Abstract
Chlorophyll biosynthesis is one of the most important cellular processes and is essential for plant photosynthesis. After germination under the soil, dark-grown seedlings are etiolated and accumulate the chlorophyll precursor protochlorophyllide (Pchlide) in cotyledons. Upon exposure to light, Pchlide is rapidly converted to chlorophyll to initiate photoautotrophic growth. In this light-regulated de-etiolation process, multiple endogenous phytohormones are also involved. Although the co-regulation of seedling greening by light and hormones has long been observed, recent studies greatly advanced our understanding of their interplay by identifying the key components connecting these pathways. The integrators, such as PHYTOCHROME-INTERACTING FACTORs, ELONGATED HYPOCOTYL 5, ETHYLENE INSENSTIVE 3 and DELLA proteins, are key transcription regulators in light or hormone signaling pathways. This review focuses on these integrators and illustrates the regulatory networks of light and hormone interactions in chlorophyll biosynthesis.
Keywords: light signaling, plant hormones, chlorophyll biosynthesis, de-etiolation, PIFs, HY5, EIN3/EIL1, DELLAs
Introduction
Crop seeds are usually buried in soil, whereby post-germinative seedlings become etiolated and grow upward heterotrophically from seed reserves. Upon reaching the soil surface, etiolated seedlings undergo a dramatic developmental transition termed de-etiolation, which includes cotyledon opening and greening (Von Arnim and Deng, 1996; Chen et al., 2004). This transition is of particular vulnerability for plant survival, requiring rapid initiation of photoautotrophic growth without causing photooxidation (Huq et al., 2004; Zhong et al., 2014). To successfully accomplish this, chlorophyll biosynthesis must be strictly controlled.
In higher plants, chlorophyll is initially biosynthesized from glutamate, which is then converted to 5-aminolevulinic acid (ALA) and further converted to protochlorophyllide (Pchlide) (Tanaka et al., 2011). When the dark-grown seedlings are exposed to light, the rate-limiting enzymes NADPH protochlorophyllide oxidoreductases (PORs) are photoactivated and catalyze the conversion of Pchlide to chlorophyllide, which is subsequently esterified to mature chlorophyll (Fujita, 1996; Reinbothe et al., 2010). In Arabidopsis, three POR enzymes, PORA, PORB and PORC have been identified, with PORA/PORB playing the main roles in young seedlings (Buhr et al., 2008). Because Pchlide is extremely phototoxic, its amount must be stoichiometrically matched with the level of POR enzymes. Over-accumulation of the free Pchlide that cannot be converted to chlorophyll in time will result in the production of reactive oxygen species (ROS) upon light exposure, causing photooxidative damage to the seedlings (op den Camp et al., 2003; Huq et al., 2004; Chen et al., 2013; Zhong et al., 2014). To survive, seedlings have evolved efficient ways to adjust the levels of Pchilde and POR enzymes to allow for rapid establishment of photosynthesis without causing photobleaching (op den Camp et al., 2003; Huq et al., 2004; Zhong et al., 2014). Moreover, carotenoid biosynthesis is also markedly upregulated to protect the etioplasts from photooxidative damage by quenching excess excitation energy when seedlings are exposed to light (Rodriguez-Villalon et al., 2009).
Light is the main environmental factor that regulates the pathway of chlorophyll biosynthesis, while plant hormones are also recruited to mediate the developmental switch of de-etiolation. Some key components in the light signaling pathway, such as PIFs and HY5, connect light signals to the signaling pathways of multiple phytohormones, including ethylene, gibberellin (GA) and cytokinin (CK). In this review, we concentrate on how chlorophyll biosynthesis is cooperatively regulated by light and endogenous hormone signals, focusing on the interplay between light and hormone signaling pathways during seedling de-etiolation.
Chlorophyll Biosynthesis Regulated by Key Components in Light Signaling Pathway
Light provides plants with energy for photosynthesis and a major source of information about their environment. Both light quality and quantity are constantly monitored by plants through a group of photoreceptors (Quail, 2002; Chen et al., 2004). Among them, phytochromes (phys, including phyA-phyE in Arabidopsis) sense far-red and red light (Quail, 2002; Chen et al., 2004). The perception of light signals by phys initiates an intracellular transduction to alter the expression of nuclear genes (Quail, 2002; Chen et al., 2004; Leivar and Quail, 2011). There are two groups of transcription factors, PIFs and HY5, that mediate light-induced responses in opposite ways (Von Arnim and Deng, 1996; Chen et al., 2004; Leivar and Quail, 2011). PIFs are negative regulators and are directly targeted by photoactivated phys for degradation (Ni et al., 1998, 2014; Huq et al., 2004; Shen et al., 2008), while HY5 is stabilized by light to promote photomorphogenesis (Ang et al., 1998; Osterlund et al., 2000). During the process of de-etiolation, both PIFs and HY5 have been shown to transcriptionally regulate the chlorophyll biosynthesis pathway.
