Abstract
Cumulative mtDNA damage occurs in aging animals, and mtDNA mutations are reported to accelerate aging in mice. We determined whether aging results in increased DNA oxidative damage and reduced mtDNA abundance and mitochondrial function in skeletal muscle of human subjects. Studies performed in 146 healthy men and women aged 18–89 yr demonstrated that mtDNA and mRNA abundance and mitochondrial ATP production all declined with advancing age. Abundance of mtDNA was positively related to mitochondrial ATP production rate, which in turn, was closely associated with aerobic capacity and glucose tolerance. The content of several mitochondrial proteins was reduced in older muscles, whereas the level of the oxidative DNA lesion, 8-oxo-deoxyguanosine, was increased, supporting the oxidative damage theory of aging. These results demonstrate that age-related muscle mitochondrial dysfunction is related to reduced mtDNA and muscle functional changes that are common in the elderly.
Keywords: sarcopenia, mtDNA, oxidative damage, mRNA, mitochondrial proteins
Many structural and functional changes occur with age in skeletal muscle in a wide range of species. In Caenorhabditis elegans, muscle changes resembling those in humans precede neuronal changes, and are a determinant of morbidity (1). Age-related muscle wasting, muscle weakness, and reduced aerobic capacity result in many metabolic disorders and diminished physical performance in humans (2–4). Reduced muscle mitochondrial function could contribute to age-related muscle dysfunction and reduced aerobic capacity. Increased prevalence of mtDNA mutations (5, 6) and decreased mtDNA abundance (7, 8) have been proposed as underlying causes of mitochondrial dysfunction in aging. This finding is based on a hypothesis that cumulative oxidative damage could be the cause of aging (9).
The rate of synthesis of contractile and mitochondrial proteins in human skeletal muscle declines with advancing age and may alter muscle metabolic capacity in older people (2–4). The activity of oxidative enzymes and content mRNA transcripts encoding mitochondrial proteins are also reduced in older muscles (3, 7, 10, 11). Reduced synthesis and activity of specific proteins can alter muscle functions. The major functional role of mitochondria is ATP generation, but it remains unclear whether mitochondrial ATP production rate (MAPR) in skeletal muscle declines with age in humans. Previous studies that attempted to address this question are not in agreement, reporting that MAPR is either unchanged with age (12–16) or declines (17–19). These differences may arise from the use in some studies of inadequate sample sizes, failure to account for wide variations in physical fitness and diet, and the inclusion of subjects with metabolic abnormalities or undergoing surgical procedures at the time of analysis. Most of the previous studies examined discrete groups of younger and older people so it is unclear whether changes in mitochondria occur continuously across the adult life span or arise more rapidly later in life. We therefore performed a comprehensive study to examine whether muscle mitochondrial function declines with age in humans by using a large group of well-characterized healthy men and women across a wide age span. We also sought to determine causes of age-related changes in mitochondrial function by examining the content of mitochondrial proteins, gene transcripts encoding mitochondrial proteins, mtDNA, and DNA oxidation.
Materials and Methods
Subjects. Healthy men and women who exercised for ≤30 min on ≤2 day/week during the previous 9 mo were recruited from the local community. Physical activity levels were confirmed by questionnaire (20). Health status was assessed by medical history, physical examination, blood chemistries (complete blood count and comprehensive chemistry panel, including liver enzymes, creatinine, electrolytes, and glucose), urine analysis, and resting electrocardiogram. Exclusion criteria included a body mass index (BMI) of >32 kg/m2, tobacco use, diabetes or other metabolic or endocrine disorders, history of alcohol or substance abuse, and use of medications that could affect the outcome measures. One hundred forty-six people (86 women and 60 men) between the ages of 19 and 89 yr met the criteria and were enrolled after providing written and oral consent. The purpose, benefits, and risks of participation were fully explained before consent was obtained. The Mayo Foundation Institutional Review Board approved these studies. Total and regional fat and fat-free masses were determined by dual x-ray absorptiometry (Lunar DPX-L, Lunar Radiation, Madison, WI) in the morning after an overnight fast.
Study Protocol. For 3 days before the study, subjects maintained their daily living activities but avoided strenuous exercise. A weight-maintaining diet containing 55% of calories from carbohydrate, 30% from fat, and 15% from protein was provided by the Mayo Clinic General Clinical Research Center. Subjects were admitted for overnight stay in the Mayo Clinic General Clinical Research Center, ate a light snack at 22:00 hours, and then consumed no other food until after study completion the next day. Muscle biopsies of the vastus lateralis were obtained under local anesthesia (10, 21). A portion of the muscle was immediately used for mitochondrial ATP production measurements and the remainder was rapidly frozen in liquid nitrogen and stored at –80 C until further analysis.
