Abstract
Plasmodium falciparum apical membrane antigen 1 (AMA1) is located in the merozoite micronemes, an organelle that contains receptors for invasion, suggesting that AMA1 may play a role in this process. However, direct evidence that P. falciparum AMA1 binds to human erythrocytes is lacking. In this study, we determined that domain III of AMA1 binds to the erythrocyte membrane protein, Kx, and that the rate of invasion of Kxnull erythrocytes is reduced, indicating a significant but not unique role of AMA1 and Kx in parasite invasion of erythrocytes. Domains I/II/III, domains I/II and domain III of AMA1 were expressed on the surface of CHO-K1 cells, and their ability to bind erythrocytes was determined. We observed that each of these domains failed to bind untreated human erythrocytes. In contrast, domain III, but not the other domains of AMA1, bound to trypsin-treated human erythrocytes. We tested the binding of AMA1 to trypsin-treated genetically mutant human erythrocytes, missing various erythrocyte membrane proteins. AMA1 failed to bind trypsin-treated Kxnull (McLeod) erythrocytes, which lack the Kx protein. Furthermore, treatmentofhumanerythrocyteswithtrypsin, followed by α-chymotrypsin, cleaved Kx and destroyed the binding of AMA1 to human erythrocytes. Lastly, the rate of invasion of Kx null erythrocytes by P. falciparum was significantly lower than Kx-expressing erythrocytes. Taken together, our data suggest that AMA1 plays an important, but not exclusive, role in invasion of human erythrocytes through a process that involves exposure or modification of the erythrocyte surface protein, Kx, by a trypsin-like enzyme.
Keywords: erythrocyte recognition, McLeod, merozoite, malaria, invasion
The complex multistep process of invasion of erythrocytes by Plasmodium falciparum begins with the binding of any surface of the merozoite, followed by reorientation to put the merozoite's apical end in close apposition to the erythrocyte surface. The apical end of merozoites contains the organelles of invasion, including the micronemes that contain receptors such as members of the Duffy binding protein family of erythrocyte-binding ligands (1). A junction forms between P. knowlesi merozoites and the erythrocyte before the merozoite is brought into the erythrocyte. The junction only forms and invasion only occurs between P. knowlesi merozoites and Duffy blood group positive human erythrocytes. Duffynull cells fail to form a junction and are not invaded by P. knowlesi merozoites (2). However, trypsin- or neuraminidase-treated human Duffynull erythrocytes could form a junction and were invaded by P. knowlesi merozoites (3) despite the fact that untreated and trypsin-treated human Duffynull cells fail to bind any of the P. knowlesi Duffy binding proteins expressed on COS-7 cells (2). This result indicates that molecules other than the parasite Duffy binding protein family are exposed by enzyme treatment to form the junction between Duffy negative human erythrocytes and P. knowlesi merozoites. One of the possible junction-forming molecules is apical merozoite antigen 1 (AMA1), which is found in the micronemes (4). Half of the cytoplasmic domain of AMA1 is highly conserved in all Plasmodium species, suggesting a role for signaling or interaction with the actin-myosin motor once the extracellular domain is engaged. Its importance for parasite survival is indicated by the fact that it was not possible to knock out the ama1 gene (5), and antibodies to AMA1 blocked invasion (6). Moreover, the expression of AMA1 from P. chabaudi, a rodent parasite, in P. falciparum merozoites increased invasion by P. falciparum into rodent erythrocytes (5). This finding indicated that P. chabaudi AMA1 alone with no other P. chabaudi genes interacted directly with rodent erythrocytes and improved invasion.
AMA1 has been divided into three domains (I, II, and III) based on the disulfide bonds (7) and the recent crystal structure (8). It is processed during late merozoite development or during invasion (9). A combined construct of domains I and II of P. yoelii AMA1, a rodent parasite gene expressed on COS-7 cells, bound rodent erythrocytes (10). However, the same domains of AMA1 of P. falciparum failed to bind human erythrocytes (J. Adams, personal communication).
To understand the role of AMA1 in erythrocyte invasion, we have explored the possibility that P. falciparum AMA1 binds human erythrocytes on its own. We found that only domain III of AMA1 binds to human erythrocytes and only if the erythrocytes are treated with trypsin. Furthermore, we found that domain III of AMA1 could not bind to Kxnull erythrocytes, indicating that Kx is the erythrocyte receptor for AMA1.