PHYTOCHROME-INTERACTING FACTORs accumulate in dark-grown seedlings and negatively regulate the tetrapyrrole metabolism of chlorophyll biosynthesis (Huq et al., 2004; Monte et al., 2004; Moon et al., 2008; Shin et al., 2009; Stephenson et al., 2009). Mutation of PIF1 and PIF3 results in an excessive amount of Pchlide in the dark and causes severe photobleaching upon light exposure (Huq et al., 2004; Monte et al., 2004; Moon et al., 2008; Shin et al., 2009; Stephenson et al., 2009). Further experiments reveal that PIF1 directly binds to the promoter of PORC, while PIF3 represses the expression of HEMA1, GUN4 and CHLH genes (Moon et al., 2008; Stephenson et al., 2009). HEMA1 is the main glutamyl-tRNA reductase that catalyzes the rate-limiting step for ALA biosynthesis, while GUN4 and CHLH promote the conversion of ALA to the chlorophyll biosynthetic branch (Stephenson and Terry, 2008; Tanaka et al., 2011). In addition, PIF5 has been shown to be involved in the negative regulation of CHLH gene expression in etiolated seedlings (Shin et al., 2009), and a large portion of nuclear-encoded chlorophyll biosynthesis genes are notably upregulated in the pifQ mutant (lacking PIF1, PIF3, PIF4 and PIF5 genes) (Leivar et al., 2009; Shin et al., 2009). PIF1 was also found to partly repress the transposase-derived transcription factor FHY3/FAR1-activated gene expression of HEMB1 that encodes the ALA dehydratase (Tang et al., 2012), and chromatin-remodeling enzyme BRM interacts with PIF1 to modulate PORC expression (Zhang et al., 2017). Moreover, PIFs have been reported to directly repress the gene expression of PSY (phytoene synthase), which is the main rate-determining enzyme of carotenoid biosynthesis (Toledo-Ortiz et al., 2010). When PIFs are degraded by light, carotenoids are rapidly synthesized to coordinate with chlorophyll biosynthesis, thus facilitating the assembly of functional photosynthetic machinery (Toledo-Ortiz et al., 2010). Therefore, PIFs play important roles in the fine tuning of tetrapyrrole metabolism, directly or indirectly regulating chlorophyll biosynthesis and photosynthetic genes to optimize the seedling greening process.
ELONGATED HYPOCOTYL 5 functions downstream of the photoreceptors and central repressors in the light signaling pathway to promote seedling photomorphogenesis. In the dark, HY5 is degraded through the COP1/DET1-mediated ubiquitination degradation pathway (Ang et al., 1998; Osterlund et al., 2000). HY5 plays a vital role in the convergence of blue, red and far-red light-signal pathways for regulating the transcription levels of HEMA1 (McCormac and Terry, 2002). Several nuclear-encoding photosynthetic and chlorophyll biosynthesis genes, such as CHLH, GUN4, PORC, CAO and CHL27, are the putative targets of HY5 (Lee et al., 2007). Although roots are heterotrophic organs, lots of chlorophyll accumulates in light-grown det1 and cop1 mutant roots, and HY5 mediates the process of chlorophyll synthesis in roots (Chory and Peto, 1990; Deng et al., 1992; Ang et al., 1998). In addition, a Myb-like transcription factor REVEILLE1 (RVE1) was recently found to act downstream of phyB to modulate chlorophyll biosynthesis by directly activating PORA expression (Xu et al., 2015; Jiang et al., 2016).