MAPR. Mitochondrial were separated from 50 mg of muscle by centrifugation, and MAPR was monitored with a bioluminescent technique (21, 22). The reaction mixture included a luciferin–luciferase ATP-monitoring reagent (BioThema, Haninge, Sweden), substrates for oxidation, and 35 μM ADP. Substrates used were: 10 mM glutamate plus 1 mM malate (GM), 20 mM succinate plus 0.1 mM rotenone (SR), 1 mM pyruvate plus 0.05 mM palmitoyl-l-carnitine plus 10 mM α-ketoglutarate plus 1 mM malate (PPKM), 1 mM pyruvate plus 1 mM malate (PM), and 0.05 mM palmitoyl-l-carnitine plus 1 mM malate (PCM) with blank tubes used for measuring background activity. All reactions for a given sample were monitored simultaneously at 25°C for 20–25 min and calibrated with addition of an ATP standard by using a BioOrbit 1251 luminometer (BioOrbit Oy, Turku, Finland). Mitochondrial integrity was monitored by measuring citrate synthase activity (3, 21) before and after membrane disruption by two freeze–thaw cycles and addition of Triton X-100. All preparations used were 90–94% intact with no differences across age or gender.
Meal Glucose Tolerance. A subgroup of 10 young (age 19–31 yr) and 10 older (65–80 yr) people consumed a mixed meal (25 kcal/kg lean body mass; 55% carbohydrate, 30% fat, 15% protein) in the morning after an overnight fast. Blood samples were collected from an arterialized hand vein before the meal and then every 15 min after meal ingestion for measurements of glucose, insulin, and fatty acids. Glucose was measured with a Beckman glucose analyzer (Beckman Coulter, Fullerton, CA). Plasma insulin was measured with a two-site immunoenzymatic assay (Access system, Beckman Coulter). Nonesterified free-fatty acids were measured by using an enzymatic colorimetric assay (NEFA C, Wako).
Proteomic Protein Quantification. Muscle samples (≈15 mg) from a subgroup of 10 younger (age 24 ± 1 yr, BMI 24 ± 1kg/m2) and 10 older (age 73 ± 2 yr, BMI 26 ± 1 kg/m2) men and women were used for analysis. Samples were homogenized in 20 vol of 50 mM Tris buffer, pH 8.5, containing 0.1% SDS and a protease inhibitor mixture (Complete Mini, Roche Applied Science, Indianapolis). Samples were centrifuged at 750 × g, and 100 mg of supernatant protein was labeled with isotope-coated affinity tags (ICATs) (cleavable ICAT reagent kit monoplex version, Applied Biosystems). Older subject samples were labeled with the “heavy” ICAT, which has nine 13C atoms, and young subject samples were labeled with the “light” ICAT, which contains 12C instead of 13C. Ten pairs of young and old samples were mixed, digested with trypsin, run over a cation exchange column, and purified with avidin to collect only the cysteine-containing peptides labeled with ICATs. The biotin tag and cleavable linker were removed with trifluoroacetic acid. Peptides were analyzed by liquid chromatography-tandem MS (QSTAR, Applied Biosystems) to sequence and identify individual peptides and to determine the relative abundance in each sample pair based on the ICAT labeling. Protein identification was based on peptide sequence by using a conservative 90% confidence threshold by using pro-icat (Applied Biosystems) and mascot (Matrix Science, available at www.matrixscience.com) databases. Proteins that were present in each of the 10 sample sets were considered for data analysis.
mtDNA and mRNA Quantification. DNA and RNA extractions were performed on frozen muscle samples from individual subjects. DNA was extracted by using a QIAamp DNA mini kit (Qiagen, Valencia, CA). RNA was extracted by Trizol method (Life Technologies, Gaithersburg, MD), treated with DNase (Life Technologies) and then reverse-transcribed by using the TaqMan reverse transcription reagents (PE Biosystems, Foster City, CA).
A real-time quantitative PCR system (PE Biosystems) was used to measure the abundance of DNA (mtDNA) and mRNAs. For mtDNA the primer and probe sequences for ND1 were as follows (GenBank accession no. NC_001807) forward primer: CCCGCCACATCTACCATCA and reverse primer: GAAGAGCGATGGTGAGAGCTAAG and probe CCTCTACATCACCGCCCCGA. Abundance of mRNA was determined for cytochrome c oxidase subunits 3 (COX3) and 4 (COX4), which are encoded by mtDNA and nuclear DNA, respectively. Samples for mtDNA and mRNA were run in duplicate and triplicate, respectively, and normalized for 28S ribosomal RNA. COX3, COX4, and 28S sequences are published elsewhere (10, 21).
DNA Oxidation. A liquid chromatography, tandem MS method was used to measure 8-oxo-deoxyguanosine (8-oxo-dG) similar to what has been reported (23). The LC uses a reverse-phase column and isocratic mobile phase (methanol:water, 70:30). With electrospray source in positive mode, 8-oxo-dG and 8-oxo-2′-deoxyadenosine (8-oxo-2dA) ions are generated and are detected with the specific transitions for m/z 284–168 and 268–152, respectively. Simultaneous monitoring of both the protonated parent ion and fragmented daughter ions increases the specificity. The method is linear from 0.05 to 10 ng/ml for 8-oxodG and 8-oxo-2dA and can detect background levels of these lesions in calf thymus DNA (85 ± 3 and 7.1 ± 0.2/106 DNA bases for 8-oxo-dG and 8-oxo-2dA, respectively).