Materials and Methods
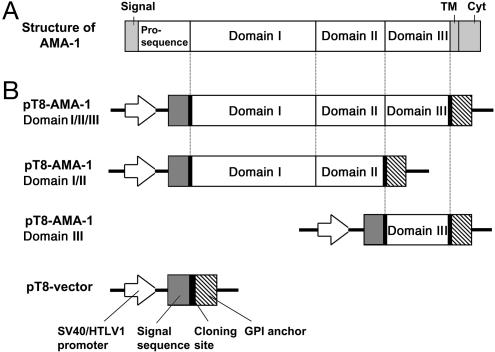
Cloning of Domains of AMA1 in the T8 Vector. Domains I/II, domain III, and domains I/II/III of AMA1 were cloned into the T8 vector (11) for expression in CHO-K1 cells. This vector contains a signal sequence and a glycosylphosphatidylinositol (GPI) anchor (Fig. 1) that allows the expression of the fusion protein on the surface of CHO-K1 cells. A plasmid that contained AMA1 of the FVO clone of P. falciparum (GenBank accession no. AY588147) was used as a template for subcloning into the T8 plasmid. The constructs containing domains I/II, III, and I/II/III were produced by PCR from this plasmid. The forward primer for domains I/II and I/II/III was (5′-GCGGATCCGCATCGAAATTGTCGAAAGA-3′; 236–255 bp) and for domain III was (5′-GCGGATCCTAGA AGT TGA ACACAACTTT-3′; 1,253–1,272 bp) with the BamHI site underlined. The reverse primer for domains I/II/III and III was (5′-GCGAATTCTTATGTTCAGGGATATCTGC-3′; 1,566–1,546 bp) and for domain I/II was (5′-GCGAATTCTCTATGGGATGACTCAGCGC-3′; 1,256–1,237 bp) with the EcoRI sites underlined.
Fig. 1.
Schematic representation of P. falciparum AMA1 and expression vectors for domains I/II/III, domains I/II, and domain III of AMA1. (A) AMA1 consists of a signal sequence, a prosequence, domains I, II, and III, a predicted transmembrane (TM), and a cytoplasmic domain (Cyt). (B) Domains I/II/III, domains I/II, and domain III of AMA1 were amplified and cloned into the T8 vector that contains a signal sequence, a multicloning site, and a glycosylphosphatidylinositol (GPI) anchor. CHO cells that are transfected with this plasmid, pT8-AMA1 domains I/II/III (Top), domains I/II (Top Middle) or domain III (Bottom Middle), express proteins for domains I/II/III, domains I/II or domain III of P. falciparum AMA1 on their surface.
Other Constructs Used. Two vectors were used in these studies, T8 and pRE4 (2). The constructs used were as follows: (i) JESEBL/EBA-181 from Dd2 in pRE4 (11); (ii) domain II of Duffy binding protein of P. vivax in pRE4 (2); (iii) EBA-175 of MC strain in pRE4 (12); (iv) DBL 2, a Duffy binding-like domain from P. falciparum erythrocyte membrane protein 1 in the T8 vector.
Transfection of CHO-K1 Cells. CHO-K1 cells (American Type Culture Collection) were placed on coverslips coated with poly-l-lysine (0.01% solution, Sigma-Aldrich) and transiently transfected as described in ref. 11. The expression of recombinant proteins on CHO cells was determined by immunofluorescence as previously described by using mAb 179 for the T8 vector and mAb 1D3 for pRE4. In addition, mAbs against AMA1 (2E3, 1C8, 1G4, and 2D9) were tested against domains of AMA1 expressed on CHO cells.
Erythrocytes Used in This Study. Normal erythrocytes were used in the binding assay as fresh samples. One Kxnull patient (McLeod 1) had chronic granulomatous disease (CGD) with a deletion in the X chromosome between the genes for Duchenne muscular dystrophy and X-linked retinitis pigmentosa, but the patient had neither of these diseases. This deletion removed both the cgd gene and the next gene XK. The other two Kxnull individuals (McLeod 2 and 3) were non-CGD patients. McLeod 3 had a point mutation (a G to C transition) in the donor site of the first intron of XK gene that led to the failure of expression. The mutation in the other patient is unknown. The Kxnull erythrocytes from McLeod 2 and 3 as well as the PP1Pk negative erythrocytes had been stored in liquid nitrogen as frozen pellets and thawed directly into PBS at 37°C (13). Duffynull 1, Kiddnull, and Bombay erythrocytes were frozen and thawed by the Red Cross Method (14) and the Duffynull 2 was from a fresh sample.
