Abstract
We present an approach called pulsed multiline excitation (PME) for measurements of multicomponent, fluorescence species and demonstrate its application in capillary electrophoresis for DNA sequencing. To fully demonstrate the advantages of PME, a fluorescent dye set has been developed whose absorption maxima span virtually the entire visible spectrum. Unlike emission wavelength-dependent approaches for identifying fluorescent species, the removal of the spectral component in PME confers a number of advantages including higher and normalized signals from all dyes present in the assay, the elimination of spectral cross-talk between dyes, and higher signal collection efficiency. Base-calling is unambiguously determined once dye mobility corrections are made. These advantages translate into significantly enhanced signal quality as illustrated in the primary DNA sequencing data and provide a means for achieving accurate base-calling at lower reagent concentrations.
Keywords: fluorescent detection, instrumentation
The advancement of DNA sequencing technology has been pivotal to the overall success of the Human Genome Project (1). The combination of enzymology (2, 3), fluorescent dye (4–6), and instrumentation developments have defined the current standard for DNA sequencing platforms with the latter having the greatest impact on increased throughput (7). The core technology of assigning a base-call from the emission wavelength of a dye's fluorescence has not changed since its introduction in 1986 (8) and remains the underlying method used in most commercial instruments today (8–13). The disadvantages of these four-color systems are (i) inefficient excitation of fluorescent dyes from a single laser source, (ii) significant spectral overlap or “cross-talk” between the dyes, and (iii) significant loss in fluorescent signal intensities from the required use of a dispersing element or band-pass filters for detection. These limitations affect the sensitivity and application of methods for analyses of multicomponent fluorescent assays directly from genomic DNA sources. Investigators have proposed alternative strategies based on nondispersing techniques such as fluorescent life-time (14–18) and radio frequency (RF) modulation (19). To date, these alternative approaches have had limited impact on the field of DNA sequencing. Thus, new technologies are needed to promote increased efficiency and cost-effectiveness for high-throughput sequencing (1, 20).
Here, we describe a simple but effective method for multifluorescence discrimination called pulsed multiline excitation (PME), which overcomes the limitations described above. This technology is based on correlating a sequence of excitation pulses from four monochromatic wavelength laser sources with detector response from the emission intensities of fluorescently labeled DNA fragments. A new fluorescent dye set has been developed whose absorption maxima span most of the visible spectrum, resulting in strong emission signals when interrogated by a matched laser and negligible signals when exposed to the remaining excitation wavelengths. The primary advantages of this simple and straightforward approach are (i) all fluorophores yield maximum signal intensities because they are excited near their absorption maxima, (ii) cross-talk between the dyes is eliminated by temporal separation, and (iii) a larger fraction of fluorescent signals are collected because no dispersing elements are required to determine a dye's identity. Effectively, the system detects different fluorescent dyes in a multicomponent assay in a “color-blind” manner. Demonstration of these advantages could translate into higher sensitivity, which would directly address the cost of fluorescent dye reagents and/or the effort in preparing template materials for sequencing reactions.
Materials and Methods
PME Detector. The lasers used and their sources are as follows: a 4-mW, 399-nm violet InGaN diode laser system from Coherent (Auburn, CA), a 20-mW, 488-nm blue Sapphire optically pumped semiconductor laser from Coherent, a 1-mW, 594-nm yellow linear polarized HeNe gas laser from Melles Griot (Carlsbad, CA), and a 35-mW SPMT, 685-nm red diode laser module from Blue Sky Research (San Jose, CA). A transistor–transistor logic stepper chip circuit was built to control the specific opening of mechanical shutters, allowing short pulses of plane polarized light in sequential order from the different laser sources. Aluminum steering mirrors (Edmund Optics, Barrington, NJ) were used to align the pulsed laser beams to a pair of equilateral dispersing prisms [25-mm edge dimension; type 2 F2 Flint glass, grade A fine annealed, coated with a single layer of HEBBAR antireflection material (Melles Griot)] to combine them by inverse dispersion. The resulting single coaxial PME beam was directed into a dark box housing. The PME-induced fluorescence was collimated by using a collection lens, and scattered laser light was rejected via a 2-mm-thick, 420-nm long-pass filter (Schott, Elmsford, NY) and 488-, 594.1-, and 685-nm holographic Notch-Plus filters (Kaiser Optical Systems, Ann Arbor, MI). Pulsed fluorescent signals were then imaged onto a R928 multialkali photomultiplier (PMT, Hamamatsu Photonics, Bridgewater, NJ). All operations and data processing were performed by using a custom-designed labview program (National Instruments, Austin, TX).