Maximal Aerobic Capacity (VO2max). A standard treadmill stress test was performed initially to assure cardiovascular health. On separate days, usually 1 or 2 wk later and at least 7 days before the inpatient study, VO2max was measured on a stationary bicycle during an incremental intensity test to fatigue (24). Expired gases, heart rate, and blood pressure were continuously monitored throughout these tests (24).
Statistical Analysis. Summarized data are reported as mean ± SEM. Regression analysis was used to measure association among selected variables. Multivariate regression was used to determine the relationship of age, body composition, MAPR, and other variables to VO2max. Differences between groups of young and older people were analyzed by using unpaired t tests. For all statistical tests, P < 0.05 was considered statistically significant.
Results
Subject Characteristics. Body weight did not significantly change with age, but older people had reduced lean body and leg mass, and body fat and BMI were increased compared with young and middle-aged people (Table 1). Reported physical activity levels during the preceding year did not differ with age.
Table 1. Participant characteristics.
| Characteristic | Younger | Middle age | Older |
|---|---|---|---|
| Age range, yr | 18-33 | 35-57 | 60-89 |
| Body weight, kg | 73.0 ± 1.3 | 72.9 ± 2.2 | 73.7 ± 2.0 |
| BMI, kg/m2 | 24.2 ± 0.3 | 25.0 ± 0.5 | 26.0 ± 0.6* |
| Lean body mass, kg | 48.8 ± 1.2 | 47.1 ± 1.9 | 44.1 ± 1.6* |
| Leg lean mass, kg | 16.9 ± 0.4 | 16.1 ± 0.7 | 14.7 ± 0.6*,** |
| Body fat, % | 27.1 ± 1.1 | 29.5 ± 1.3 | 35.3 ± 1.5*,** |
| Trunk fat, kg | 10.5 ± 0.5 | 11.6 ± 0.6 | 14.1 ± 0.8*,** |
| Physical activity, units/wk | 216 ± 28 | 205 ± 20 | 226 ± 24 |
Values are mean ± SEM. n = 76 for younger, 36 for middle age, 34 for older.
, different from younger, P < 0.05; **, different from middle age, P < 0.05.
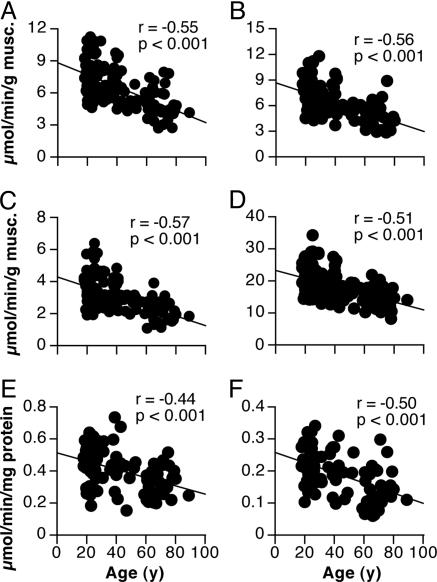
MAPR and Citrate Synthase Activity. MAPR per gram of muscle declined with age ≈8% per decade by using substrates that supply electrons primarily to complex I (e.g., glutamate plus malate) or complex II (succinate plus rotenone), respectively, of the respiratory chain (Fig. 1). Similar results were obtained for other substrates tested (not shown). After normalizing MAPR per milligram of mitochondrial protein, there was still a ≈5% decline per decade (Fig. 1).
Fig. 1.
Decline in muscle MAPR and citrate synthase activity with age. (A–C) MAPR is shown by using glutamate plus malate (A), pyruvate plus palmitoyl-Lcarnitine plus ketoglutarate plus malate (B), and succinate plus rotenone as substrates (C), respectively. (D) Citrate synthase activity. (E and F) MAPR by using glutamate plus malate and succinate plus rotenone, respectively, after normalization for mitochondrial protein. n = 146 for all measurements.