Ethylene Is Crucial for Cotyledon Greening and Survival of Seedling Soil Emergence
Plant hormones are small molecules that mediate a myriad of cellular responses. Many hormones are involved in light-induced seedling greening. One prominent factor affecting chlorophyll biosynthesis is ethylene, which dramatically represses Pchlide accumulation and induces the gene expression of both PORA and PORB in etiolated seedlings (Zhong et al., 2009, 2010, 2014). Thus, ethylene plays a critical role in protecting cotyledons from photooxidative damage when the seedlings are exposed to light. The effects of ethylene are mediated by EIN3/EIL1, the master transcription factors in the ethylene signaling pathway (Chao et al., 1997; Guo and Ecker, 2004). EIN3/EIL1 markedly repress the accumulation of Pchlide and directly bind to the promoters of PORA and PORB to activate their gene expression (Zhong et al., 2009, 2010, 2014). Genetic studies reveal that EIN3/EIL1 cooperate with PIF1 and act downstream of COP1 in promoting seedling greening (Zhong et al., 2009). The protein levels of EIN3 are enhanced by COP1 but are decreased by light (Zhong et al., 2009; Shi et al., 2016a,b). In addition, overexpressing EIN3 rescues the far-red light-triggered cotyledon greening defects (Zhong et al., 2009).
After germination in soil, the mechanical impedance of soil boosts ethylene production to adjust seedling morphogenesis to enhance the lifting capacity and protect against mechanical injuries (Zhong et al., 2014; Shen et al., 2016; Shi et al., 2016a). EIN3/EIL1 directly activate two independent pathways, an ERF1 pathway to slow down cell elongation and a PIF3 pathway to control Pchlide biosynthesis (Zhong et al., 2012, 2014). These two pathways are coupled to maintain a suitable amount of Pchilde to rapidly initiate photoautotrophic growth without causing photooxidation upon emergence (Zhong et al., 2014). When seedlings penetrate their way toward the surface, the dim light under the soil increases gradually and represses COP1 protein activity (Shi et al., 2016a). COP1 has been found to be the E3 ligase of EBF1 and EBF2, the F-box proteins of the E3 ligases for EIN3 degradation (Shi et al., 2016a). Therefore, COP1 and ethylene mediate the soil-imposed light and mechanical stress signals, respectively, to adjust EIN3 protein levels in response to soil condition changes when seedlings grow upward in the soil (Shi et al., 2016a). Interestingly, EIN3 also promotes the nuclear enrichment of COP1 protein to generate a positive feedback for EIN3 stability regulation (Yu et al., 2013, 2016). At the moment of emergence and reaching sunlight, photoactivated photoreceptor phyB directly interacts with EIN3 and rapidly degrades EIN3 by bringing it to the E3 ligases EBF1 and EBF2 (Shi et al., 2016b). As a result, the repression of photomorphogenesis by EIN3 and ethylene is rapidly lifted to initiate de-etiolation effectively.
Gibberellin Regulates Chlorophyll Biogenesis Partially via the Light Signaling Pathway
Seedling de-etiolation is also subject to gibberellin (GA) regulation, as inhibiting gibberellin signaling can induce partial photomorphogenesis in the dark (Alabadi et al., 2004, 2008). DELLAs are a subfamily of the GRAS transcriptional regulators and negatively regulate gibberellin signaling to repress GA-mediated responses (Jiang and Fu, 2007). Moreover, DELLAs inhibit the transcription activity of PIF3 and PIF4 through direct blocking of the DNA-recognition domain of these factors (de Lucas et al., 2008; Feng et al., 2008). In dark-grown seedlings, DELLAs accumulate and regulate the biosynthetic pathways of both carotenoid and chlorophyll (Cheminant et al., 2011). DELLAs upregulate the expression of genes involved in chlorophyll biosynthesis (CHLH, PORC and CAO) and photosynthesis (LHCB2.2, PSAG and PSAE-1) in a PIF-dependent manner (Cheminant et al., 2011). In addition, DELLAs also positively regulate PORA and PORB gene expression independently of PIFs and repress ROS-induced photooxidative damage during de-etiolation (Cheminant et al., 2011). However, the regulation of HY5 on gibberellin-mediated chlorophyll biosynthesis seems more moderate than that of PIFs in dark conditions (Cheminant et al., 2011).
