Abstract
Severe burn trauma is generally followed by a catabolic response that leads to muscle wasting and weakness affecting skeletal musculature. Here, we perform whole-genome expression and in vivo NMR spectroscopy studies to define respectively the full set of burn-induced changes in skeletal muscle gene expression and the role of mitochondria in the altered energy expenditure exhibited by burn patients. Our results show 1,136 genes differentially expressed in a mouse hind limb burn model and identify expression pattern changes of genes involved in muscle development, protein degradation and biosynthesis, inflammation, and mitochondrial energy and metabolism. To assess further the role of mitochondria in burn injury, we performed in vivo 31P NMR spectroscopy on hind limb skeletal muscle, to noninvasively measure high-energy phosphates and the effect of magnetization transfer on inorganic phosphate (Pi) and phosphocreatine (PCr) resonances during saturation of γATP resonance, mediated by the ATP synthesis reactions. Although local burn injury does not alter high-energy phosphates or pH, apart from PCr reduction, it does significantly reduce the rate of ATP synthesis, to further implicate a role for mitochondria in burn trauma. These results, in conjunction with our genomic results showing down-regulation of mitochondrial oxidative phosphorylation and related functions, strongly suggest alterations in mitochondrial-directed energy expenditure reactions, advancing our understanding of skeletal muscle dysfunction suffered by burn injury patients.
Keywords: mitochondria, mitochondrial oxidative phosphorylation, muscle dysfunction, nuclear magnetic resonance
Severe thermal injuries are typically followed by a catabolic state (1, 2) that renders the body susceptible to infection, and leads to muscle wasting and weakness affecting skeletal musculature (3–6). In addition, alterations in liver mitochondrial function accompany burn trauma (7, 8), suggesting that mitochondrial dysfunction may play a role in the progression of thermal injury. To date, altered mitochondrial function, which accounts for up to 90% of ATP production in respiring cells (9), has not been well studied in skeletal muscle after burn injury.
Mitochondrial functions are altered by environmental stimuli via coordinated changes in gene expression (10). For instance, specific members of the peroxisomal proliferator activator receptor γ coactivator (PGC1) gene family respond to physiological stimuli to regulate genes that encode mitochondrial biogenesis functions (11), nuclear and mitochondrial oxidative metabolism proteins, tricarboxylic acid (TCA) cycle enzymes, and lipid oxidation and electron transport complexes (12).
Relating physiological variables at the organism level to metabolic function within the intracellular environment is difficult, because the majority of experimental techniques require the removal and/or destruction of the tissue to be examined. In vivo NMR spectroscopy overcomes this limitation (13, 14), allowing measurements of physiological biomarkers in intact systems. In addition, the advent of GeneChip microarrays has greatly advanced physiological studies by providing a snapshot of the transcriptome in a specific organ or group of cells in response to experimentally defined physiological conditions. Combining microarray and NMR data provides the opportunity to perform “functional genomics” to advance our knowledge of organismal physiology by integrating multiple levels of biological function, from genes to proteins, enzymatic pathways to cellular responses, individual organs to the whole organism.
The single hind limb burn mouse model exhibits increased protein degradation and aberrant insulin-mediated glucose utilization in muscle underlying the burn (15–17). To this end, we hypothesize that mitochondrial function is altered in skeletal muscle after burn. Furthermore, because skeletal muscle is the major tissue of energy metabolism, such burn-induced mitochondrial dysfunction could underlie the severe catabolic dysgfunction produced by thermal injury. Here we evaluate mitochondrial dysfunction in skeletal muscle after burn trauma in the mouse hind limb burn model by characterizing the concomitant gene expression patterns in burn versus control muscle tissue, and by employing in vivo NMR on intact mice to assess alterations in mitochondrial function. Our results indicate that mitochondrial dysfunction in skeletal muscle accompanies burn pathogenesis. As such dysfunction likely contributes in whole or in part to the muscle atrophy observed in burn patients, future clinical treatments might be targeted to limit this dysfunction.
Materials and Methods
Animal Studies. BALB/c mice (20–25 g) (The Jackson Laboratory) were used as a representative inbred stock and reliable population for the microarray studies. Only male mice were used, to avoid the variability that can result from the female oestrous cycle. Mice were anesthetized by i.p. injection of 40 mg/kg pentobarbital sodium and were randomized into hind limb burn or control unburned groups. We chose to use hind limbs from unburned animals as our control. Although at first it might seem more logical to use the unburned limb of each experimental mouse as the control for the burned limb, such a control will display the systemic effects of burn injury and thus will not accurately reflect the gene expression levels present in a naive nonburned limb. The ideal control limb would be both unburned and immobilized, to better mirror the burned limb. However, such a control is difficult to achieve because it would have to lack isotonic contractions, yet be normally innervated. Because the experimental mice and the control mice exhibit similar overall activity levels, the limbs from unburned animals represent a suitable control for the present experiments.