PME Coaxial Beam Alignment. To ensure that the four pulsed laser beams were collimated and coaxial, and that the collection optics were transmitting maximum signal intensities for all four dyes, the following four-step alignment procedure was developed: (i) a knife edge test was applied to the beam waist of each laser to provide coincidence for x,y coordinates, (ii) beams were made to overlap in the far field to assure angular coincidence, (iii) fluorescent emission was visually centered by using an aperture in front of the PMT, and (iv) a static dye mixture was introduced into the capillary to maximize fluorescent signal intensities for each of the four PME waveform channels by fine alignment of the steering mirrors while in full operational mode.
Fluorescently Labeled Oligonucleotides. The amino-reactive fluorescent dyes Pacific blue, 7-aminocoumarin, Alexa Fluor (AF) 405, NBD-X, 5-FAM, BODIPY-FL, 6-JOE, BODIPY R6G, 6-HEX, BODIPY 576/589, BODIPY 581/591, Texas Red-X, 6-ROX, BODIPY 630/650-X, AF 633, BODIPY 650/665, and AF 680 were purchased from Molecular Probes, and Cy3.5, Cy5.5, and Cy7 dyes were purchased from Amersham Pharmacia Biotech. DNA synthesis reagents were purchased from Glen Research (Sterling, VA). The R931 universal sequencing primer (5′-TTGTAAAACGACGGCCAGT) was synthesized by using a DNA synthesizer (model 394, Applied Biosystems). Preparation and HPLC purification of fluorescently labeled primers were performed as described in ref. 5. Spectroscopic measurements for all dye–primer solutions (4 × 10–6 M in 20 mM triethylammonium acetate, pH 7.0) were performed by using a fluorescent spectrophotometer (model F-4010, Hitachi).
Response Index. To further characterize the four dye–primer solutions, a spectrometer was coupled to the PME detector by using a Jarrell-Ash (Waltham, MA) grating with 1,800 grooves per meter and blazed at 450 nm. For each dye–primer solution (10–8 M), an emission spectrum was obtained by excitation with a single continuous-wave laser source (e.g., without pulsing). For direct comparison, laser powers were adjusted to equivalent levels, which were ≈1.0 mW. The integrated area for each emission spectrum was obtained and normalized to the 685-nm laser. Response values were calculated by setting diagonal elements to one and normalizing off-diagonal row elements accordingly.
Color-Calling Experiment. The four dye–primer solutions (1 × 10–9 M) and all six possible (50:50) dye–primer mixtures (0.5 × 10–9 M each) were analyzed in optical glass fluorescence cuvettes (Starna Cells, Atascadero, CA) in a blinded manner. The order of the dye–primer solutions was predetermined by a random number generator. The 10 solutions were arbitrarily numbered 1 through 10, and the code for each solution was blinded from technical personnel. The 10 solutions were then analyzed according to the predetermined sequence order. To reduce the possibility of photobleaching due to repetitive exposure of dye–primer solutions to the PME beam, the solutions were stirred during all fluorescent measurements. To make a dye-call, the PME waveform for each sample was analyzed by using Boolean logic and sorted into 1 of the 10 waveform categories. Once the dye-call sequence was determined, the codes for the numbered dye solutions were revealed and compared with the experimental data.
Capillary Electrophoresis (CE) Device. The PME detector was designed as a modular system. For the electrophoresis experiments, the dark box housing was replaced by a custom-built single CE box. A 30-kV power supply (Glassman High Voltage, High Bridge, NJ) was used for all experiments. The collimating lens for the cuvette experiments was replaced by a standard microscope objective [Deutsche Industrie Norm ×10, 0.25 N.A. (Edmund Optics)]. Synthetic fused silica capillaries (75-μm i.d.) were purchased from Polymicro Technologies (Phoenix), and POP-7 polymer and 10× running buffer were purchased from Applied Biosystems.
Before electrophoresis, the POP-7-loaded capillary was conditioned by prerunning at 15 kV for 10 min. The loading parameters evaluated were injection voltages ranging from 2 to 8 kV and injection times ranging from 10 to 160 sec. The separation parameters evaluated were running voltages ranging from 6 to 16.7 kV and capillary lengths of 40 and 80 cm. The experimental conditions used for the data shown in Fig. 4 are an 80-cm capillary length with an injection voltage of 3.2 kV, an injection time of 80 sec, and a running voltage of 16.7 kV.
Fig. 4.
Overlapping fluorescent waveforms for 50:50 mixtures (0.5 × 10–9 M each) of all possible six dye-labeled primer combinations. See the legend of Fig. 3 for details.