Cytokinin Plays an Important Role in Chlorophyll Biosynthesis and Chloroplast Development
Exogenous cytokinin treatment induces cotyledon expansion and chloroplast partial differentiation (Chory et al., 1994; Vandenbussche et al., 2007). Two GATA family transcription factors, GNC and CGA1/GNL, are induced by cytokinin and regulate the expression of many chloroplast-related genes (Hudson et al., 2011; Chiang et al., 2012). Dark-grown seedlings display small etioplasts with prolamellar bodies in the absence of cytokinin, while large lens-shaped plastids contain some prothylakoid membranes in the presence of cytokinin (Chory et al., 1994). Recent reports indicate that cytokinin mediates the etioplast-to-chloroplast transition by promoting characteristic ultrastructural changes (Cortleven and Schmulling, 2015; Cortleven et al., 2016). Cytokinin signal is perceived by the receptors AHK2 and AHK3 and transduced to B-type ARR transcription factors (Argyros et al., 2008). ARRs directly regulate the expression of genes in chlorophyll biosynthesis and the light harvesting complex, such as HEMA1 and LHCB6 (Cortleven and Schmulling, 2015; Cortleven et al., 2016). As cytokinin has been reported to increase the protein levels of HY5 (Vandenbussche et al., 2007), it is possible that HY5 is a point of convergence between light and cytokinin signaling pathways.
The Function of Other Plant Hormones in Regulating Seedling Greening
In addition to the well-documented hormones just described, other hormones are also important in regulating seedling greening. Auxin represses HY5 protein accumulation via IAA14 and its regulatory target ARFs in roots (Kobayashi et al., 2012). Moreover, chlorophyll synthesis genes are markedly activated in detached roots via cytokinin but are repressed by auxin (Kobayashi et al., 2017), suggesting that auxin signaling is also involved in the regulation of chlorophyll biosynthesis in the root greening response. However, further analyses are required to elucidate the regulatory network of auxin and light signals in regulating chlorophyll biosynthesis. Brassinosteroid (BR) is known to be involved in the process of de-etiolation. Many chlorophyll biosynthesis genes are upregulated from the microarray data of BR-insensitive bri1-116 seedlings in darkness (Sun et al., 2010). The key transcriptional factor GATA2 has been identified in mediating the crosstalk between BR and light signaling pathways (Luo et al., 2010). Recently, ABI4 was found to activate COP1 expression to repress seedling de-etiolation (Xu et al., 2016). In addition, strigolactones are reported to also be involved in light signaling via regulating the nuclear localization of COP1 (Tsuchiya et al., 2010), and jasmonate inhibits COP1 activity to promote photomorphogenesis (Zheng et al., 2017). However, the signaling pathway of ABA, strigolactones and jasmonate in regulating chlorophyll biosynthesis remains largely unknown.
Conclusion and Perspectives
Involvement of plant hormones in light-regulated seedling greening has been known for decades. However, we have not identified the molecular links connecting light signaling to the multiple hormonal pathways until recent years. The key transcription factors of both light and hormone signaling pathways appear to be the integrators (Figure 1). EIN3 directly activates the gene expression of PORA/PORB and represses Pchlide accumulation to optimize the greening process. The repression of EIN3 in synthesizing Pchlide is through activating PIF3 transcription, whereas both phyB and COP1 predominantly regulate the protein levels of EIN3. PIFs play a pivotal role in integrating light and GA signals, and DELLAs directly sequester the transcription activity of PIFs. In addition, HY5 protein stability is regulated by auxin and cytokinin to coordinate these signals in mediating root greening, while COP1 could be new integrator as its nuclear localization can be regulated by ethylene, strigolactone and jasmonate hormones. Further studies, such as identifying additional integrators in light and hormonal signaling pathways and addressing how these components are integrated in regulating seedling greening, are needed. Moreover, we are only beginning to address the regulation of chloroplast development. Whether and how plant hormones regulate the etioplast-chloroplast differentiation process is critical in filling the gaps of greening. In summary, although we have not obtained a detailed network depicting how seedling greening is regulated by light and all the hormonal signals, the identification of key transcription regulators as signaling integrators has created a great starting point.
FIGURE 1.