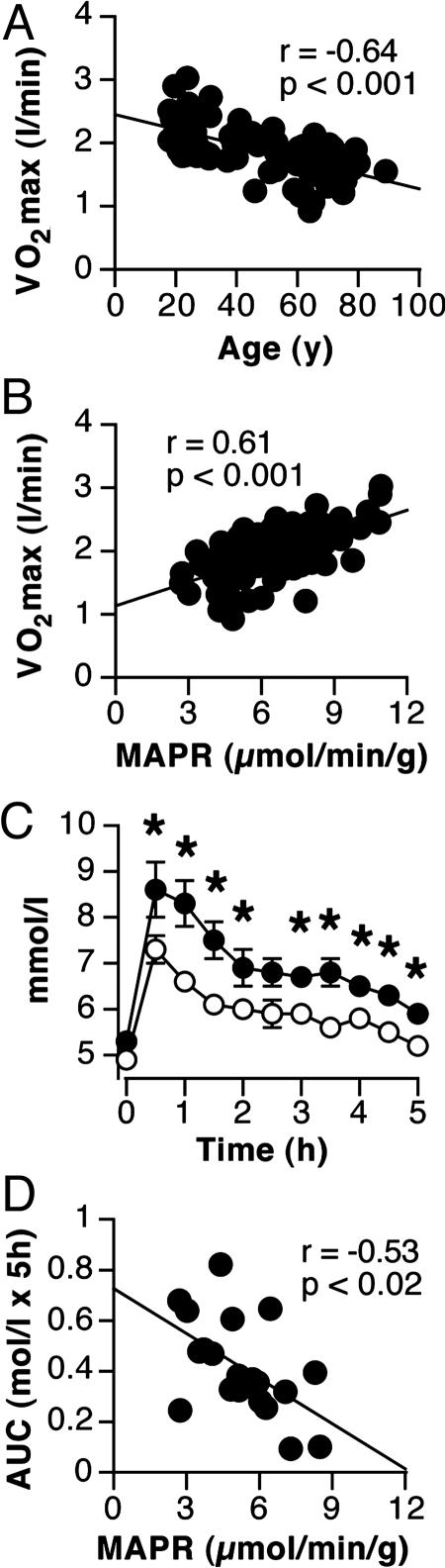
VO2max and Meal Glucose Tolerance. VO2max while cycling declined with age ≈8% per decade in both men and women (data not shown). VO2max was positively related to lean mass of the legs (r = 0.88, P < 0.001) and MAPR (r = 0.54, P < 0.001). After covariate adjustment for leg lean mass, which decreased 3% per decade (P < 0.01), the decline in VO2max was still 5% per decade (P < 0.001, Fig. 2). Furthermore, VO2max remained positively correlated with MAPR (Fig. 2). Together, leg lean mass and MAPR explained 86% of the variance in VO2max.
Fig. 2.
MAPR is related to VO2max and meal glucose tolerance. (A and B) VO2max, after covariate adjustment for leg lean mass, declined with age (A) and was positively associated with MAPR, n = 91 (B). (C) Fasting plasma glucose was not different between younger and older people (n = 10/group), but glucose excursion after a mixed meal was higher in older people (*, P < 0.05). (D) Post meal glucose area under the curve (AUC) was inversely related to MAPR.
Despite normal fasting glucose levels (4.9 ± 0.1 mM in young, 5.2 ± 0.1 in older), older people displayed higher glucose excursion after the meal indicating lower glucose tolerance (Fig. 2). Fasting and postprandial insulin and free fatty acid levels did not differ significantly between young and older people (not shown). The net area under the glucose curve (glucose area under the curve, 314 ± 49 mmol/l × 5 h for young; 508 ± 58 older, P < 0.025) was negatively correlated with MAPR (Fig. 2).
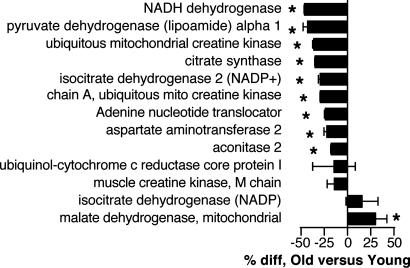
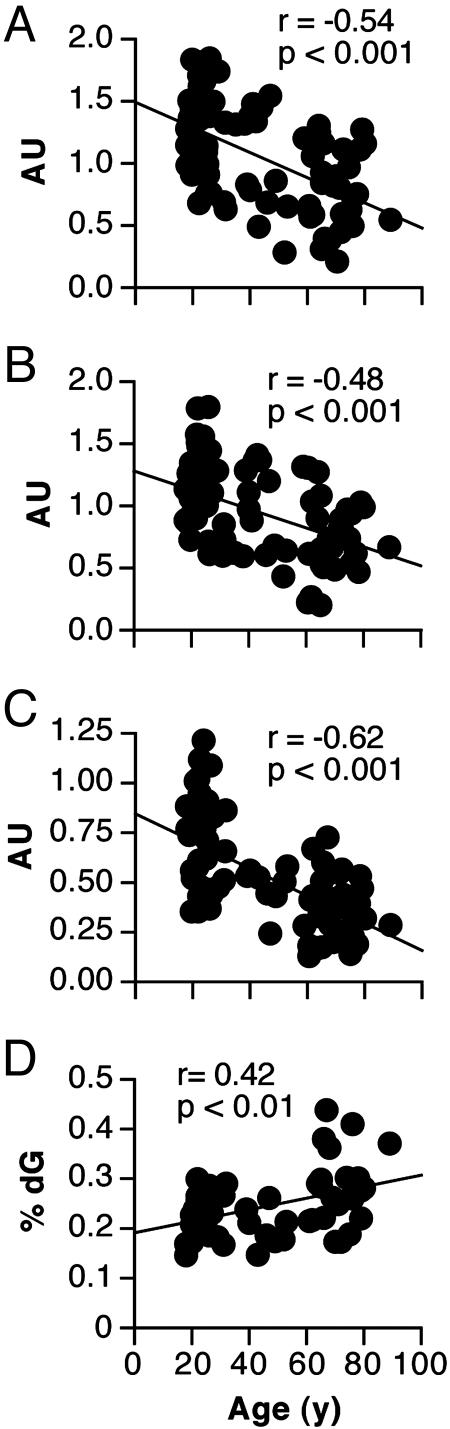
Protein and mRNA Contents. Of the 13 mitochondrial proteins found in all samples, one was significantly higher in older people whereas nine were significantly lower in older people (Fig. 3). The remaining three proteins tended to be lower in older people but showed higher variability. The abundance of COX3 and COX4 (Fig. 4) in 74 subjects declined by 10% and 8% per decade, respectively (P < 0.001).