Enzyme Treatment of Erythrocytes. Normal human erythrocytes were enzyme treated as described in ref. 11. Treatment with trypsin, followed by neuraminidase, was performed as described for each enzyme. Mutant erythrocytes (e.g., Kellnull, Kxnull, Duffynull, PP1Pk negative) were treated only once with 1 mg/ml of trypsin for 1 h at 37°C instead of 2 h. This step was followed by treatment with 1 mg/ml of soybean trypsin inhibitor for 10 min at room temperature. Normal human erythrocytes were treated by the same procedures as the mutant erythrocytes.
Erythrocyte Binding Assay. The binding assay was as described in ref. 11 with some modifications. The glutaraldehyde-fixed cells were incubated with 0.25 ng/μl of 4′,6-diamidino-2-phenylindole (DAPI) (Molecular Probes) to observe the nuclei of CHO-K1 cells to which erythrocytes have bound. The number of CHO-K1 cells in a 495 μm × 660 μm field were captured by a camera attached to the microscope (LEICA DMIRE2, Germany). The number of rosettes on one coverslip was determined and expressed as the number of rosettes per 2.0 × 105 CHO-K1 cells. To eliminate bias, the person counting the coverslips did not know which DNA had been transfected on each slide or which erythrocytes were used.
Invasion Assay into Kxnull Erythrocytes. Prewashed normal human erythrocytes and McLeod1 (Kxnull) human erythrocytes were tested for invasion by P. falciparum clones HB3 and D10 as described in ref. 15. The identity of the P. falciparum clones was confirmed by DNA fingerprinting (16).
Immunoblotting of Enzyme-Treated Human Erythrocytes. Normal human erythrocytes were treated with enzymes as described above. They were suspended in TE (10 mM Tris, pH 8.0/1 mM EDTA) and centrifuged at 18,000 × g for 10 min at 4°C. The supernatant was discarded, and this step was repeated twice by resuspending the pellet with TE. The proteins separated on a SDS gel were transferred to a poly(vinylidene difluoride) membrane (Invitrogen). The membrane was blocked as described in the ECL chemiluminescent system (Amersham Pharmacia). The membranes were immunoblotted with monoclonal antibody against Kx (17, 18) and probed with peroxidase-linked goat anti-mouse IgG (Jackson ImmunoResearch). The antibody was detected with the ECL system as described by the manufacturer.
Results and Discussion
Domain III of AMA1 Binds Trypsin-Treated Human Erythrocytes. To determine whether P. falciparum AMA1 binds human erythrocytes, AMA1 domains I/II/III, domains I/II, and domain III were expressed on the surface of CHO-K1 cells (Fig. 1). Domains I/II/III and domain III expressed on CHO cells bound mAbs 2E3 and 1C8 and domains I/II and I/II/III bound mAbs 2D9 and 1G4. The transfection efficiency (as determined by the percent of positive CHO cells stained by mAbs 179 or 1D3) of all constructs into CHO cells was 80–90%. None of the domains of AMA1 bound to untreated human erythrocytes (Fig. 2).
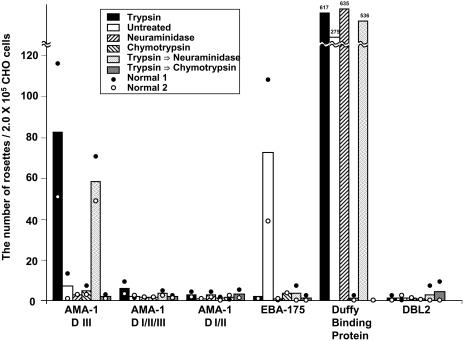
Fig. 2.
AMA1 domain III binds erythrocytes that are trypsin treated. Domains I/II/III, domains I/II, domain III, EBA-175, domain II of Duffy antigen binding protein from P. vivax, domain II of JESEBL from the Dd2 clone of P. falciparum, and DBL2 were expressed in CHO-K1 cells. Normal and enzyme-treated erythrocytes from two individuals were either untreated or treated with trypsin, neuraminidase, α-chymotrypsin, trypsin followed by neuraminidase, or trypsin followed by α-chymotrypsin before addition to transfected cells. Transfected cells with five or more attached erythrocytes were counted as rosettes, and the number of rosettes per 2.0 × 105 CHO cells was calculated after standardization for the number of CHO cells. The transfection efficiency of all constructs into CHO cells was 80–90%. The numbers that are above the bar of Duffy protein represent the number of rosettes for the average of two experiments. The height of the bar are the average of two experiments (white and black circles). D, domain.