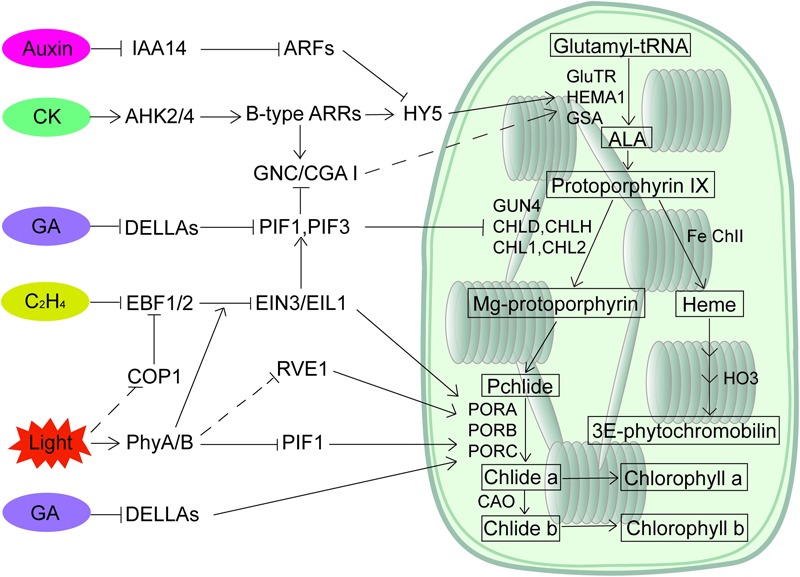
A regulatory network of chlorophyll biosynthesis by light and plant hormones. PIFs and HY5 are major transcription factors in mediating light-regulated chlorophyll biosynthesis. Multiple hormones participate in chlorophyll biosynthesis through both transcriptional and post-transcriptional regulation of PIFs and HY5, while light regulates the action of key components such as EIN3/EIL1 in hormone signaling pathways to modulate the hormonal responses.
Author Contributions
SZ proposed the topic. SZ, XL, and YL collected the literature and critically assessed the information. XL and SZ wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize to our colleagues whose work could not be included because of space constraints.
Footnotes
Funding. This work was supported by grants from the National Key Research and Development Program of China (2016YFA0502900) and the National Science Foundation of China (31570188) to SZ. XL was supported by a China Post-doctoral Science Foundation Grant (2016M600857) and the Outstanding Post-doctoral Fellowship of Peking-Tsinghua Center for Life Sciences.
References
- Alabadi D., Gallego-Bartolome J., Orlando L., Garcia-Carcel L., Rubio V., Martinez C., et al. (2008). Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J. 53 324–335. 10.1111/j.1365-313X.2007.03346.x [DOI] [PubMed] [Google Scholar]
- Alabadi D., Gil J., Blazquez M. A., Garcia-Martinez J. L. (2004). Gibberellins repress photomorphogenesis in darkness. Plant Physiol. 134 1050–1057. 10.1104/pp.103.035451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang L. H., Chattopadhyay S., Wei N., Oyama T., Okada K., Batschauer A., et al. (1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1 213–222. 10.1016/S1097-2765(00)80022-2 [DOI] [PubMed] [Google Scholar]
- Argyros R. D., Mathews D. E., Chiang Y. H., Palmer C. M., Thibault D. M., Etheridge N., et al. (2008). Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20 2102–2116. 10.1105/tpc.108.059584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhr F., El Bakkouri M., Valdez O., Pollmann S., Lebedev N., Reinbothe S., et al. (2008). Photoprotective role of NADPH: protochlorophyllide oxidoreductase A. Proc. Natl. Acad. Sci. U.S.A. 105 12629–12634. 10.1073/pnas.0803950105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Q. M., Rothenberg M., Solano R., Roman G., Terzaghi W., Ecker J. R. (1997). Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89 1133–1144. 10.1016/S0092-8674(00)80300-1 [DOI] [PubMed] [Google Scholar]
- Cheminant S., Wild M., Bouvier F., Pelletier S., Renou J. P., Erhardt M., et al. (2011). DELLAs regulate chlorophyll and carotenoid biosynthesis to prevent photooxidative damage during seedling deetiolation in Arabidopsis. Plant Cell 23 1849–1860. 10.1105/tpc.111.085233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Xu G., Tang W., Jing Y., Ji Q., Fei Z., et al. (2013). Antagonistic basic helix-loop-helix/bZIP transcription factors form transcriptional modules that integrate light and reactive oxygen species signaling in Arabidopsis. Plant Cell 25 1657–1673. 10.1105/tpc.112.104869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Chory J., Fankhauser C. (2004). Light signal transduction in higher plants. Annu. Rev. Genet. 38 87–117. 10.1146/annurev.genet.38.072902.092259 [DOI] [PubMed] [Google Scholar]
- Chiang Y. H., Zubo Y. O., Tapken W., Kim H. J., Lavanway A. M., Howard L., et al. (2012). Functional characterization of the GATA transcription factors GNC and CGA1 reveals their key role in chloroplast development, growth, and division in Arabidopsis. Plant Physiol. 160 332–348. 10.1104/pp.112.198705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J., Peto C. A. (1990). Mutations in the DET1 gene affect cell-type-specific expression of light-regulated genes and chloroplast development in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 87 8776–8780. 10.1073/pnas.87.22.8776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J., Reinecke D., Sim S., Washburn T., Brenner M. (1994). A role for cytokinins in de-etiolation in Arabidopsis (det mutants have an altered response to cytokinins). Plant Physiol. 