duncan, M. W., Moody, C. J., Amzel, L. M. & Ross, D. (2001) Biochemistry 40, 15135–15142. [DOI] [PubMed] [Google Scholar]
- 47.Radjendirane, V., Joseph, P., Lee, Y. H., Kimura, S., Klein-Szanto, A. J., Gonzalez, F. J. & Jaiswal, A. K. (1998) J. Biol. Chem. 273, 7382–7389. [DOI] [PubMed] [Google Scholar]