The left hind limb of anesthetized animals was subjected to a nonlethal scald injury, representing 3–5% total body surface area, by immersion in 90°C water for 3 s. This model (18) is considered clinically relevant to common burn injuries, as the tissue temperature between the soleus muscle and fibula reaches 46–54°C, similar to that at which human tissues show thermal damage (19). Analgesia was provided as s.c. buprenorphine at 0.05–0.1 mg/kg, as required. The gastrocnemius muscle was excised at 6 h, 1 day, and 3 days after burn (n = 2 for each time point). Gastrocnemius muscles excised at 6 h, 1 day, and 3 days from unburned animals served as controls. Burned and unburned muscle appeared morphologically indistinguishable by histological examination of hematoxylin and eosin stained sections (data not shown). All animal experiments were approved by the Subcommittee on Research Animal Care of Massachusetts General Hospital, Boston.
Total RNA Extraction. Muscle was homogenized for 60 s by using a Brinkman Polytron PT3000, followed by Trizol RNA extraction (GIBCO/BRL). Total RNA was further purified by using the RNeasy kit (Qiagen, Valencia, CA).
Gene Array Hybridization and Analysis. Biotinylated cRNA was generated from 10 μg of total RNA, and hybridized onto MOE430A oligonucleotide arrays, which were subsequently stained, washed, and scanned. All procedures followed standard Affymetrix protocols (Santa Clara, CA).
The data files of scanned image files hybridized with probes from RNA extracted from different time points from burned or unburned mice were converted to cell intensity (.CEL) files by using the microarray suite 5.0 (mas, Affymetrix). The data were scaled to a target intensity of 500, and all possible pairwise array comparisons of the replicates to control unburned mouse were performed for each time point (e.g., four combinations when two arrays from each time point were compared to the two arrays hybridized to control RNA), using a mas 5.0 change call algorithm. In each comparison, probe sets with a signal value difference >100 and in which one of the two samples being compared was not called “Absent” were scored as differentially modulated when: (i) the number of change calls in the same direction were at least three, four, and six when the number of comparisons were four, six, and nine, respectively; and (ii) the other comparisons were unchanged, to partially compensate for biological stochasticity and technical variation. An additional constraint of a minimum ratio of 1.65 was applied to control the known false positives at 5% [based on the ratios of 100 genes determined as invariant in most conditions tested (Affymetrix)].
Probe sets representing the same transcript were ordered on its corresponding unigene, and the 3′ most probe set was selected from combined lists of all probe sets selected in both experiments. A collection of genes with experimental evidence or annotated as mitochondrial was compiled by using annotation of MOE430A chip [Affymetrix, retrieved December, 2003; and genespider function, (Gene Spring, Silicon Genetics, Redwood City, CA), compiled from GenBank, Locuslink and Unigene (20)]. The overrepresentation statistics were calculated as hypergeometric probability by using all genes selected in each experiment that had Gene Ontology annotation for biological process (21), using information compiled for MOE430A chip [Affymetrix and from KEGG pathways (22) (retrieved March, 2004)]. P values were calculated by using the r statistical package (version 1.7.1 for Windows; ref. 23). Functional categories that did not have at least three genes at one time point and overlapping categories were removed.
NMR Spectroscopy. Data acquisition. The theoretical basis of saturation transfer experiments has been described by Forsen and Hoffman (24) (supporting calculations are provided in Supporting Text, which is published as supporting information on the PNAS web site).
Animals were studied with in vivo 31P NMR spectroscopy 3 days after burn trauma. The mice were transiently anesthetized by using a nose cone and vaporizer with a halothane/oxygen mixture (5% for induction, 1% for maintenance), and placed in a customized restraining tube. Their left hind limb was placed into a solenoid coil (four turns; length, 2 cm; diameter, 1 cm) tuned to 31P frequency (162.1 MHz). The rectal body temperature was maintained at 37 ± 1°C by using heated water blankets. All in vivo 31P NMR experiments were performed in a horizontal bore magnet (proton frequency at 400 MHz, 21 cm diameter, Magnex Scientific) using a Bruker Advance console. Field homogeneity was adjusted by using the 1H signal of tissue water. A 90° pulse was optimized for detection of phosphorus spectra (repetition time 2 s, 400 averages, 4,000 data points). Saturation 90° selective pulse trains (duration, 36.534 ms; bandwidth, 75 Hz) followed by crushing gradients were used to saturate the γATP peak. The same saturation pulse train was also applied downfield of the inorganic phosphate (Pi) resonance, symmetrically to the γATP resonance. T1 relaxation times of Pi and phosphocreatine (PCr) were measured by using an inversion recovery pulse sequence in the presence of the γATP saturation. An adiabatic pulse (400 scans; sweep width, 10 KHz; 4,000 data points) was used to invert the Pi and the PCr, with an inversion time between 152 and 7,651 ms.