104 339–347. 10.1104/pp.104.2.339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortleven A., Marg I., Yamburenko M. V., Schlicke H., Hill K., Grimm B., et al. (2016). Cytokinin regulates the etioplast-chloroplast transition through the two-component signaling system and activation of chloroplast-related genes. Plant Physiol. 172 464–478. 10.1104/pp.16.00640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortleven A., Schmulling T. (2015). Regulation of chloroplast development and function by cytokinin. J. Exp. Bot. 66 4999–5013. 10.1093/jxb/erv132 [DOI] [PubMed] [Google Scholar]
- de Lucas M., Daviere J. M., Rodriguez-Falcon M., Pontin M., Iglesias-Pedraz J. M., Lorrain S., et al. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451 480–484. 10.1038/nature06520 [DOI] [PubMed] [Google Scholar]
- Deng X. W., Matsui M., Wei N., Wagner D., Chu A. M., Feldmann K. A., et al. (1992). COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell 71 791–801. 10.1016/0092-8674(92)90555-Q [DOI] [PubMed] [Google Scholar]
- Feng S., Martinez C., Gusmaroli G., Wang Y., Zhou J., Wang F., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451 475–479. 10.1038/nature06448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y. (1996). Protochlorophyllide reduction: a key step in the greening of plants. Plant Cell Physiol. 37 411–421. 10.1093/oxfordjournals.pcp.a028962 [DOI] [PubMed] [Google Scholar]
- Guo H., Ecker J. R. (2004). The ethylene signaling pathway: new insights. Curr. Opin. Plant Biol. 7 40–49. 10.1016/j.pbi.2003.11.011 [DOI] [PubMed] [Google Scholar]
- Hudson D., Guevara D., Yaish M. W., Hannam C., Long N., Clarke J. D., et al. (2011). GNC and CGA1 modulate chlorophyll biosynthesis and glutamate synthase (GLU1/Fd-GOGAT) expression in Arabidopsis. PLoS ONE 6:e26765 10.1371/journal.pone.0026765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E., Al-Sady B., Hudson M., Kim C., Apel K., Quail P. H. (2004). Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305 1937–1941. 10.1126/science.1099728 [DOI] [PubMed] [Google Scholar]
- Jiang C., Fu X. (2007). GA action: turning on de-DELLA repressing signaling. Curr. Opin. Plant Biol. 10 461–465. 10.1016/j.pbi.2007.08.011 [DOI] [PubMed] [Google Scholar]
- Jiang Z., Xu G., Jing Y., Tang W., Lin R. (2016). Phytochrome B and REVEILLE1/2-mediated signalling controls seed dormancy and germination in Arabidopsis. Nat. Commun. 7:12377 10.1038/ncomms12377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K., Baba S., Obayashi T., Sato M., Toyooka K., Keranen M., et al. (2012). Regulation of root greening by light and auxin/cytokinin signaling in Arabidopsis. Plant Cell 24 1081–1095. 10.1105/tpc.111.092254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K., Ohnishi A., Sasaki D., Fujii S., Iwase A., Sugimoto K., et al. (2017). Shoot removal induces chloroplast development in roots via cytokinin signaling. Plant Physiol. 173 2340–2355. 10.1104/pp.16.01368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., He K., Stolc V., Lee H., Figueroa P., Gao Y., et al. (2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19 731–749. 10.1105/tpc.106.047688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Quail P. H. (2011). PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 16 19–28. 10.1016/j.tplants.2010.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Tepperman J. M., Monte E., Calderon R. H., Liu T. L., Quail P. H. (2009). Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 21 3535–3553. 10.1105/tpc.109.070672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X. M., Lin W. H., Zhu S., Zhu J. Y., Sun Y., Fan X. Y., et al. (2010). Integration of light- and brassinosteroid-signaling pathways by a GATA transcription factor in Arabidopsis. Dev. Cell 19 872–883. 10.1016/j.devcel.2010.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormac A. C., Terry M. J. (2002). Light-signalling pathways leading to the co-ordinated expression of HEMA1 and Lhcb during chloroplast development in Arabidopsis thaliana. Plant J. 32 549–559. 10.1046/j.1365-313X.2002.01443.x [DOI] [PubMed] [Google Scholar]