Fig. 3.
Relative abundance of mitochondrial proteins in muscle from young and older subjects. The percentage difference (*, P < 0.05) of older relative to young is shown (n = 10/group). Negative values indicate less protein in older subjects.
Fig. 4.
Age-related changes in mitochondrial gene transcripts, mtDNA, and DNA oxidation. (A and B) Abundance of mRNA transcripts encoding COX3 and COX4, respectively, declined with age, n = 74. (C) Abundance of mtDNA by using ND1 gene probe declined with age, n = 74. (D) The level of 8-oxo-dG, relative to 2-deoxyguanisine (dG) increased with age, n = 60.
mtDNA Content and DNA Oxidation. mtDNA content, relative to nuclear DNA, was inversely related to age in 74 subjects (Fig. 4). The mtDNA content was positively related to both MAPR (r = 0.47, P < 0.01) and VO2max (r = 0.48, P < 0.01) in these subjects. Similar results were obtained when using a separate mtDNA probe targeting the ND4 gene locus (not shown). The relative content of 8-oxo-dG in 55 people (Fig. 4) increased with age so that average values for people 65–80 years of age were ≈25% higher than people 20–35 years of age (P < 0.01). The amount of 8-oxo-2dA did not change with age (data not shown).
Discussion
The current study demonstrates that older people have significantly higher oxidative damage to DNA and that mtDNA abundance decreases with age. This decreased mtDNA abundance is associated with lower content of mRNA transcripts that encode mitochondrial proteins. We found that mitochondrial protein content and activity of a key oxidative enzyme (citrate synthase) are reduced in skeletal muscle from older people. There was also a continuous decline in mitochondrial capacity for oxidative phosphorylation (ATP production) with advancing age in skeletal muscle samples from a large number of healthy men and women between the ages of 18 and 87 years. The change in mitochondrial ATP production was closely related to VO2max and glucose tolerance after a mixed meal.
A major finding in the current study was that MAPR declined with age in a well characterized group of healthy adults when expressed either per unit of muscle mass or after normalization for mitochondrial protein. These results indicate that the decline in MAPR in older muscles is due to a combination of reduced mitochondrial content and a functional alteration in the existing mitochondrial population. An in vitro MAPR assay was used so it is possible that mitochondrial function could be affected during the preparation process. However, it is unlikely that this would cause a systematic age-related effect. Moreover, our results agree with recent findings that mitochondrial function is reduced in older people, assessed by either in vitro mitochondrial respiration (19) or in vivo NMR spectroscopy (17, 18). The present data were obtained from a larger number of people than used in the previous studies and there were no systematic differences in the quality of the mitochondrial preparations detected so the observed changes in MAPR appear to reflect a true change with age.
Many studies have shown that aerobic exercise enhances muscle mitochondrial biogenesis (25). We therefore controlled for physical activity by including only subjects who were not regularly performing vigorous physical activity. We also kept the participants on a standard diet for 3 days, confirmed that they were free of overt cardiovascular or metabolic disease, and were using few if any medications. However, as in nearly all studies of aging, it is impossible to exclude the possibility that there are undetected differences in daily physical activity between younger and older people that may be important, or that some of the observed changes could be due to an interaction between aging and sedentary behavior. Discrepancies among previous reports that addressed whether MAPR declines with age in human muscle may be related to the variable control over activity levels and diet. The inclusion of subjects who were undergoing limb surgery or suspected of having metabolic disorders (15, 16) may have also affected previous findings. Additionally, most investigations have been performed on either the quadriceps or calf muscles but the deltoid (15) or the tibialis anterior (12) were examined in two of the studies that reported no effect of age on MAPR, raising the possibility that age effects may vary among muscle groups. It was reported that the decline with age in citrate synthase activity differs between the vastus lateralis and gastrocnemius muscles (26), but additional comparisons among muscles in humans are needed to confirm this finding.