Data analysis. The xwin NMR software package (paravision x, Bruker) was used for peak quantitation. Free induction decays were zero filled to 8,000 points and apodized with exponential multiplication (20 Hz) before Fourier transformation. The spectra were then manually phased and corrected for baseline broad features. The Levenberg–Marquardt algorithm was used to least-square-fit a model of mixed Gaussian/Lorentzian functions to the data. Similarly, the T1app relaxation time for Pi and PCr was calculated by fitting the function y = A1(1 – A2e–(t/T1app)), where y is the z magnetization and t is the inversion time, to the inversion recovery data.
Results
Transcriptome Studies Reveal That Burn Injury Leads to Mitochondrial Dysfunction in Skeletal Muscle. Analysis of Affymetrix microarrays identified 1,136 genes as differentially expressed in skeletal muscle in burned versus unburned control animals (Table 3, which is published as supporting information on the PNAS web site). A total of 96 of these genes are mitochondrial-specific (Table 4, which is published as supporting information on the PNAS web site).
We compared the 1,136 differentially expressed genes to annotations in the Gene Ontology consortium (21) and Kyoto Encyclopedia of Genes and Genomes (22) to identify functionally related sets of genes and statistically significant (P < 0.05) biological processes associated with burn injury. The top-ranked processes identified include muscle development, inflammatory response, glycolysis, immune response, regulation of muscle contraction, and the TCA cycle.
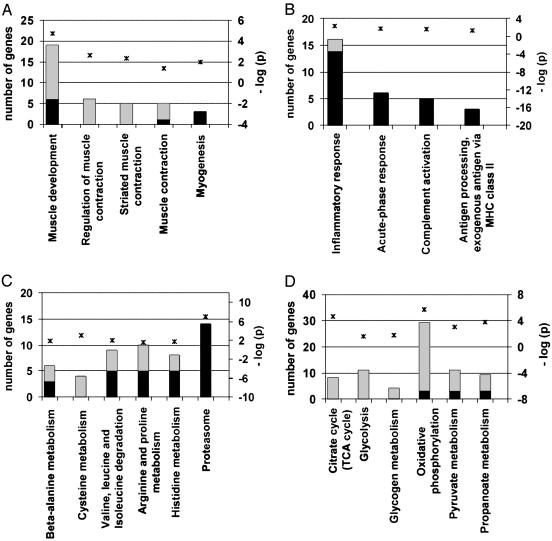
We group these functions into four categories (Fig. 1; additional information is given in Table 5, which is published as supporting information on the PNAS web site). The first set acts in muscle development and function, including contraction, and these genes are typically down-regulated in response to burn trauma (Fig. 1 A). The second set mediates inflammation and acute-phase immune responses (Fig. 1B), consistent with the pathophysiology of burn trauma. The inflammatory changes are dramatic, with >20-fold induction in some cases, such as for serum amyloid A3. Although we expect significant changes with larger burns, our results show that limited and direct burn injury has significant localized effects and consequences. The third set of genes (Fig. 1C) functions in amino acid and protein biosynthesis and degradation. The up-regulation of many components of proteasome-mediated protein degradation is consistent with well documented protein catabolism in muscle (25), further validating our approach. The fourth set of burn-associated genes (Fig. 1D) encodes several mitochondrial and energy-related functions. Extensive down-regulation is observed for TCA cycle, glycolytic, and fatty acid and glycogen metabolic activities. We placed these mitochondrial and metabolism genes into three distinct groups to begin to identify biological pathways that mediate skeletal muscle dysfunction and weakness after thermal trauma, (discussed further below; see also Fig. 2): group A encodes carbohydrate metabolism functions, including gluconeogenesis, glycolysis and glucose transport activities; group B encodes fatty acid oxidation and biosynthesis functions; and group C encodes CoA biosynthesis, electron transport, and the TCA cycle activities, which all act in oxidative phosphorylation.
Fig. 1.
Several differentially expressed genes for hind limb burn belong to one of four functional categories, as identified by using Gene Ontology and KEGG metabolic pathways at P ≤ 0.05. Black bars indicate number of up-regulated genes; gray bars correspond to down-regulated genes in the hind limb burn model versus control mice (left y axis). The negative log10 of P values represented by asterisks are indicated in the right y axis.
Fig. 2.
Differentially expressed genes at three time points after hind limb burn. Group A includes genes involved in carbohydrate metabolism, group B represents fatty acid metabolism genes, and group C includes genes involved in oxidative metabolism. Red boxes represent up-regulation of the gene, and blue boxes represent down-regulation of the gene compared with normal unburned muscle.
Group A: Carbohydrate Metabolism. Gluconeogenesis. These genes are up-regulated at the 6-h and 1-day time points after burn trauma; and include the putative transcription factors mATF4 and the proglucagon activator atf3.