- Monte E., Tepperman J. M., Al-Sady B., Kaczorowski K. A., Alonso J. M., Ecker J. R., et al. (2004). The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc. Natl. Acad. Sci. U.S.A. 101 16091–16098. 10.1073/pnas.0407107101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J., Zhu L., Shen H., Huq E. (2008). PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 105 9433–9438. 10.1073/pnas.0803611105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M., Tepperman J. M., Quail P. H. (1998). PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95 657–667. 10.1016/S0092-8674(00)81636-0 [DOI] [PubMed] [Google Scholar]
- Ni W., Xu S. L., Tepperman J. M., Stanley D. J., Maltby D. A., Gross J. D., et al. (2014). A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science 344 1160–1164. 10.1126/science.1250778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- op den Camp R. G. L., Przybyla D., Ochsenbein C., Laloi C., Kim C. H., Danon A., et al. (2003). Rapid induction of distinct stress responses after the release of singlet oxygen in arabidopsis. Plant Cell 15 2320–2332. 10.1105/tpc.014662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund M. T., Hardtke C. S., Wei N., Deng X. W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405 462–466. 10.1038/35013076 [DOI] [PubMed] [Google Scholar]
- Quail P. H. (2002). Photosensory perception and signalling in plant cells: new paradigms? Curr. Opin. Cell Biol. 14 180–188. 10.1016/S0955-0674(02)00309-5 [DOI] [PubMed] [Google Scholar]
- Reinbothe C., El Bakkouri M., Buhr F., Muraki N., Nomata J., Kurisu G., et al. (2010). Chlorophyll biosynthesis: spotlight on protochlorophyllide reduction. Trends Plant Sci. 15 614–624. 10.1016/j.tplants.2010.07.002 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Villalon A., Gas E., Rodriguez-Concepcion M. (2009). Phytoene synthase activity controls the biosynthesis of carotenoids and the supply of their metabolic precursors in dark-grown Arabidopsis seedlings. Plant J. 60 424–435. 10.1111/j.1365-313X.2009.03966.x [DOI] [PubMed] [Google Scholar]
- Shen H., Zhu L., Castillon A., Majee M., Downie B., Huq E. (2008). Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell 20 1586–1602. 10.1105/tpc.108.060020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Li Y., Pan Y., Zhong S. (2016). Activation of HLS1 by mechanical stress via ethylene-stabilized EIN3 is crucial for seedling soil emergence. Front. Plant Sci. 7:1571 10.3389/fpls.2016.01571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Liu R., Xue C., Shen X., Wei N., Deng X. W., et al. (2016a). Seedlings transduce the depth and mechanical pressure of covering soil using COP1 and ethylene to regulate EBF1/EBF2 for soil emergence. Curr. Biol. 26 139–149. 10.1016/j.cub.2015.11.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Shen X., Liu R., Xue C., Wei N., Deng X. W., et al. (2016b). The red light receptor phytochrome B directly enhances substrate-E3 ligase interactions to attenuate ethylene responses. Dev. Cell 39 597–610. 10.1016/j.devcel.2016.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Kim K., Kang H., Zulfugarov I. S., Bae G., Lee C. H., et al. (2009). Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc. Natl. Acad. Sci. U.S.A. 106 7660–7665. 10.1073/pnas.0812219106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson P. G., Fankhauser C., Terry M. J. (2009). PIF3 is a repressor of chloroplast development. Proc. Natl. Acad. Sci. U.S.A. 106 7654–7659. 10.1073/pnas.0811684106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson P. G., Terry M. J. (2008). Light signalling pathways regulating the Mg-chelatase branchpoint of chlorophyll synthesis during de-etiolation in Arabidopsis thaliana. Photochem. Photobiol. Sci. 7 1243–1252. 10.1039/b802596g [DOI] [PubMed] [Google Scholar]