Sanger Sequencing Reactions. The forward (5′-CGTTGTAAAACGACGGCCAGTGTGTTTGACTCAGCCTAGC) and reverse (5′-GCTATGACCATGATTACGCCATGAACAGGCTTTGCTC) primers for PCR were purchased from Integrated DNA Technologies (Coralville, IA). The underlined portion of the DNA sequence represents unique sequences to the HNF-1α gene, and the remaining 5′-end sequences are complementary to the universal forward and reverse sequencing primers. Hot-start PCR was performed by using FastStart Taq DNA polymerase and the GC-rich solution according to the manufacturer's protocol (Roche Applied Science). PCR cycle conditions were as follows: 95°C for 6 min and 35 cycles of 95°C for 30 sec, 57°C for 45 sec, and 72°C for 90 sec, followed by a final extension at 72°C for 7 min. DNA sequencing reactions were performed by using TaqFS DNA polymerase in 80 mM Tris·HCl (pH 9.0)/2.5 mM MgCl2 buffer. Four separate dye–primer (0.4 pmol) reactions were thermocycled in the presence of 500 μM dNTPs and one ddNTP (6.4 μM) by using the following conditions: 20 cycles of 95°C for 10 sec, 57°C for 20 sec, and 70°C for 60 sec. The separate reactions were then pooled, purified by ethanol precipitation, and dried under vacuum centrifugation. Sequencing reactions were resuspended in 100 μl of HPLC water, heated to 65°C for 3 min, and cooled to room temperature before injection into the PME-CE instrument.
Results
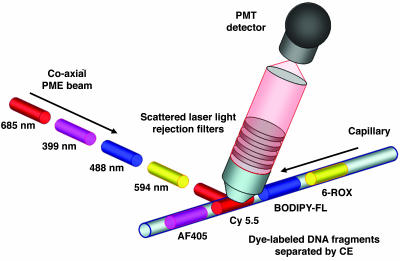
PME Technology. To test the PME strategy, a detector was constructed. Four laser beams (399, 488, 594, and 685 nm), originating from fixed locations, were chopped sequentially in exclusive order by mechanical shutters under the control of a transistor–transistor logic stepper chip. This custom-built chip controls the frequency of the laser cycle, which was cycled at ≈5 Hz. The streams of pulsed beams were then directed to a dual prism assembly, converging the four beams into a single coaxial beam by inverse dispersion. The resulting coaxial PME beam was focused into a detection housing containing either an optical glass cuvette (not shown) or a single, synthetic fused silica capillary (Fig. 1). Laser-specific rejection filters were used in the detection optical train to remove scattered laser light before the dye's fluorescence was imaged directly onto the PMT detector. The correlation between the sequential laser pulses and PMT detector response is the underlying principle of the PME technology.
Fig. 1.
Drawing of the PME technology. Here, each laser operates in a continuous-wave mode with mechanical shutters pulsing the different excitation beams in sequential order. An arrangement of steering mirrors directs the pulsed laser beams into a dual prism assembly, where the prisms are positioned at a 45° angle relative to one another. This allows efficient convergence in overlapping the four beams into a single beam by the process of inverse dispersion. The single coaxial PME beam then interrogates the fluorescently labeled DNA fragments, which are separated by capillary gel electrophoresis. Scattered laser light is rejected via specific long-pass or wavelength notch filters, with pulsed emission signals from the dye-labeled DNA fragments being imaged onto the photomultiplier (PMT) detector without use of any dispersing elements. The fluorescence is detected directly from the cuvette in an orthogonal geometry (not shown) or from the capillary, which is positioned 45° relative to the optical axis of the detection train to further reduce scattered laser light.
Identification of PME Dyes. Sets of fluorescent dyes form the basis of multicomponent DNA sequencing detection. Current DNA sequencing applications, using multicolor dye sets, exhibit substantial overlap in emission spectra (4–6, 8, 9) because of the close proximity of the laser's excitation wavelength required to achieve sufficient emission signals. The introduction of energy-transfer cassettes has resulted in an increase in acceptor dye emission signals (4–6), although not sufficient to increase the spectral range necessary to reduce spectral overlap. The multiple laser approach described here utilizes virtually the entire visible spectrum by distributing the four excitation sources between 399 and 685 nm, significantly reducing the spectral overlap exhibited in multifluorescent assays.
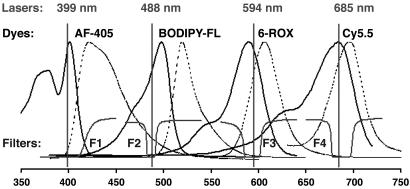
To identify a set of four fluorescent dyes for DNA sequencing applications, fluorophores were selected on the basis of spectral proximity of their absorption maxima to the excitation wavelengths of the different lasers evaluated. We HPLC-purified and spectroscopically characterized a number of commercially available dyes, which were synthetically coupled to the R931 universal sequencing primer (5). Overlapping excitation spectra for all dye-labeled primers were then compared with the laser excitation wavelengths 399, 488, 594, and 685 nm to identify a set of four fluorescent dyes exhibiting strong emission signals for the matched laser source and minimal signals for the remaining lasers (data not shown). From these analyses, we have identified a set of four fluorescent dyes (AF-405, BODIPY-FL, 6-ROX, and Cy5.5). The absorption spectra for these dyes are shown in Fig. 2 (solid lines). From these spectra, the λmax for each dye nearly matches one of the four laser wavelengths while showing substantially less overlap from the remaining lasers.