Because it was possible that the erythrocyte receptor for AMA1 was hidden, we treated the erythrocytes with various enzymes and tested them for binding. Domain III bound to trypsin-treated erythrocytes (Fig. 2). Domains I/II and I/II/III did not bind under the conditions studied. Why does only domain III of AMA1 bind to human erythrocytes and not domains I/II/III that contains domain III? P. falciparum AMA1 is enzymatically cleaved around the time of invasion (9). Because of the inability to obtain invasive merozoites, we are unable to determine whether the processing of AMA1 is required for optimal invasion. It is possible that one of the cleavage products of AMA1 contains domain III and that this fragment acts as the parasite receptor.
The effectiveness of the trypsin treatment on erythrocytes was determined by the inability of EBA-175, the glycophorin A ligand, to bind these erythrocytes, because glycophorin A is cleaved by trypsin. A control transfectant, DBL 2, which does not bind erythrocytes, also did not bind the trypsin-treated erythrocytes.
Domain III failed to bind either neuraminidase- or α-chymotrypsin-treated erythrocytes (Fig. 2). The failure to bind erythrocytes after treatment with these enzymes could be due to either the destruction of the receptor or the failure to expose it. To resolve this question, we treated the erythrocytes with trypsin that permits bindingandfollowedthistreatmentwithneuraminidaseorα-chymotrypsin. We found that domain III of AMA1 bound to human erythrocytes treated with trypsin followed by neuraminidase (Fig. 2), indicating that sialic acid residues are not part of the AMA1 receptor. Furthermore, the binding to trypsin-treated erythrocytes is not related to a reduced negative charge due to the removal of glycophorin A because erythrocytes treated with neuraminidase alone did not bind to domain III. In contrast, the binding of domain III of AMA1 was abolished after human erythrocytes were treated with trypsin followed by α-chymotrypsin, indicating that the receptor for AMA1 is a protein or glycoprotein modified by trypsin and destroyed by α-chymotrypsin (Fig. 2).
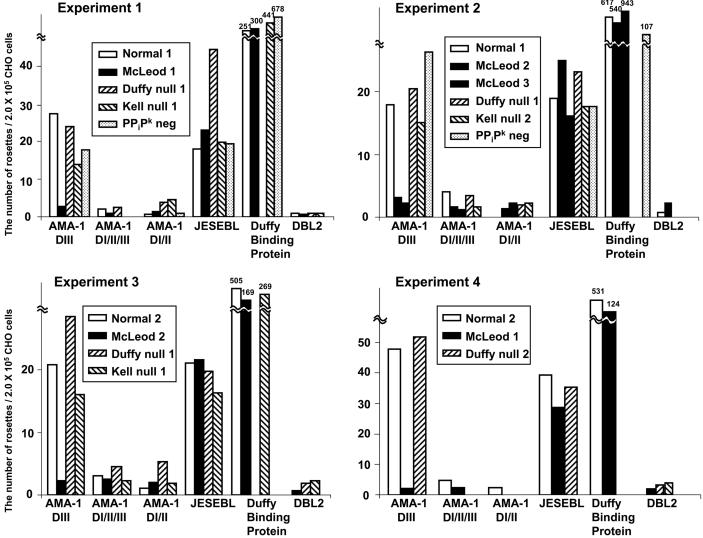
AMA1 Domain III Fails to Bind Kxnull Human Erythrocytes. To identify the erythrocyte protein that serves as the receptor for domain III of AMA1, we determined the binding of domain III to trypsin-treated erythrocytes lacking various surface proteins (Bombay negative, Kiddnull, Kellnull, Duffynull, Kxnull, and PPiK negative). Of the negative and null cells tested, only Kxnull erythrocytes showed no binding to domain III of AMA1. In four different experiments using three different Kxnull (McLeod) erythrocytes, we found that trypsin-treated Kxnull erythrocytes had extremely low binding to domain III of AMA1 (Fig. 3). Kxnull erythrocytes demonstrated lower binding than normal controls, but the difference in binding was not significant. The Kell protein is bound by a disulfide bond to the Kx protein (19), Kellnull erythrocytes have a reduced expression of the Kx protein on the erythrocyte surface (20).
Fig. 3.