MAPR was closely associated with VO2max, even after adjusting for differences in leg lean mass. This finding suggests that muscle mitochondrial function is a determinant of VO2max in untrained individuals (27) and contributes to the decline in VO2max with advancing age. This finding is in contrast to exercise-trained individuals, in which VO2max is more limited by blood supply to the working muscle (28). In support of our findings, it was shown that oxygen uptake in contracting hindlimb muscles is reduced in old vs. young sedentary rats, even after matching for convective oxygen delivery, indicating that oxygen utilization in the mitochondria is reduced (29).
Another potential effect of mitochondrial dysfunction in older people is impaired glucose tolerance and diabetes (30, 31). Reduced MAPR in older people or first-degree relatives of people with type 2 diabetes was hypothesized to cause accumulation of intramuscular lipids and insulin resistance (17, 31). Consistent with that hypothesis, we found an inverse relationship between MAPR and the glycemic response to a mixed meal. However, glucose area under the curve was also positively related to trunk fat mass, measured by dual x-ray absorptiometry (r = 0.52, P < 0.01), and there is evidence that abdominal fat may be more important than either age or mitochondrial function for determining the increase in insulin resistance with aging (10, 32). Further, we recently showed that insulin stimulates transcription and translation of mitochondrial genes and proteins and increases MAPR, but this response is blunted in people with type 2 diabetes (21). Thus, we remain open to the possibility that insulin resistance contributes to mitochondrial dysfunction rather than the reverse. Further validation studies are therefore needed to establish the relationships between mitochondrial dysfunction, muscle lipid content, and insulin resistance.
We used a tandem MS method in conjunction with isotope labeling to quantify the content of multiple mitochondrial proteins in muscle samples from younger and older people. Previously, the activity of citrate synthase has been used as a measure of mitochondrial content (3, 10, 26). The decline in citrate synthase activity with age was supported by the proteomic analysis, in which citrate synthase and eight other mitochondrial proteins were reduced in older muscles. It should be noted that we focused our proteomic analysis on the 13 mitochondrial proteins that were present in all 10 pairs of samples from representative young and older people. Approximately 600 proteins have been identified in mitochondria from human heart muscle (33). However, the purpose of that investigation was to catalog as many proteins as possible in mitochondria isolated from several grams of tissue and the relative content of individual proteins was not reported. In contrast, our intent was to quantify multiple proteins from individual subjects. We therefore measured mitochondrial protein abundance within a whole-tissue homogenate to minimize the selective or variable loss of individual proteins during mitochondrial purification, and because using equal amounts of protein, as required for ICAT labeling, would minimize or eliminate the ability to detect the overall reduction in mitochondrial protein content. To our knowledge, this is the first demonstration of lower mitochondrial protein content in older human muscle by using this approach.
Content and function of specific proteins in muscle depends on protein synthesis and breakdown. Mitochondrial protein synthesis declines with age in human muscle (3). This decline may be due to reduced mRNA template availability because both COX3 and COX4 transcript levels declined significantly with age, in agreement with earlier work in rats and humans (7, 10, 11). Mitochondrial gene transcripts from both mtDNA and nuclear DNA are similarly reduced with age but it is not yet clear how coordinated expression between the genomes is controlled. We recently reported that mRNA abundance of three nuclear-derived transcription factors that regulate mitochondrial biogenesis [peroxisome-proliferator receptor coactivator 1 α (PGC-1α), nuclear respiratory factor 1 (NRF-1), and mitochondrial transcription factor A (TFAM)] did not change with age in human muscle (10). Thus, further work on the effect of age on the action of these and other nuclear signals that regulate mitochondrial biogenesis is needed.
Another important finding was that mtDNA content in muscle declined with age. This decline could contribute to mitochondrial dysfunction by reducing template availability for transcription and translation of key mitochondrial proteins. Collectively the present findings help explain the reduction in mitochondrial protein synthesis, protein content, activity of individual enzymes, and ultimately the ability of mitochondria to perform oxidative phosphorylation in older muscle.
The reduction in muscle mtDNA with aging could be attributable to accumulated oxidative damage (9). Our finding that DNA oxidation increased with age is consistent with this oxidative theory of aging and is in agreement with rodent studies that found higher DNA oxidation in older animals (34, 35). We used total DNA for this measurement to avoid introducing artifacts when separating nuclear and mtDNA, and therefore the measurement does not distinguish how the DNA damage is distributed in the nuclear and mitochondrial genomes. The level of oxidized bases is reported to be two to three times higher in mtDNA than nuclear DNA despite the fact that capacity to repair these lesions may actually be higher in mitochondria (36). The activity of antioxidant enzymes is also higher in older rat muscles (37). Together these results indicate that production rate of reactive oxygen species increases with age, exceeding the capacity of antioxidant defense enzymes and DNA repair. Oxidative damage is associated with increased mtDNA mutations and deletions in older muscles (5, 38). The importance of mtDNA damage was recently demonstrated in mice in which accumulation of mtDNA mutations resulted in accelerated aging and shorter lifespan (39). Oxidative damage to proteins, lipids, and other cellular components may also adversely affect the function of aging cells (40, 41).