Glycolysis. Several genes encoding glycolysis enzymes are down-regulated 3 days after burn, including 2,3-bisphosphoglycerate mutase, lactate dehydrogenase 1 and 2, enolase 3, and subunits of the pyruvate dehydrogenase complex, such as pyruvate dehydrogenase E1 α 1 and dihydrolipoamide S-acetyltransferase. Down-regulation of these glycolytic enzyme genes suggests decreased glucose utilization occurs in muscle after burn injury. Glucose transport. The glucose-6-phosphatase transport protein 1 gene and GLUT 4, the dominant insulin-responsive glucose transporter in muscle, are down-regulated at 3 days, indicating that glucose uptake is reduced after hind limb burn injury.
Group B: Lipid Metabolism. Burn trauma also down-regulates fatty acid oxidation genes at 3 days after burn, suggesting accumulation of muscle triglycerides. Affected genes include dodecenoyl-CoA delta isomerase; acetyl-CoA acyltransferase 2; adipocyte complement related protein, which acts in fatty acid β-oxidation; carnitine palmitoyltransferase 1, which is the rate-controlling enzyme of the long-chain fatty acid β-oxidation pathway in muscle mitochondria; and lipoprotein lipase, which increases the cytosolic pool of nonesterified fatty acids.
Group C: Oxidative Phosphorylation. These activities interconnect glycolysis, gluconeogenesis, and fatty acid biosynthesis with mitochondrial functions via the pyruvate dehydrogenase (PDH) complex, the TCA cycle, and the electron transport respiratory chain.
TCA cycle. Fuel enters the TCA cycle mainly as acetyl-CoA, which is generated from carbohydrate sources, and is a major control point. At 6 h to 1 day, burn trauma up-regulates the gene for PDH kinase 4. This enzyme catalyzes PDH phosphorylation, and helps control the activation state of the PDH complex, which is inactive in its phosphorylated state. Also, downstream TCA cycle genes are down-regulated, including PDH (lipoamide) β, succinate dehydrogenase subunits, and citrate synthase. Furthermore, malate dehydrogenase, the final enzyme of the TCA cycle, is down-regulated at 3 days after hind-limb burn.
Electron transport. Many components of the mitochondrial respiratory chain are down-regulated at 3 days, including subunits of NADH ubiquinone dehydrogenases (complex I), succinate dehydrogenase complex (complex II), subunits of cytochrome c oxidase (complex IV), and ATP synthase (F1F0 ATPase or complex V). This finding suggests that burn trauma results in reduced ATP by muscle mitochondria, which we test below by in vivo NMR spectroscopy. Mitochondrial creatine kinase, which is responsible for the transfer of high-energy phosphate from mitochondria to creatine, is also reduced by burn injury, to suggest diminished muscle energy metabolism, as observed during fasting (26) and in diabetic mouse models (27). Finally, PGC-1β is continuously down-regulated 6 h after burn.
In Vivo 31P NMR Spectroscopy Shows That Burn Injury Reduces ATP Synthesis but Does Not Significantly Alter High-Energy Phosphates or pH. We used in vivo 31P NMR to assess whether burn trauma alters muscle concentrations of high-energy phosphates via perturbed mitochondrial functions, to validate our gene expression results. Significantly, although both the PCr concentration and PCr/ATP concentration ratio are altered by thermal injury (Table 1), the Pi/PCr concentration ratio and the intramyocellular pH are essentially unchanged between the control versus the burn mice. For instance, the intramyocellular pH, calculated from the chemical shift of the Pi peak (28), is 7.25 ± 0.07 and 7.05 ± 0.11, respectively (P = 0.181).
Table 1. Concentrations of high-energy phosphates in hind limb skeletal muscle of normal and burned mice.
| Metabolite | Control (n = 5) | Burn (n = 6) (%) | P value |
|---|---|---|---|
| Phosphomonoestes | 6.47 ± 2.86* | 6.27 ± 3.28 (-3.09)† | 0.92 |
| Pi | 6.81 ± 2.98 | 5.61 ± 2.28 (-17.62) | 0.48 |
| PCr | 40.20 ± 2.24 | 35.64 ± 3.51 (-11.34) | 0.03‡ |
| γATP | 13.01 ± 1.40 | 15.45 ± 2.66 (+18.75) | 0.09 |
| αATP | 18.51 ± 1.29 | 21.32 ± 4.30 (+15.18) | 0.18 |
| βATP | 15.01 ± 2.84 | 15.70 ± 2.14 (+4.60) | 0.67 |
| Mean ATP | 15.51 ± 1.39 | 17.49 ± 1.62 (+12.77) | 0.06 |
| PCr/ATP | 2.61 ± 0.23 | 2.06 ± 0.3 (-21.07) | 0.01‡ |
| Pi/PCr | 0.17 ± 0.08 | 0.16 ± 0.07 (-5.88) | 0.87 |
Values are concentrations of high-energy-related phosphates calculated from the relative peak areas (fraction of total integral) of phosphates in NMR spectra. NMR spectral values are calculated as mean ± SE, indicated in % of the total integral. Mean ATP is the average of the three ATP peaks.