Fig. 2.
Spectral scheme for the PME technology illustrating the combination of different lasers, fluorescent dyes, and filters. The four lasers are described in the PME detector section. Dye–primer spectra were obtained experimentally (solid lines are excitation, and dotted lines are emission scans). F1, 420-nm long-pass filter; F2, F3, and F4, 488-, 594.1-, and 685-nm Notch-Plus filters.
To characterize the degree of spectral overlap between the PME dyes, the detector was configured with a spectrophotometer to obtain the entire emission spectra for each dye-labeled primer by excitation with the different laser sources at equivalent powers. Integrated areas for each spectrum were obtained and normalized against the diagonal elements to estimate the relative contributions of the off-diagonal values (Table 1). To distinguish our approach from a four-color, cross-talk matrix, we refer to Table 1 as the response matrix, with the response index values indicating the relative response of a given dye to a given laser. These data show that the off-diagonal responses are typically <5% with the notable exception being the Cy5.5 dye response to the 594-nm laser (18%). These observations are consistent with the excitation spectra and laser position in Fig. 2.
Table 1. PME response matrix.
| Lasers, nm
|
||||
|---|---|---|---|---|
| 399 | 488 | 594 | 685 | |
| AF-405 | 1 | 0.01 | -0.01 | 0.00 |
| BODIPY-FL | 0.04 | 1 | -0.01 | 0.01 |
| 6-ROX | 0.02 | 0.04 | 1 | 0.00 |
| Cy5.5 | 0.03 | 0.00 | 0.18 | 1 |
This composite table is the result of multiple emission scans for AF-405 (six replicates), BODIPY-FL (three replicates), 6-ROX (seven replicates), and Cy5.5 (seven replicates) derived from the PME detector coupled with a spectrophotometer. Standard deviations for the data set were ≤0.02. Solutions of each dye—primer (10-8 M) were scanned by using a 399-, 488-, 594-, or 685-nm laser at equivalent laser powers of ≈1.0 mW. Integrated areas for each emission scan were obtained and normalized to the 685-nm laser power. The boldface values represent the asymmetry of the response matrix, which is predicted by the Franck—Condon potential energy curves.
No Cross-Talk Between Fluorescent Dyes in the PME System. For conventional DNA sequencing, transformation of “raw,” multi-component, fluorescent signals into “processed,” four-color sequence data involves a number of algorithmic manipulations such as removal of cross-talk, baseline adjustment, mobility dye correction, emission signal intensity normalization, and base-calling. Of these, determination of the cross-talk matrix, which allows for the conversion of mixed-fluorescent signals into concentration estimates for the dye-labeled DNA fragments at a given time point during a sequencing experiment, is likely the most relevant operation (21). Numerous research groups have described methods to determine a representative cross-talk matrix for DNA sequencing data, albeit with limited success (22–28). For comparison, we present an Applied Biosystems cross-talk matrix for the BigDye v3.1 terminator chemistry derived from a model 377 DNA sequencer (Table 2) that shows significantly higher off-diagonal values.
Table 2. Cross-talk matrix.
| Detection windows, nm
|
||||
|---|---|---|---|---|
| 530 | 560 | 580 | 610 | |
| G-dye 1* | 1 | 0.75 | 0.39 | 0.13 |
| A-dye 2 | 0.08 | 1 | 0.44 | 0.19 |
| T-dye 3 | 0.02 | 0.19 | 1 | 0.38 |
| C-dye 4 | 0.01 | 0.01 | 0.18 | 1 |
This table was obtained from a BigDye v3.1 Matrix Standard by using the Applied Biosystems datautility tool for the ABI PRISM model 377 DNA sequencer according to the manufacturer's protocol.
The chemical structures for BigDye v3.1 dyes have not been published to date. BigDye v1.1 structures are described in ref. 30.
From this comparison, we describe two important findings. First, the shaded triangle in Table 1, which shows approximately zero response values, illustrates the asymmetrical nature of the response matrix. This observation is consistent with the Franck–Condon potential energy curves for fluorescent compounds, which predicts that “redder” lasers should not excite “bluer” dyes. The shaded response index values, therefore, should be zero. Secondly, because of the sequential pulsing of the lasers, the raw fluorescent signals derived from the matched laser–dye combination are intrinsically free from contaminating off-diagonal responses. That is, because the laser pulses are separated in time, the responding fluorescent signals for a given dye are also separated in time, eliminating the spectral cross-talk of conventional DNA sequencing technology.