AMA1 domain III fails to bind trypsin-treated Kxnull erythrocytes. Two erythrocytes from normal individuals (normal 1 and 2), Duffynull blood group (Duffy null 1 and 2), Kellnull blood group (Kell null 1 and 2), McLeod blood group, which lacks Kx protein and has reduced Kell protein, (McLeod 1, 2, and 3), PPiPk negative blood group were treated with trypsin before addition to transfected CHO-K1 cells. McLeod 1 is from a patient of CGD, and McLeod 2 and 3 are from non-CGD donors with point mutation in the XK gene. The data of four independent experiments are shown. Kell null 2 RBC was also Duffy null. D, domain.
As a positive control to demonstrate that trypsin-treated null erythrocytes did not destroy the ability of Kxnull cells to bind parasite proteins expressed on CHO-K1 cells, we used region II of JESEBL from the P. falciparum clone Dd2 and region II of the P. vivax Duffy binding protein, both of which bind to trypsin-treated erythrocytes (2, 11). The inclusion of JESEBL region II in addition to region II of the Duffy binding protein was chosen because it had a similar binding efficiency as domain III of AMA1, whereas Duffy bound at a higher frequency (Fig. 3). As a further control for effective trypsin treatment, we demonstrated that trypsin-treated Kxnull erythrocytes failed to bind to EBA-175 (data not shown), a parasite ligand for trypsin-sensitive glycophorin A (12).
Because one of the patients who had Kxnull erythrocytes also had CGD, other gene deletions on the X chromosome in addition to the XK gene encoding the Kx protein may have occurred and affected binding to domain III of AMA1. The fact that the other two Kxnull donors are non-CGD patients provides evidence that the absence of the Kx protein itself was responsible for the failure to bind domain III. A point mutation in one of the non-CGD patients has been identified as a mutation in the donor site of the first intron leading to the Kxnull phenotype. This finding indicates that the absence of the Kx protein itself, and not some other deletion on the X chromosome, is responsible for the failure of domain III of AMA1 to bind to Kxnull erythrocytes.
The Kx protein has 7–10 transmembrane domains, depending on whether the first domain is a signal sequence or a transmembrane domain. The next two domains have a low score on the tmhmm 2.0 program for prediction of transmembrane helices in proteins (Center for Biological Sequence Analysis, Technical University of Denmark, Lyngby). The proof for these transmembrane domains must be determined experimentally. The loop between the next to last transmembrane domain contains a cysteine that was shown by mutagenesis to bind to the extracellular cysteine of the Kell protein (19). From this result, we could identify the last three extracellular loops that contain 11–16 amino acids. The proximity of these extracellular domains to the erythrocyte membrane would be consistent with the requirement for treatment with trypsin to expose them for binding to domain III of AMA1.
Enzyme Treatment of Erythrocytes Modifies the Kx Protein. To determine the effect of trypsin and other enzymes on the Kx protein, we determined the fate of the Kx protein in normal and enzyme-treated erythrocytes by using a mAb against the Kx protein (17, 18). We detected three bands on the immunoblot from normal, untreated erythrocytes between the 41- and 26-kDa molecular mass markers. The highest molecular mass band also appears in the Kxnull erythrocytes, although the intensity is less in Kxnull. In other experiments, the upper band is present in Kxnull and of the same mobility (data not shown). Thus, the top band may either be encoded by the XK gene or a background band. As described in ref. 19, two unique bands missing from Kxnull erythrocytes were detected in normal erythrocytes. Upon treatment with trypsin, there was a downward shift in the mobility of the upper unique Kx band (Fig. 4), confirmed by scanning and analysis of the protein peaks in two independent experiments by using the imagequant software (Amersham Pharmacia) (data not shown). The Kx protein may be cleaved by trypsin, but the protein is held together in the SDS-gel electrophoresis by the hydrophobic transmembrane domains. Thus, binding of domain III of AMA1 depends on trypsin treatment of erythrocytes for either exposure of the Kx binding site or cleavage of the Kx protein. It is known that a protease is required for invasion by merozoites of P. chabaudi into mouse erythrocytes (21). Could a trypsin-like protease be involved in the modification of human erythrocyte surface by P. falciparum during invasion? Alternatively, other merozoite receptors may bring AMA1 in close apposition with the membrane during invasion, allowing the binding to the Kx protein without requiring enzymatic modification of the erythrocyte surface.
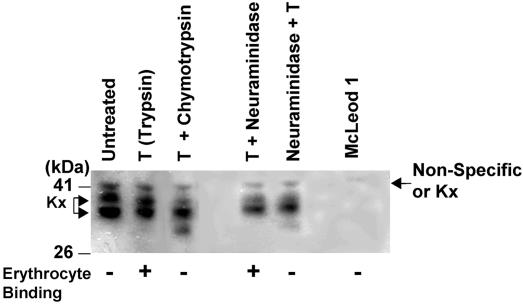
Fig. 4.