Values are percent differences between burn and control mice.
P < 0.05 burn versus control values (Student's t test).
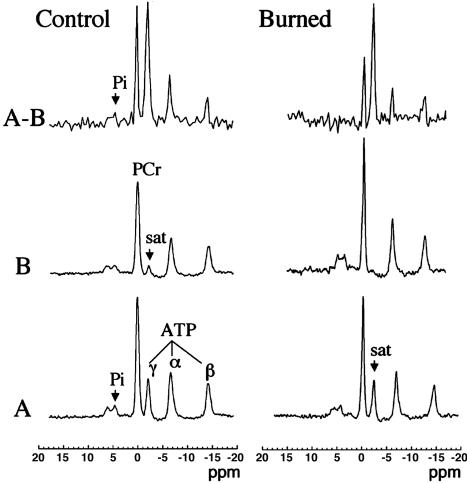
We also performed 31P NMR saturation-transfer experiments to determine rates of mitochondrial ATP synthesis. Fig. 3 shows representative 31P NMR spectra acquired from normal and burn mice before and after saturation of the γATP resonance, and their difference spectrum (Fig. 3 A–B), with the mean results presented in Table 1. On irradiation of the γATP resonance, the signal intensities of PCr, Pi, αATP, and βATP resonances all decrease, either by magnetization transfer or direct off resonance saturation. These results show the Pi and PCr signal intensities decrease by 82.8% and 43.6%, respectively (percent change in ΔM/Mo, Table 2). The fractional change ΔM/Mo and the observed spin lattice relaxation time (T1obs) were used to calculate the rate constant from Eq. 5 (see Supporting Text), which when multiplied by the Pi concentration, gives the ATP synthesis flux. The unidirectional flux of the reaction Pi → γATP is significantly reduced by 95.5% in burned mice. Although burn also is seen to reduce the flux of the reaction PCr → γATP by 42.3%, this reduction is statistically insignificant.
Fig. 3.
NMR spectra of in vivo 31P NMR saturation-transfer performed on the hind limb skeletal muscle of awake mice. Representative summed 31P-NMR spectra acquired from normal and burned mice before (A) and after (B) saturation of the γATP resonance, with the difference spectrum between the two (A–B). The arrow indicates the position of the saturation (sat) by rf irradiation (-2.4 ppm). Pi, inorganic phosphate; ppm, chemical shift in parts per million.
Table 2. Rate of ATP synthesis in hind limb skeletal muscle of awake mice, as determined by in vivo31P NMR saturation transfer.
| Control (n = 5) | Burn (n = 6) | P value* | |
|---|---|---|---|
| ATP synthesis flux (reaction Pi→γATP) | |||
| ΔM/Mo | 0.261 ± 0.047† | 0.045 ± 0.01 (-82.8%)‡ | 0.007 |
| T1obs (s) | 1.31 | 6.62 | |
| κ (s-1) | 0.200 ± 0.036 | 0.007 ± 0.002 | 0.005 |
| Pi§ (μmol/g) | 0.379 ± 0.032 | 0.461 ± 0.090 | 0.377 |
| ATP synthesis (μmol/g/s) | 0.089 ± 0.021 | 0.004 ± 0.002 (-95.5%)‡ | 0.015 |
| ATP synthesis flux (reaction PCr→γATP) | |||
| ΔM/Mo | 0.234 ± 0.084† | 0.132 ± 0.096 (-43.6%)‡ | 0.446 |
| T1obs (S) | 2.3 | 1.9 | |
| κ (s-1) | 0.121 ± 0.044 | 0.070 ± 0.051 | 0.464 |
| PCr§ (μmol/g) | 3.324 ± 0.132 | 3.347 ± 0.316 | 0.947 |
| ATP synthesis (μmol/g/s) | 0.390 ± 0.142 | 0.225 ± 0.168 (-42.3%)‡ | 0.472 |
ΔM/Mo, the fractional change in Pi or PCr magnetization as a result of saturation transfer; T1obs, observed spin lattice relaxation time of Pi or PCr during γATP saturation in seconds; κ, rate constant (s-1). ATP synthesis is calculated as Pi (or PCr) concentration × κ. A bioluminescence assay kit was used to assess ATP concentration (see supporting information).
P value burn versus control values (Student's t test).
Values are Mean ± SE from five and six animals per group.
Values are percent differences between burn and control animals.
Values are extrapolated from the baseline NMR spectrum (comparing peak integrals from Pi or PCr and [ATP] = 3.18295 μmol/g (or 3.16694 μmol/g) of normal (or burned) tissue measurement by using the assay described in the Materials and Methods.