In summary, the current study demonstrates that age-related reduction in muscle mtDNA and increased DNA oxidation is associated with reduced levels of mitochondrial gene transcripts and proteins. These changes are closely related to declining capacity for mitochondrial ATP production in skeletal muscle and collectively may contribute to lower physical function and higher insulin resistance that are common in older people.
Acknowledgments
We thank the Mayo Clinic General Clinical Research Center staff and our laboratory associates for assistance in performing these studies. This work was supported by National Institutes of Health Grants MO1-RR00585, RO1-AG09531, RO1-DK41973, and T32-DK07352, the Mayo Foundation, and the Dole–Murdock Professorship in Nutrition.
Abbreviations: 8-oxo-dG, 8-oxo-deoxyguanosine; 8-oxo-2dA, 8-oxo-2′-deoxyadenosine; COX, cytochrome c oxidase; MAPR, mitochondrial ATP production rate; VO2max, maximum aerobic capacity; ICAT, isotope-coated affinity tag.
References
- 1.Herndon, L. A., Schmeissner, P. J., Dudaronek, J. M., Brown, P. A., Listner, K. M., Sakano, Y., Paupard, M. C., Hall, D. H. & Driscoll, M. (2002) Nature 419, 808–814. [DOI] [PubMed] [Google Scholar]
- 2.Short, K. R. & Nair, K. S. (1999) J. Endocrinol. Invest. 22, 95–105. [PubMed] [Google Scholar]
- 3.Rooyackers, O. E., Adey, D. B., Ades, P. A. & Nair, K. S. (1996) Proc. Natl. Acad. Sci. USA 93, 15364–15369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balagopal, P., Rooyackers, O. E., Adey, D. B., Ades, P. A. & Nair, K. S. (1997) Am. J. Physiol. 273, E790–E800. [DOI] [PubMed] [Google Scholar]
- 5.Melov, S., Shoffner, J. M., Kaufman, A. & Wallace, D. C. (1995) Nucleic Acids Res. 23, 4122–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee, C. M., Weindruch, R. & Aiken, J. M. (1997) Free Radic. Biol. Med. 22, 1259–1269. [DOI] [PubMed] [Google Scholar]
- 7.Barazzoni, R., Short, K. R. & Nair, K. S. (2000) J. Biol. Chem. 275, 3343–3347. [DOI] [PubMed] [Google Scholar]
- 8.Welle, S., Bhatt, K., Shah, B., Needler, N., Delehanty, J. M. & Thornton, C. A. (2003) J. Appl. Physiol. 94, 1479–1484. [DOI] [PubMed] [Google Scholar]
- 9.Harman, D. (1956) J. Gerontol. 11, 298–300. [DOI] [PubMed] [Google Scholar]
- 10.Short, K. R., Vittone, J. L., Bigelow, M. L., Proctor, D. N., Rizza, R. R., Coenen-Schimke, J. M. & Nair, K. S. (2003) Diabetes 52, 1888–1896. [DOI] [PubMed] [Google Scholar]
- 11.Welle, S., Bhatt, K. & Thornton, C. A. (2000) J. Appl. Physiol. 89, 297–304. [DOI] [PubMed] [Google Scholar]
- 12.Kent-Braun, J. A. & Ng, A. V. (2000) J. Appl. Physiol. 89, 1072–1078. [DOI] [PubMed] [Google Scholar]
- 13.Taylor, D. J., Kemp, G. J., Thompson, C. H. & Radda, G. K. (1997) Mol. Cell. Biochem. 174, 321–324. [PubMed] [Google Scholar]
- 14.Rasmussen, U. F., Krustrup, P., Kjaer, M. & Rasmussen, H. N. (2003) Pflugers Arch. 446, 270–278. [DOI] [PubMed] [Google Scholar]
- 15.Chretien, D., Gallego, J., Barrientos, A., Casademont, J., Cardellach, F., Munnich, A., Rotig, A. & Rustin, P. (1998) Biochem. J. 329, 249–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brierley, E. J., Johnson, M. A., Bowman, A., Ford, G. A., Reed, J. W., James, O. F. W. & Turnbull, D. M. (1997) Ann. Neurol. 41, 114–116. [DOI] [PubMed] [Google Scholar]