Dye-Calling in a Color-Blind Manner. The basis of existing four-color DNA sequencing is the spectral resolution of fluorescent signals into different colors in order to illuminate the DNA sequence under investigation (8, 9). In contrast, the PME approach of color-blind detection proceeds without the aid of fluorescence band-pass filters, gratings, prisms, or any other dispersing elements. To test the concept that color-blind detection can determine the identities of fluorescent species under conditions used in DNA sequencing, 10–9 M solutions were prepared corresponding to each of the four dye-labeled primers and all six possible 50:50 mixtures. With the identities of the 10 solutions blinded before conducting the dye-calling experiment, measurements of PME waveforms were made for 500 random selections of these solutions. Because the fluorescent dyes were matched to the lasers (Fig. 2), a PME waveform for a given dye was expected to yield a strong fluorescent signal when interrogated by the matched laser pulse, and a minimal signal with the remaining laser pulses.
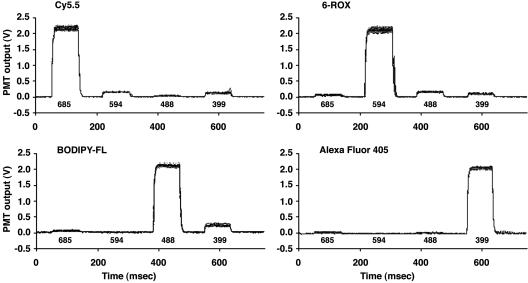
Figs. 3 (single-dye runs) and 4 (mixed-dye runs) show the raw fluorescent data of overlapping PME waveforms from a 500 dye-call experiment. The raw fluorescent data show that the single and mixed dye–primer solutions exhibited unique and unambiguous PME waveforms with a high degree of reproducibility. From these data, the identity of dye(s) could be determined from the correlation of the dominant fluorescent signal(s) to the timing of laser pulses. The order of the dye-calls was then determined and found to be in 100% agreement with the blinded sequence order.
Fig. 3.
Overlapping fluorescent waveforms for individual dye-labeled primer solutions (1.0 × 10–9 M). Bold numbers under the waveforms correspond to the sequential pulsing of the 685-, 594-, 488-, and 399-nm lasers. Their powers are 0.72, 0.28, 0.17, and 0.5 mW, respectively. Each waveform represents an average of 10 laser cycles, which were normalized to their initial laser power measurement to account for power drift during the experiment, subtracted against an initial water run to remove Raman scattering, and baseline corrected. A total of seven different blinded experiments were conducted, ranging from 25 to 500 dye-calls, resulting in a total of 2,075 calls. Dye-calling was performed by using a Boolean logic approach, and in all cases, dyes were called correctly when compared with the reference sequence.
The mixed waveforms in Fig. 4 are approximately one-half the signal intensities of those of the single dye–primer waveforms in Fig. 3, in accord with the corresponding dye concentrations. These data illustrate the potential application for heterozygote identification in resequencing studies. Although the data show that dye identification can be performed directly from the raw fluorescent data, inversion of the response matrix is required for quantitative analyses (e.g., measurement of 50:50 mixtures from heterozygous bases). The diagonally dominant nature of the response matrix (resulting from the matching of dyes to laser wavelengths) ensures the stability of the inversion process. Thus, these data strongly support our contention that a given dye can be identified in a multicomponent assay in a color-blind manner.
Sanger Sequencing with PME-CE. To evaluate the application of DNA sequencing by using the PME fluorescent detector, we constructed a single CE instrument. A number of parameters were investigated, including run voltage, injection voltage and time, and length of capillary to optimize the separation of the Sanger reactions. The powers for the four lasers were adjusted to balance the signal intensities of the four dye-labeled fragments. Data acquisition consisted of recording the average PMT voltage at the mid-region of the PME waveforms (Fig. 3) for each laser cycle. Averaged PMT voltages corresponding to each of the four laser time intervals were then plotted against electrophoretic run time to yield an electropherogram (Fig. 5).
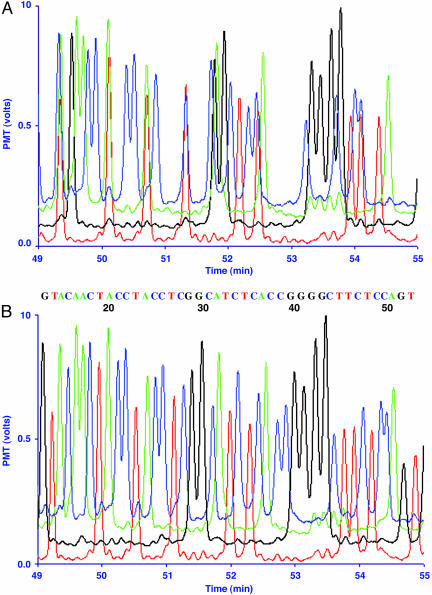
Fig. 5.