Discussion
The first mitochondrial myopathy was identified just 45 years ago, which led to the advent of mitochondrial medicine (29), and defects in mitochondrial function are now implicated in a growing list of diseases, including Parkinson's disease, Alzheimer's disease, and Huntington's disease. Here, by analyzing coordinated changes in gene expression and measuring ATP synthesis reactions with in vivo 31P NMR, we show that perturbed mitochondrial function also plays a role in skeletal muscle burn trauma. Furthermore, we identify metabolic and mitochondrial pathways that may mediate medically significant alterations in muscle metabolism after thermal injury, and thus might serve as novel targets for burn therapy.
In healthy individuals, insulin induces muscle and adipose tissue uptake of glucose, in conjunction with decreased net production of liver glucose, and also promotes muscle protein synthesis via increased amino acid uptake (30). The absence of the anabolic effects of insulin after burn trauma might underlie increased protein catabolism and muscle wasting (31, 32), and alterations in postreceptor insulin signaling could promote such insulin resistance (31). Here, we demonstrate that reduced glucose utilization in the skeletal muscle of burned mice is at least in part due to coordinated changes in the expression of enzymes and transporters of glucose uptake and metabolism. Regulation of glycolysis and the TCA cycle depend on substrate entry into the cycle and on key cycle reactions. For instance, the PDH complex is crucial for pyruvate conversion into acetyl CoA and TCA cycle entry. Our observed up-regulation of PDH kinase likely inhibits reactions catalyzed by the PDH complex, which will limit acetyl-CoA production from carbohydrates.
We also observe that several genes down-regulated after burn trauma encode glycolytic enzymes and transporters, as well as PDH complex enzymes. These results suggest that the muscle has decreased glucose levels, which could in turn result in an increased demand for fatty acid oxidation to fulfill its energy requirements. Nonetheless, our results show that fatty acid oxidation gene expression is actually reduced in the hind limb burn model. This reduction should result in triglyceride accumulation. Such accumulation suggests a mitochondrial dysfunction, and to this end, the burn tissue shows a significant decrease in the expression of genes encoding key proteins in oxidative metabolism, and a concomitant reduction of ATP production.
Our in vivo 31P NMR analysis indicates that burn trauma does not alter high-energy phosphates, including ATP, nor intramyocellular pH, in agreement with previous NMR and conventional biopsy results for trauma (33). Although we have limited this initial analysis to 3 days after burn and changes in muscle energetics and/or pH levels could occur before this time point, previous NMR results show high-energy phosphate and pH levels remain largely unchanged from 1 to 5 days after injury (33). Our in vivo 31P NMR saturation–transfer spectra data also show that burn trauma reduces ATP synthesis, suggesting a significant reduction in the rate of mitochondrial phosphorylation, because in vivo 31P NMR saturation–transfer can noninvasively measure fast enzyme reaction exchange rates (34). Furthermore, under appropriate conditions, this technique (35, 36) can measure the net skeletal muscle rate of oxidative ATP synthesis catalyzed by mitochondrial ATPase, which by definition is proportional to the oxygen consumption rate by the P/O ratio (the ratio of the net rate of ATP synthesis by oxidative phosphorylation to the rate of oxygen consumption) (37, 38). Other investigators have proposed that the NMR-measured unidirectional ATP synthesis flux primarily reflects flux through the F1F0-ATP synthase, with negligible coupled glyceraldehyde-3-phosphate dehydrogenase and phosphoglycerate kinase reactions (36). Although the net glycolytic contribution via glyceraldehyde-3-phosphate dehydrogenase and phosphoglycerate kinase to ATP production is small, compared with that of oxidative phosphorylation, these enzymes occur at near equilibrium, and thus the unidirectional production of ATP can be high. Because burn injury down-regulates glycolytic enzyme gene expression, we assume that the contribution of glycolytic reactions to the unidirectional ATP synthesis flux is negligible.
The reduced expression of glucose oxidation and transport genes observed here, as well as the down-regulated expression of TCA cycle, β-oxidation, and electron transport genes, supports a picture that burn trauma results in impaired mitochondrial energy production. This altered mitochondrial function is possibly mediated by PGC-1β, because the PGC family, and in particular, PGC-1α, regulate several components of energy metabolism by coactivating transcription factors, including nuclear receptors (11, 12, 39–44). Further studies are required to elucidate the mechanism used by PGC-1β to alter mitochondrial metabolism in burns.
The transcriptional changes that occur in response to burn injury likely also contribute to altered glucose and fatty acid metabolism. The burn-mediated reduced expression of oxidative phosphorylation genes probably further impairs metabolism, to mediate the metabolic pathology that accompanies burn. We note that burn injury likely both directly and indirectly exerts its effects on the limb by heat-induced trauma and the associated contraction and immobilization of the injured limb, respectively. Although diminished contractile force in the burned limb could also affect gene expression, it is not possible here to discern between cause and effect. Nonetheless, the observed changes reveal that burn injury results in dramatic dysfunction of skeletal muscle, which likely contributes to its altered catabolic state and weakness.