- 17.Petersen, K. F., Befoy, D., Dufour, S., Dziura, J., Ariyan, C., Rothman, D. L., DiPietro, L., Cline, G. W. & Shulman, G. I. (2003) Science 300, 1140–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conley, K. E., Jubrias, S. A. & Esselman, P. C. (2000) J. Physiol. 526, 203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tonkonogi, M., Fernstrom, M., Walsh, B., Ji, L. L., Rooyackers, O., Hammarqvist, F., Wernerman, J. & Sahlin, K. (2003) Pflugers Arch. 446, 261–269. [DOI] [PubMed] [Google Scholar]
- 20.Taylor, H. L., Jacobs, D. R., Shucker, B., Knudsen, J., Leon, A. S. & DeBacker, G. (1978) J. Chron. Dis. 31, 741–755. [DOI] [PubMed] [Google Scholar]
- 21.Stump, C. S., Short, K. R., Bigelow, M. L., Schimke, J. M. & Nair, K. S. (2003) Proc. Natl. Acad. Sci. USA 100, 7996–8001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wibom, R. & Hultman, E. (1990) Am. J. Physiol. Endocrinol. Metab. 259, E204–E209. [DOI] [PubMed] [Google Scholar]
- 23.Hua, Y., Wainhaus, S. B., Yang, Y., Shen, L., Xiong, Y., Xu, X., Zhang, F., Bolton, J. L. & van Breemen, R. B. (2000) Amer. Soc. Mass Spectrom. 12, 80–87. [DOI] [PubMed] [Google Scholar]
- 24.Proctor, D. N. & Beck, K. C. (1996) J. Appl. Physiol. 81, 2495–2499. [DOI] [PubMed] [Google Scholar]
- 25.Booth, F. W. (1991) Physiol. Rev. 71, 541–585. [DOI] [PubMed] [Google Scholar]
- 26.Houmard, J. A., Weidner, M. L., Gavigan, K. E., Tyndall, G. L., Hickey, M. S. & Alshami, A. (1998) J. Appl. Physiol. 85, 1337–1341. [DOI] [PubMed] [Google Scholar]
- 27.Haseler, L. J., Lin, A. P. & Richardson, R. S. (2004) J. Appl. Physiol. 97, 1077–1081. [DOI] [PubMed] [Google Scholar]
- 28.Richardson, R. S., Grassi, B., Gavin, T. P., Haseler, L. J., Tagore, K., Roca, J. & Wagner, P. D. (1999) J. Appl. Physiol. 86, 1048–1053. [DOI] [PubMed] [Google Scholar]
- 29.Hepple, R. T., Hagan, J. L., Krause, D. J. & Jackson, C. C. (2002) J. Appl. Physiol. 94, 744–751. [DOI] [PubMed] [Google Scholar]
- 30.Kelley, D. E., He, J., Menshikova, E. V. & Ritov, V. B. (2002) Diabetes 51, 2944–2950. [DOI] [PubMed] [Google Scholar]
- 31.Petersen, K. F., Dufour, S., Befroy, D., Garcia, R. & Shulman, G. I. (2004) N. Engl. J. Med. 350, 664–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khort, W. M., Kirwin, J. P., Staten, M. A., Bourey, R. E., King, D. S. & Holloszy, J. O. (1993) Diabetes 42, 273–281. [PubMed] [Google Scholar]
- 33.Taylor, S. W., Fahy, E., Zhang, B., Glenn, G. M., Warnock, D. E., Wiley, S., Murphy, A. N., Gaucher, S. P., Capaldi, R. A., Gibson, B. W., et al. (2003) Nat. Biotechnol. 21, 281–286. [DOI] [PubMed] [Google Scholar]
- 34.Hamilton, M. L., Van Remmen, H., Drake, J. A., Yang, H., Guo, Z. M., Kewitt, K., Walter, C. A. & Richardson, A. (2001) Proc. Natl. Acad. Sci. USA 98, 10469–10474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei, Y.H. (1998) Proc. Soc. Exp. Biol. Med. 217, 53–63. [DOI] [PubMed] [Google Scholar]
- 36.Stevnsner, T., Thorslund, T., de Souza-Pinto, N. C. & Bohr, V. A. (2002) Exp. Gerontol. 37, 1189–1196. [DOI] [PubMed] [Google Scholar]
- 37.Hollander, J., Bemja, J., Ookawara, T. & Ji, L. L. (2000) Mech. Ageing Dev. 116, 33–45. [DOI] [PubMed] [Google Scholar]
- 38.Michikawa, Y., Mazzucchelli, F., Bresolin, N., Scarlato, G. & Attardi, G. (1999) Science 286, 774–779. [DOI] [PubMed] [Google Scholar]
- 39.Trifunovic, A., Wrendenberg, A., Falkenberg, M., Spelbrink, J. N., Rovio, A., T., Bruder, C. E., Bohlooly-Y, M., Gidlof, S., Oldfors, A., Wibom, R., et al. (2004) Nature 429, 417–423. [DOI] [PubMed] [Google Scholar]
- 40.Grune, T., Shringarpure, R., Sitte, N. & Davies, K. (2001) J. Gerontol. A Biol. Sci. Med. Sci. 56, B459–B467. [DOI] [PubMed] [Google Scholar]
- 41.Bassett, C. N. & Montine, T. J. (2003) J. Nutr. Health Aging 7, 24–29. [PubMed] [Google Scholar]