The Kx protein is proteolytically modified. Immunoblot of ghosts from normal erythrocytes, Kxnull (McLeod) erythrocytes from a patient who had CGD, and the following treatment of normal erythrocytes: trypsin (T), T followed by α-chymotrypsin, T followed by neuraminidase, and neuraminidase followed by T. The erythrocyte binding data are from Figs. 2 and 3 (+, binding; –, no binding). Molecular masses of the standards (kDa) are shown on the left.
As direct evidence that modification of the Kx protein affects binding, we found that enzyme treatments, which eliminated binding, cleaved a portion of the Kx protein. An additional lower molecular mass band appeared in the lysate from erythrocytes that were treated with trypsin followed by α-chymotrypsin, indicating that the Kx protein was cleaved by α-chymotrypsin. This cleavage is associated with the failure of erythrocytes treated with trypsin followed by α-chymotrypsin to bind domain III of AMA1 (Fig. 2).
Furthermore, the lower band, similar to the one in samples treated with trypsin followed by α-chymotrypsin, appeared in erythrocytes that have been treated with neuraminidase followed by trypsin. Again, the latter treatment is associated with the failure of domain III of AMA1 to bind these erythrocytes (data not shown). Despite the appearance of this band under these two conditions, the upper two bands are still intact. Although the upper Kx band is of lower intensity in Fig. 4, this finding was not consistent in other experiments (data not shown). Because XK is a single gene, we speculate that the two bands may indicate alternative splicing or posttranslational modification. There could be multiple modifications that are superimposed on the two bands. One of these bands may be cleaved and eliminate binding to domain III of AMA1.
The Kxnull (McLeod) phenotype is characterized by mild hemolytic anemia. It is possible that the failure of domain III of AMA1 to bind Kxnull erythrocytes is due to the irregular shape of the erythrocytes, although both JESEBL and the Duffy-binding protein bound these erythrocytes. Furthermore, the absence of the Kx protein on the erythrocyte surface might lead to compensatory changes in other erythrocyte proteins that might affect the binding of domain III of AMA1 to Kxnull erythrocytes. However, the finding that enzymatic treatments of erythrocytes produce an additional fragment of the Kx protein and that these enzymatically treated erythrocytes fail to bind domain III of AMA1 is consistent with the direct involvement of Kx in binding domain III of AMA1.
Reduced Invasion into Kxnull Erythrocytes. To analyze the invasion rate of P. falciparum into Kxnull erythrocytes, invasion assays were performed by using two P. falciparum clones, HB3 and D10. For HB3, we carried out three independent experiments done in duplicate by using normal human erythrocytes and fresh Kxnull erythrocytes. The rate of invasion for P. falciparum clone HB3 into Kxnull erythrocytes was reduced to 33.3–48.3% compared with the invasion into normal erythrocytes (Table 1). In one experiment, the invasion rate of P. falciparum clone D10 showed a similar level of reduction as observed for HB3.
Table 1. Invasion rate of P. falciparum clone HB3 into McLeod erythrocytes.
| Invasion rate, % ring-infected erythrocytes
|
|||
|---|---|---|---|
| Experiment | Normal | McLeod | % Control |
| 1 | 17.5 | 8.2 | 46.9 |
| 2 | 5.7 | 1.9 | 33.3 |
| 3 | 4.4 | 2.1 | 48.3 |
If Kx is the only receptor for AMA1, then why is there invasion of Kxnull erythrocytes, albeit at a reduced rate? First, the redundancy of invasion pathways have been described in P. falciparum, P. knowlesi, and P. yoelii (1). Deletion of EBA-175 does not eliminate invasion, possibly because there are multiple other DBL genes and potentially other receptors (22). P. knowlesi cannot invade human Duffy blood group negative erythrocytes, but if the Duffy negative erythrocytes are treated with trypsin or neuraminidase (3, 23), invasion occurs in the absence of the receptor. P. yoelii can invade mouse Duffynull reticulocytes at a normal rate but poorly invades into Duffynull normocytes (24), indicating an alternative pathway for invasion of reticulocytes. Thus, redundancies are a part of the strategy of Plasmodium to survive variation or deletion of proteins on erythrocytes. The only known exception is the elimination of P. vivax from West Africa by the lack of expression of the Duffy blood group antigen on erythrocytes.