Electropherogram of the HNF-1α exon 10 gene region using PME dye–primers. (A) Unprocessed fluorescent data obtained during the electrophoretic run from the PME detector. Blue, green, black, and red traces are AF-405, BODIPY-FL, 6-ROX, and Cy5.5 dye–primers terminated with ddCTP, ddATP, ddGTP, and ddTTP, respectively. Laser powers were 1.4, 1.0, 0.30, and 1.2 mW for the 399-, 488-, 594-, and 685-nm lasers, respectively. Baselines were shifted to space the four laser traces in a nonoverlapping manner. (B) Transformation of the raw trace data derived from the experiment described in A into readable DNA sequence data by using mobility software correction. Base-calling was determined based on the highest fluorescent signals for each fragment peak. The numbers below the DNA sequence correspond to the base number following the PCR primer position.
PCR was used to amplify different exons of hepatic nuclear factor 1α (HNF-1α). Mutations in this gene are known to cause maturity-onset diabetes of the young (29). Cycle sequencing was performed on purified PCR products, using AF-405, BODIPY-FL, 6-ROX, and Cy5.5 dye–primers, to produce fluorescently labeled Sanger reactions. To illustrate the quality of the electrophoretically separated, dye-labeled DNA fragments, Fig. 5A shows the raw fluorescent signals derived from the PME-CE prototype instrument for a portion of the PCR amplicon for HNF-1α exon 10. These data clearly show well defined peaks with high signal-to-noise ratios. The electrophoretic mobility differences of the PME dye-labeled fragments caused several peaks to overlap. We observed that AF-405, Cy5.5, and 6-ROX dye–primers alone showed elution time differences of +1.6, +0.2, and –0.8 min with respect to the BODIPY-FL dye–primer, under the conditions described in Materials and Methods. Longer extension fragments exhibited nonlinear behavior throughout the electrophoretic run (data not shown). Unambiguous DNA sequence data were produced from the raw data in Fig. 5A by applying only mobility software corrections, guided by the observed mobility differences of the PME dyes (Fig. 5B).
Discussion
Collectively, these data illustrate the primary advantages of PME fluorescence detection. That is, the dye-call and DNA sequence data show (i) that high and uniform fluorescent signals can be obtained for all dyes present in the assay under investigation, (ii) the absence of cross-talk between the dyes, and (iii) that base-calling can be performed in a color-blind manner. The first two features, uniform signals and elimination of cross-talk, remove two central processing steps for conversion of raw fluorescent data into DNA sequence information. Only mobility software correction is needed to transform the PME raw fluorescent signals into readable DNA sequence data. Thus, the enhancement of the data quality, as measured by the reduction in software processing steps, should significantly improve standard DNA sequence metrics (e.g., read lengths and pass rates). Efforts are currently focused on quantifying the PME sensitivity of DNA sequencing reactions with respect to decreased fluorescent dye reagent use and/or template input.
The flexibility of normalizing signal intensities in the raw data are another unique aspect of the PME system that has not been accomplished in conventional DNA sequencing technology (4–6, 30). Unlike the data presented in Table 1, the laser powers were adjusted before conducting the 500 dye-call experiment to yield equivalent fluorescent signal intensities for all dye–primers. When the laser powers are adjusted, the off-diagonal elements are scaled proportionally, creating a new response matrix. For example, the smaller response of Cy5.5 in Figs. 3 and 4 compared to that reported in Table 1 is due to the ≈3-fold reduction in the 594-nm laser power compared to the 685-nm laser power. We believe that the color-blind detection method described here represents an enabling technology that differs significantly from the spectral resolution method for base-calling DNA sequencing data.
Other multiple laser systems have been reported for DNA sequencing applications, which resolve fluorescent light into color for base-calling purposes (17, 31–33). In a recent report, Alaverdian et al. (19) also used four lasers, which were all operated in a continuous-wave mode but modulated at different RFs. They illustrated the RF approach using the Beckman sequencing chemistry. This is a cyanine-based fluorescent dye set that spans a narrower region of the wavelength spectrum, resulting in significant cross-talk values. In this method, the emission intensity pattern for a given dye is a mixture of different RFs, which are demodulated to determine the true signal intensity from each laser. Scaling the RF method to multiple capillary arrays poses major obstacles. For example, the RF method requires a PMT channel and a separate discriminator/count recorder for each capillary. Moreover, the RF scheme imposes a computational load of demodulation for each of the resulting capillary signal channels. Charge-coupled device detector arrays, which can image at least 96 capillaries, are not suitable for photon counting and are in general not capable of the frame readout rates of hundreds of hertz required for the RF scheme. Conversely, the PME technology is readily scalable for imaging of high-density capillary arrays with a charge-coupled device detector. Thus, the PME technology described here is fundamentally different from any of the multiple laser methods reported.