In summary, we have used microarray and in vivo NMR data to explore the components of the mitochondrial enzymatic machinery, as well as the contributing regulatory factors, that mediate the aberrant metabolic state that typically accompanies thermal trauma. Our results demonstrate that burn injury results in large scale changes in several components that mediate mitochondrial function, muscle development, and inflammation and identifies potential regulatory candidates for new therapeutic strategies to improve both short- and long-term care of burn patients.
Supplementary Material
Acknowledgments
We thank Dr. Scott Stachel for comments and editing, and Julie Wilhelmy for technical assistance. This work was supported in part by National Institutes of Health (NIH) Center Grant P50GM021700, NIH Glue Grant U54GM062119, and Shriners Grant 8590 (to L.G.R.). Q.Z. is a Shriners Research Fellow.
Author contributions: L.G.R. and A.A.T. designed research; L.G.A., Q.Z., G.D., and M.N.M. performed research; K.E.P., L.G.A., S.G., L.G.R., and A.A.T. analyzed data; K.E.P., L.G.R., and A.A.T. wrote the paper; L.G.A. wrote only sections pertaining the NMR methodology; and R.G.T. was senior consultant on data interpretation.
Abbreviations: PCr, phosphocreatine; Pi, inorganic phosphate; TCA, tricarboxylic acid; PDH, pyruvate dehydrogenase.
References
- 1.Yu, Y. M., Tompkins, R. G., Ryan, C. M. & Young, V. R. (1999) J. Parenter. Enteral. Nutr. 23, 160–168. [DOI] [PubMed] [Google Scholar]
- 2.Tredget, E. E. & Yu, Y. M. (1992) World J. Surg. 16, 68–79. [DOI] [PubMed] [Google Scholar]
- 3.Wilmore, D. W., Long, J. M., Mason, A. D., Jr., Skreen, R. W. & Pruitt, B. A., Jr. (1974) Ann. Surg. 180, 653–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tomera, J. F., Martyn, J. & Hoaglin, D. C. (1988) J. Trauma 28, 1499–1504. [DOI] [PubMed] [Google Scholar]
- 5.Kim, C., Fuke, N. & Martyn, J. A. (1988) Anesthesiology 68, 401–406. [DOI] [PubMed] [Google Scholar]
- 6.Yasuhara, S., Kanakubo, E., Perez, M. E., Kaneki, M., Fujita, T., Okamoto, T. & Martyn, J. A. (1999) J. Burn Care Rehab. 20, 462–470. [DOI] [PubMed] [Google Scholar]
- 7.Hu, H., Greif, R. L. & Goodwin, C. W. (1994) Metabolism 43, 913–916. [DOI] [PubMed] [Google Scholar]
- 8.Vemula, M., Berthiaume, F., Jayaraman, A. & Yarmush, M. L. (2004) Physiol. Genomics 18, 87–98. [DOI] [PubMed] [Google Scholar]
- 9.Mootha, V. K., Bunkenborg, J., Olsen, J. V., Hjerrild, M., Wisniewski, J. R., Stahl, E., Bolouri, M. S., Ray, H. N., Sihag, S., Kamal, M., et al. (2003) Cell 115, 629–640. [DOI] [PubMed] [Google Scholar]
- 10.St-Pierre, J., Lin, J., Krauss, S., Tarr, P. T., Yang, R., Newgard, C. B. & Spiegelman, B. M. (2003) J. Biol. Chem. 278, 26597–26603. [DOI] [PubMed] [Google Scholar]
- 11.Scarpulla, R. C. (2002) Biochim. Biophys. Acta 1576, 1–14. [DOI] [PubMed] [Google Scholar]
- 12.Kelly, D. P. & Scarpulla, R. C. (2004) Genes Dev. 18, 357–368. [DOI] [PubMed] [Google Scholar]
- 13.Ackerman, J. J., Grove, T. H., Wong, G. G., Gadian, D. G. & Radda, G. K. (1980) Nature 283, 167–170. [DOI] [PubMed] [Google Scholar]
- 14.Hitzig, B. M., Prichard, J. W., Kantor, H. L., Ellington, W. R., Ingwall, J. S., Burt, C. T., Helman, S. I. & Koutcher, J. (1987) FASEB J. 1, 22–31. [DOI] [PubMed] [Google Scholar]
- 15.Odessey, R. & Parr, B. (1982) Metabolism 31, 82–87. [PubMed] [Google Scholar]
- 16.Nelson, K. M. & Turinsky, J. (1981) J. Surg. Res. 31, 404–414. [DOI] [PubMed] [Google Scholar]
- 17.Nelson, K. M. & Turinsky, J. (1981) J. Surg. Res. 31, 288–297. [DOI] [PubMed] [Google Scholar]
- 18.Tomera, J. F. & Martyn, J. (1988) Burns Incl. Therm. Inj. 14, 210–219. [DOI] [PubMed] [Google Scholar]