From the crystal structure of AMA1 (8), it was shown that domains I and II consist of two PAN domains that may mediate protein–protein or protein–carbohydrate interactions (25). However, Plasmodium has been shown to use AMA1 for both erythrocyte invasion by merozoites (5) and hepatocyte invasion by sporozoites (26). Because AMA1 is a single-copy gene, it is possible that different regions of AMA1 are involved in invasion at different stages of the parasite lifecycle. Although an invasion blocking monoclonal antibody was mapped to domain II (8), monoclonal antibodies to domain III also block invasion (ref. 27; L.H.M., unpublished data). Domain III has no homology to protein structures in other organisms (8). Domain III, like other malarial domains (e.g., the Duffy binding domain), may be a structure specific to the Apicomplexa or Plasmodium. These domains might play an important role in extracellular interactions with host molecules.
This article begins to describe the molecular interactions of P. falciparum AMA1 with erythrocytes. First, binding of AMA1 to erythrocytes requires the proteolytic cleavage of the erythrocyte. Previous data on P. chabaudi merozoites showed that diisopropylfluorophosphate-treated merozoites could not invade erythrocytes unless the erythrocytes were pretreated with α-chymotrypsin or proteases extracted from parasites (21). This observation suggests that the protease modifies the erythrocyte surface during invasion. Second, the findings suggest that proteolytic cleavage of AMA1 is required for invasion. Proteolytic digestion of AMA1 has been described (9) and may explain why domain III of AMA1 binds but not domains I/II/III. Third, Kxnull erythrocytes are invaded at a reduced rate, but the invasion still occurs, suggesting that there is a redundancy in the requirement for AMA1 in the invasion sequence. Redundancy during many steps in invasion has been described for P. falciparum (1). The only other study on AMA1's role in invasion is the demonstration that AMA1 of the rodent parasite expressed in P. falciparum increases invasion of mouse erythrocytes (5). Taken together, these data reveal the role of AMA1 in merozoite invasion and open up the possibility that parasite proteases might play a role in the process.
Acknowledgments
We thank Dr. Olivier Bertrand, Director of Research, Institut National de la Santé et de la Recherche Médicale U665, for the generous gift of the mAb against the Kx protein; Dr. James Dvorak, Laboratory of Malaria and Vector Research (LMVR), National Institute of Allergy and Infectious Diseases (NIAID), for help in setting up the microscope for counting the cells; Drs. Owen Schwartz and Juraj Kabat (NIAID) for help with microscopy; Dr. Jiang-bing Mu, (LMVR, NIAID) for finger-printing the parasites; Michael Fay (NIAID) for help with statistical analysis; Angela Picket, the Blood Bank, National Institutes of Health, for some null erythrocytes; Kelly Rausch, Malaria Vaccine Development Branch (NIAID) for imagequant analysis of the immunoblots of enzyme-treated erythrocytes. M.R. was supported in part by National Institutes of Health Specialized Center of Research Grant HL54459 in transfusion medicine and biology.
Abbreviations: AMA1, apical membrane antigen 1; CGD, chronic granulomatous disease.
References
- 1.Miller, L. H., Baruch, D. I., Marsh, K. & Doumbo, O. K. (2002) Nature 415, 673–679. [DOI] [PubMed] [Google Scholar]
- 2.Chitnis, C.E. & Miller, L. H. (1994) J. Exp. Med. 180, 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller, L. H., Aikawa, M., Johnson, J. G. & Shiroishi, T. (1979) J. Exp. Med. 149, 172–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bannister, L. H., Hopkins, J. M., Dluzewski, A. R., Margos, G., Williams, I. T., Blackman, M. J., Kocken, C. H., Thomas, A. W. & Mitchell, G. H. (2003) J. Cell Sci. 116, 3825–3834. [DOI] [PubMed] [Google Scholar]
- 5.Triglia, T., Healer, J., Caruana, S. R., Hodder, A. N., Anders, R. F., Crabb, B. S. & Cowman, A. F. (2000) Mol. Microbiol. 38, 706–718. [DOI] [PubMed] [Google Scholar]
- 6.Crewther, P. E., Matthew, M. L., Flegg, R. H. & Anders, R. F. (1996) Infect. Immun. 64, 3310–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hodder, A. N., Crewther, P. E., Matthew, M. L., Reid, G. E., Moritz, R. L., Simpson, R. J. & Anders, R. F. (1996) J. Biol. Chem. 271, 29446–29452. [DOI] [PubMed] [Google Scholar]
- 8.Pizarro, J. C., Vulliez-Le Normand, B., Chesne-Seck, M. L., Collins, C. R., Withers-Martinez, C., Hackett, F., Blackman, M. J., Faber, B. W., Remarque, E. J., Kocken, C. H. M., et al., Science, in press. [DOI] [PubMed]
- 9.Howell, S. A., Withers-Martinez, C., Kocken, C. H., Thomas, A.W. & Blackman, M. J. (2001) J. Biol. Chem. 276, 31311–31320. [DOI] [PubMed] [Google Scholar]
- 10.Fraser, T. S., Kappe, S. H., Narum, D. L., VanBuskirk, K. M. & Adams, J. H. (2001) Mol. Biochem. Parasitol. 117, 49–59. [DOI] [PubMed] [Google Scholar]
- 11.Mayer, D. C. G., Mu, J.-B., Kaneko, O., Duan, J., Su, X.-Z. and Miller, L. H. (2004) Proc. Natl. Acad. Sci. USA 101, 2518–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sim, B. K., Chitnis, C. E., Wasniowska, K., Hadley, T. J. & Miller, L. H. (1994) Science 264, 1941–1944. [DOI] [PubMed] [Google Scholar]
- 13.Judd, W. J., ed. (1994) in Methods in Immunohematology (Montgomery Scientific, Durham, NC), pp. 188–190.