In recent years, dye-labeled, energy-transfer dideoxynucleotide terminators (6, 30) have been widely used in the genome community for high-throughput DNA sequencing, primarily because of their simplified liquid handling for reaction set-up. The use of the multiple lasers described here allowed us the flexibility to explore alternative dyes for DNA sequencing purposes, many of which are not commercially available as dye-terminators. In this study, we identified a set of four dyes, AF-405, BODIPY-FL, 6-ROX, and Cy5.5, that matched the excitation wavelengths of the lasers described. To demonstrate the feasibility of the PME technology for DNA sequencing, we chose the alternative approach of the dye–primer method. Although the reaction set-up requires more liquid handling steps, the labeling of a universal sequencing primer with this unique dye set is straightforward while unambiguously demonstrating the unique features of PME method. We note that because the absorption maxima of the dyes are matched to the laser excitation sources, the signal enhancement observed for energy transfer dyes is negated because their fluorescent intensities are already near maximum levels. Thus, our efforts in syntheses of singly labeled dye-terminators will be tremendously simplified.
PME is suitable for development into a compact DNA sequencing instrument using small solid-state lasers, laser diodes, and microfluidic separation devices for field-use applications. Removal of the spectral component for PME detection provides additional advantages for other applications as well. For example, microarray analyses are limited to scanning instruments for imaging chips because these devices also use both spatial and spectral components for fluorescent detection. The strategy of one-dimensional scanning of two-dimensional chips reduces the speed and sensitivity of these instruments. The PME approach of bathing the entire chip surface with pulsed lasers is ideally suited for whole imaging of fluorescent, high-density, oligonucleotide arrays because both x and y spatial components would be used. The two-dimensional, spatial approach would allow the possibility of simultaneous imaging of all features on a high-density chip. Thus, we believe that the PME technology presented here illustrates tremendous potential benefits beyond current DNA sequencing applications.
Acknowledgments
We thank Dr. James Kinsey for providing space for construction of the PME device and Claudia Gomez and Travis Kemper for technical support. This work was supported by National Institutes of Health Grants 1 R21 HG002443 and 1 R41 HG003265 and National Science Foundation Grant REU/PHY 0139202.
Author contributions: E.K.L., F.N., D.A.H., G.B.I.S., C.K., B.R.J., R.F.C., and M.L.M. designed research; E.K.L., W.C.H., F.N., D.A.H., M.J.A., R.R.M., C.S.B., B.W., L.A.B., and C.K. performed research; W.C.H., C.K., B.R.J., R.F.C., and M.L.M. contributed new reagents/analytic tools; E.K.L., W.C.H., C.K., B.R.J., R.F.C., and M.L.M. analyzed data; and C.K., B.R.J., R.F.C., and M.L.M. wrote the paper.
Abbreviations: AF, Alexa Fluor; CE, capillary electrophoresis; PME, pulsed multiline excitation; RF, radio frequency.
References
- 1.Collins, F. S., Green, E. D., Guttmacher, A. E. & Guyer, M. S. (2003) Nature 422, 835–847. [DOI] [PubMed] [Google Scholar]
- 2.Tabor, S. & Richardson, C. C. (1987) Proc. Natl. Acad. Sci. USA 84, 4767–4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tabor, S. & Richardson, C. C. (1995) Proc. Natl. Acad. Sci. USA 92, 6339–6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ju, J., Ruan, C., Fuller, C., Glazer, A. & Mathies, R. (1995) Proc. Natl. Acad. Sci. USA 92, 4347–4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Metzker, M. L., Lu, J. & Gibbs, R. A. (1996) Science 271, 1420–1422. [DOI] [PubMed] [Google Scholar]
- 6.Lee, L., Spurgeon, S., Heiner, C., Benson, S., Rosenblum, B., Menchen, S., Graham, R., Constantinescu, A., Upadhya, K. & Cassel, J. (1997) Nucleic Acids Res. 25, 2816–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marshall, E. & Pennisi, E. (1998) Science 280, 994–995. [DOI] [PubMed] [Google Scholar]