- Sun Y., Fan X. Y., Cao D. M., Tang W., He K., Zhu J. Y., et al. (2010). Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell 19 765–777. 10.1016/j.devcel.2010.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka R., Kobayashi K., Masuda T. (2011). Tetrapyrrole metabolism in Arabidopsis thaliana. Arabidopsis Book 9:e0145 10.1199/tab.0145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Wang W., Chen D., Ji Q., Jing Y., Wang H., et al. (2012). Transposase-derived proteins FHY3/FAR1 interact with PHYTOCHROME-INTERACTING FACTOR1 to regulate chlorophyll biosynthesis by modulating HEMB1 during deetiolation in Arabidopsis. Plant Cell 24 1984–2000. 10.1105/tpc.112.097022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Huq E., Rodriguez-Concepcion M. (2010). Direct regulation of phytoene synthase gene expression and carotenoid biosynthesis by phytochrome-interacting factors. Proc. Natl. Acad. Sci. U.S.A. 107 11626–11631. 10.1073/pnas.0914428107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya Y., Vidaurre D., Toh S., Hanada A., Nambara E., Kamiya Y., et al. (2010). A small-molecule screen identifies new functions for the plant hormone strigolactone. Nat. Chem. Biol. 6 741–749. 10.1038/nchembio.435 [DOI] [PubMed] [Google Scholar]
- Vandenbussche F., Habricot Y., Condiff A. S., Maldiney R., Van Der Straeten D., Ahmad M. (2007). HY5 is a point of convergence between cryptochrome and cytokinin signalling pathways in Arabidopsis thaliana. Plant J. 49 428–441. 10.1111/j.1365-313X.2006.02973.x [DOI] [PubMed] [Google Scholar]
- Von Arnim A., Deng X. W. (1996). Light control of seedling development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47 215–243. 10.1146/annurev.arplant.47.1.215 [DOI] [PubMed] [Google Scholar]
- Xu G., Guo H., Zhang D., Chen D., Jiang Z., Lin R. (2015). REVEILLE1 promotes NADPH: protochlorophyllide oxidoreductase A expression and seedling greening in Arabidopsis. Photosynth. Res. 126 331–340. 10.1007/s11120-015-0146-5 [DOI] [PubMed] [Google Scholar]
- Xu X., Chi W., Sun X., Feng P., Guo H., Li J., et al. (2016). Convergence of light and chloroplast signals for de-etiolation through ABI4-HY5 and COP1. Nat. Plants 2:16066 10.1038/nplants.2016.66 [DOI] [PubMed] [Google Scholar]
- Yu Y., Wang J., Shi H., Gu J., Dong J., Deng X. W., et al. (2016). Salt stress and ethylene antagonistically regulate nucleocytoplasmic partitioning of COP1 to control seed germination. Plant Physiol. 170 2340–2350. 10.1104/pp.15.01724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Wang J., Zhang Z., Quan R., Zhang H., Deng X. W., et al. (2013). Ethylene promotes hypocotyl growth and HY5 degradation by enhancing the movement of COP1 to the nucleus in the light. PLoS Genet. 9:e1004025 10.1371/journal.pgen.1004025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Li Y., Zhang X., Zha P., Lin R. (2017). The SWI2/SNF2 chromatin-remodeling ATPase BRAHMA regulates chlorophyll biosynthesis in Arabidopsis. Mol. Plant 10 155–167. 10.1016/j.molp.2016.11.003 [DOI] [PubMed] [Google Scholar]
- Zheng Y., Cui X., Su L., Fang S., Chu J., Gong Q., et al. (2017). Jasmonate inhibits COP1 activity to suppress hypocotyl elongation and promote cotyledon opening in etiolated Arabidopsis seedlings. Plant J. 90 1144–1155. 10.1111/tpj.13539 [DOI] [PubMed] [Google Scholar]
- Zhong S., Shi H., Xi Y., Guo H. (2010). Ethylene is crucial for cotyledon greening and seedling survival during de-etiolation. Plant Signal. Behav. 5 739–742. 10.4161/psb.5.6.11698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Shi H., Xue C., Wang L., Xi Y., Li J., et al. (2012). A molecular framework of light-controlled phytohormone action in Arabidopsis. Curr. Biol. 22 1530–1535. 10.1016/j.cub.2012.06.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Shi H., Xue C., Wei N., Guo H., Deng X. W. (2014). Ethylene-orchestrated circuitry coordinates a seedling’s response to soil cover and etiolated growth. Proc. Natl. Acad. Sci. U.S.A. 111 3913–3920. 10.1073/pnas.1402491111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Zhao M., Shi T., Shi H., An F., Zhao Q., et al. (2009). EIN3/EIL1 cooperate with PIF1 to prevent photo-oxidation and to promote greening of Arabidopsis seedlings. Proc. Natl. Acad. Sci. U.S.A. 106 21431–21436. 10.1073/pnas.0907670106 [DOI] [PMC free article] [PubMed] [Google Scholar]