- 14.Mallory, D., ed. (1993) Immunohematology Methods and Procedures (Am. Red Cross, Nat. Ref. Lab., Rockville, MD), pp. 121–125.
- 15.Kaneko, O., Soubes, S. C. & Miller, L. H. (1999) Exp. Parasitol. 93, 116–119. [DOI] [PubMed] [Google Scholar]
- 16.Mu, J., Duan, J., Makova, K. D., Joy, D. A., Huynh, C. Q., Branch, O. H., Li, W. H. & Su, X. Z. (2002) Nature 418, 323–326. [DOI] [PubMed] [Google Scholar]
- 17.Carbonnet, F., Blanchard, D., Hattab, C., Cochet, S., Petit-Leroux, Y., Loirat, M. J., Cartron, J. P. & Bertrand, O. (2000) Transfus. Med. 10, 145–154. [DOI] [PubMed] [Google Scholar]
- 18.Hattab, C., Blanchard, D., Gane, P., Verkarre, V., Petit-Leroux, Y., Loirat, M. J., Cartron, J. P. & Bertrand, O. (2003) Transfus. Med. 13, 43–48. [DOI] [PubMed] [Google Scholar]
- 19.Russo, D., Redman, C. & Lee, S. (1998) J. Biol. Chem. 273, 13950–13956. [DOI] [PubMed] [Google Scholar]
- 20.Carbonnet, F., Hattab, C., Collec, E., Le Van Kim, C., Cartron, J. P. & Bertrand, O. (1997) Br. J. Haematol. 96, 857–863. [DOI] [PubMed] [Google Scholar]
- 21.Braun-Breton, C., Rosenberry, T. L. & da Silva, L. P. (1988) Nature 332, 457–459. [DOI] [PubMed] [Google Scholar]
- 22.Kaneko, O., Fidock, D. A., Schwartz, O. M. & Miller, L. H. (2000) Mol. Biochem. Parasitol. 110, 135–146. [DOI] [PubMed] [Google Scholar]
- 23.Mason, S. J., Miller, L. H., Shiroishi, T., Dvorak, J. A. & McGinniss, M. H. (1977) Br. J. Haematol. l36, 327–335. [DOI] [PubMed] [Google Scholar]
- 24.Swardson-Olver, C. J., Dawson, T. C., Burnett, R. C., Peiper, S. C., Maeda, N. & Avery, A. C. (2002) Blood 99, 2677–2684. [DOI] [PubMed] [Google Scholar]
- 25.Tordai, H., Banyai, L. & Patthy, L. (1999) FEBS Lett. 461, 63–67. [DOI] [PubMed] [Google Scholar]
- 26.Silvie, O., Franetich, J. F., Charrin, S., Mueller, M. S., Siau, A., Bodescot, M., Rubenstein, E., Hannoun, L., Charoenvit, Y., Kochen, C. H., et al. (2004) J. Biol. Chem. 279, 9490–9496. [DOI] [PubMed] [Google Scholar]
- 27.Mueller, M. S., Renard, A., Boato, F., Vogel, D., Naegeli, M., Zurbriggen, R., Robinson, J. A. & Pluschke, G. (2003) Infect. Immun. 71, 4749–4758. [DOI] [PMC free article] [PubMed] [Google Scholar]