- 8.Smith, L., Sanders, J., Kaiser, R., Hughes, P., Dodd, C., Connell, C., Heiner, C., Kent, S. & Hood, L. (1986) Nature 321, 674–679. [DOI] [PubMed] [Google Scholar]
- 9.Prober, J., Trainor, G., Dam, R., Hobbs, F., Robertson, C., Zagursky, R., Cocuzza, A., Jensen, M. & Baumeister, K. (1987) Science 238, 336–341. [DOI] [PubMed] [Google Scholar]
- 10.Brumley, R. L., Jr., & Smith, L. M. (1991) Nucleic Acids Res. 19, 4121–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi, S., Murakami, K., Anazawa, T. & Kambara, H. (1994) Anal. Chem. 66, 1021–1026. [Google Scholar]
- 12.Ueno, K. & Yeung, E. S. (1994) Anal. Chem. 66, 1424–1431. [Google Scholar]
- 13.Kheterpal, I., Scherer, J., Clark, S., Radhakrishnan, A., Ju, J., Ginther, C., Sensabaugh, G. F. & Mathies, R. A. (1996) Electrophoresis 17, 1852–1859. [DOI] [PubMed] [Google Scholar]
- 14.Nunnally, B. K., He, H., Li, L.-C., Tucker, S. A. & McGown, L. B. (1997) Anal. Chem. 69, 2392–2397. [DOI] [PubMed] [Google Scholar]
- 15.Lieberwirth, U., Arden-Jacob, J., Drexhage, K. H., Herten, D. P., Muller, R., Neumann, M., Schulz, A., Siebert, S., Sagner, G., Klingel, S., et al. (1998) Anal. Chem. 70, 4771–4779. [DOI] [PubMed] [Google Scholar]
- 16.Lassiter, S. J., Stryjewski, W., Benjamin, L., Legendre, J., Erdmann, R., Wahl, M., Wurm, J., Peterson, R., Middendorf, L. & Soper, S. A. (2000) Anal. Chem. 72, 5373–5382. [DOI] [PubMed] [Google Scholar]
- 17.Zhu, L., Stryjewski, W., Lassiter, S. & Soper, S. A. (2003) Anal. Chem. 75, 2280–2291. [DOI] [PubMed] [Google Scholar]
- 18.Zhu, L., Stryjewski, W. J. & Soper, S. A. (2004) Anal. Biochem. 330, 206–218. [DOI] [PubMed] [Google Scholar]
- 19.Alaverdian, L., Alaverdian, S., Bilenko, O., Bogdanov, I., Filippova, E., Gavrilov, D., Gorbovitski, B., Gouzman, M., Gudkov, G., Domratchev, S., et al. (2002) Electrophoresis 23, 2804–2817. [DOI] [PubMed] [Google Scholar]
- 20.Kling, J. (2003) Nat. Biotechnol. 21, 1425–1427. [DOI] [PubMed] [Google Scholar]
- 21.Smith, L. M., Kaiser, R. J., Sanders, J. Z. & Hood, L. E. (1987) Methods Enzymol. 155, 260–301. [DOI] [PubMed] [Google Scholar]
- 22.Giddings, M. C., Jr., Brumley, R. L., Haker, M. & Smith, L. M. (1993) Nucleic Acids Res. 21, 4530–4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin, Z., Severin, J., Giddings, M. C., Huang, W. A., Westphall, M. S. & Smith, L. M. (1996) Electrophoresis 17, 1143–1150. [DOI] [PubMed] [Google Scholar]
- 24.Berno, A. J. (1996) Genome Res. 6, 80–91. [DOI] [PubMed] [Google Scholar]
- 25.Huang, W., Yin, Z., Fuhrmann, D. R., States, D. J. & Thomas, L. J., Jr. (1997) Electrophoresis 18, 23–25. [DOI] [PubMed] [Google Scholar]
- 26.O'Brien, K. M., Schageman, J. J., Major, T. H., Evans, G. A. & Garner, H. R. (1998) BioTechniques 24, 1014–1016. [DOI] [PubMed] [Google Scholar]
- 27.Li, L. & Speed, T. P. (1999) Electrophoresis 20, 1433–1442. [DOI] [PubMed] [Google Scholar]
- 28.Domnisoru, C., Zhan, X. & Musavi, M. (2000) Electrophoresis 21, 2983–2989. [DOI] [PubMed] [Google Scholar]
- 29.Yamagata, K., Oda, N., Kaisaki, P. J., Menzel, S., Furuta, H., Vaxillaire, M., Southam, L., Cox, R. D., Lathrop, G. M., Boriraj, V. V., et al. (1996) Nature 384, 455–458. [DOI] [PubMed] [Google Scholar]
- 30.Rosenblum, B., Lee, L., Spurgeon, S., Khan, S., Menchen, S., Heiner, C. & Chen, S. (1997) Nucleic Acids Res. 25, 4500–4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carson, S., Cohen, A., Belenkii, A., Ruiz-Martinez, M., Berka, J. & Karger, B. (1993) Anal. Chem. 65, 3219–3226. [DOI] [PubMed] [Google Scholar]
- 32.Wiemann, S., Stegemann, J., Grothues, D., Bosch, A., Estivill, X., Schwager, C., Zimmermann, J., Voss, H. & Ansorge, W. (1995) Anal. Biochem. 224, 117–121. [DOI] [PubMed] [Google Scholar]
- 33.Zhang, J., Voss, K. O., Shaw, D. F., Roos, K. P., Lewis, D. F., Yan, J., Jiang, R., Ren, H., Hou, J. Y., Fang, Y., et al. (1999) Nucleic Acids Res. 27, e36. [DOI] [PMC free article] [PubMed] [Google Scholar]