- 19.Shangraw, R. E. & Turinsky, J. (1982) J. Surg. Res. 33, 345–355. [DOI] [PubMed] [Google Scholar]
- 20.Wheeler, D. L., Church, D. M., Federhen, S., Lash, A. E., Madden, T. L., Pontius, J. U., Schuler, G. D., Schriml, L. M., Sequeira, E., Tatusova, T. A. & Wagner, L. (2003) Nucleic Acids Res. 31, 28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., Davis, A. P., Dolinski, K., Dwight, S. S., Eppig, J. T., et al. (2000) Nat. Genet. 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanehisa, M. & Goto, S. (2000) Nucleic Acids Res. 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ihaka R, G. R. (1996) J. Comput. Graph. Stat. 5, 299–314. [Google Scholar]
- 24.Forsen, S. & Hoffman, R. (1963) J. Chem. Phys. 39, 2892–2901. [Google Scholar]
- 25.Mitch, W. E. & Goldberg, A. L. (1996) N. Engl. J. Med. 335, 1897–1905. [DOI] [PubMed] [Google Scholar]
- 26.Jagoe, R. T., Lecker, S. H., Gomes, M. & Goldberg, A. L. (2002) FASEB J. 16, 1697–1712. [DOI] [PubMed] [Google Scholar]
- 27.Yechoor, V. K., Patti, M. E., Saccone, R. & Kahn, C. R. (2002) Proc. Natl. Acad. Sci. USA 99, 10587–10592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bailey, I. A., Williams, S. R., Radda, G. K. & Gadian, D. G. (1981) Biochem. J. 196, 171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luft, R., Ikkos, D., Palmieri, G., Ernster, L. & Afzelius, B. (1962) J. Clin. Invest. 41, 1776–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carter, E. A. (1998) Nutr. Rev. 56, S170–S176. [DOI] [PubMed] [Google Scholar]
- 31.Ikezu, T., Okamoto, T., Yonezawa, K., Tompkins, R. G. & Martyn, J. A. (1997) J. Biol. Chem. 272, 25289–25295. [DOI] [PubMed] [Google Scholar]
- 32.Hasselgren, P. O. & Fischer, J. E. (1992) Nutrition 8, 434–439. [PubMed] [Google Scholar]
- 33.Sprague, D. B., Gadian, D. G., Williams, S. R., Proctor, E. & Goode, A. W. (1987) J. R. Soc. Med. 80, 495–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alger, J. R. & Shulman, R. G. (1984) Q. Rev. Biophys. 17, 83–124. [DOI] [PubMed] [Google Scholar]
- 35.Brindle, K. M., Blackledge, M. J., Challiss, R. A. & Radda, G. K. (1989) Biochemistry 28, 4887–4893. [DOI] [PubMed] [Google Scholar]
- 36.Jucker, B. M., Dufour, S., Ren, J., Cao, X., Previs, S. F., Underhill, B., Cadman, K. S. & Shulman, G. I. (2000) Proc. Natl. Acad. Sci. USA 97, 6880–6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sako, E. Y., Kingsley-Hickman, P. B., From, A. H., Foker, J. E. & Ugurbil, K. (1988) J. Biol. Chem. 263, 10600–10607. [PubMed] [Google Scholar]
- 38.Kingsley-Hickman, P. B., Sako, E. Y., Ugurbil, K., From, A. H. & Foker, J. E. (1990) J. Biol. Chem. 265, 1545–1550. [PubMed] [Google Scholar]
- 39.Wu, Z., Puigserver, P., Andersson, U., Zhang, C., Adelmant, G., Mootha, V., Troy, A., Cinti, S., Lowell, B., Scarpulla, R. C. & Spiegelman, B. M. (1999) Cell 98, 115–124. [DOI] [PubMed] [Google Scholar]
- 40.Scarpulla, R. C. (2002) Gene 286, 81–89. [DOI] [PubMed] [Google Scholar]
- 41.Gopalakrishnan, L. & Scarpulla, R. C. (1995) J. Biol. Chem. 270, 18019–18025. [DOI] [PubMed] [Google Scholar]
- 42.Puigserver, P., Wu, Z., Park, C. W., Graves, R., Wright, M. & Spiegelman, B. M. (1998) Cell 92, 829–839. [DOI] [PubMed] [Google Scholar]
- 43.Lehman, J. J., Barger, P. M., Kovacs, A., Saffitz, J. E., Medeiros, D. M. & Kelly, D. P. (2000) J. Clin. Invest. 106, 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baar, K., Wende, A. R., Jones, T. E., Marison, M., Nolte, L. A., Chen, M., Kelly, D. P. & Holloszy, J. O. (2002) FASEB J. 16, 1879–1886. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.