Abstract
Despite their versatility and power in controlling gene regulation in nature, nuclear hormone receptors (NHRs) have largely eluded utility in heterologous gene regulation applications such as gene therapy and metabolic engineering. The main reason for this void is the pleiotropic interference of the receptor–ligand combination with regulatory networks in the host organism. In recent years, numerous strategies have been developed to engineer ligand–receptor pairs that do not cross-interact with host regulatory pathways. However, these strategies have either met with limited success or cannot be readily extended to other ligand–receptor pairs. Here, we present a simple, effective, and readily generalizable strategy for reengineering NHRs to respond specifically to a selected synthetic ligand. The method involves generation of genetic diversity by stepwise individual site saturation mutagenesis of a fixed set of ligand-contacting residues and random point mutagenesis, followed by phenotypic screening based on a yeast two-hybrid system. As a test case, this method was used to alter the specificity of the NHR human estrogen receptor α in favor of the synthetic ligand 4,4′-dihydroxybenzil, relative to the natural ligand 17β-estradiol, by >107-fold. The resulting ligand–receptor pair is highly sensitive to the synthetic ligand in human endometrial cancer cells and is essentially fully orthogonal to the wild-type receptor–natural ligand pair. This method should provide a powerful, broadly applicable tool for engineering receptors/enzymes with improved or novel ligand/substrate specificity.
Keywords: gene therapy, nuclear hormone receptor, protein engineering, orthogonal ligand–receptor pairs
The ability to manipulate naturally occurring proteins to bind and respond to synthetic ligands in a manner independent, or orthogonal, from the influence of natural proteins and ligands, constitutes a significant challenge in protein engineering (1). Such a tool has important utility in the creation of gene switches for the control of heterologous gene expression in applications such as gene therapy and metabolic engineering (2, 3), as well as in the selective regulation of cellular processes such as apoptosis, genetic recombination, signal transduction, and motor protein function (4).
To date, numerous synthetic ligand-mutant receptor pairs have been created that are orthogonal to the analogous natural interaction to varying degrees. Among the proteins used for this work, nuclear hormone receptors (5), naturally occurring transcription factors, have served as a prime target, owing largely to their “gene switch-like” attributes: rapid induction kinetics (6–8), dose-dependent ligand response, and readily interchangeable functional modules (9, 10). Despite the extensive work carried out to engineer new specific ligand–receptor pairs from nuclear hormone receptors, the absence of a conceptually simple, generally applicable engineering approach remains a concern. Rational design of ligands to rematch interaction with a given mutant of human estrogen receptor (hER) α or hERβ having weakened response to the natural ligand 17β-estradiol (E2), for example, has been applied with limited success (11–13). Rational design involving the targeted replacement of residues in the human retinoid acid receptor γ (14) and human retinoid X receptor α (hRXRα) (15) with specific amino acids based on structure–function considerations has produced ligand–receptor pairs with altered specificity, although the chosen protein engineering approach in both cases cannot be readily generalized to any receptor–ligand system. A purely directed evolution approach based on random point mutagenesis of the hERα ligand binding domain (LBD) and selection of variants with altered selectivity for a target ligand has also been used (16, 17). Although the latter engineering approach can be applied to any receptor protein, the degree of selectivity enhancement toward the target ligand demonstrated in these instances was only moderate. Recently, a combined rational design/combinatorial approach involving simultaneous randomization of a selected set of residues in the ligand-binding pocket of the hRXRα and phenotypic selection against a target ligand led to the identification of a number of receptor variants with significantly altered selectivity for the target ligand compared with natural ligand (18). However, the presented library creation strategy cannot be readily extended to other receptors because it allows only a limited number of amino acid substitutions at chosen receptor sites, and these allowable substitutions must be rationally selected.
Here, we present a systematic strategy for the identification of variant receptor proteins with significantly altered selectivity for a target synthetic ligand. The demonstrated approach combines structure-based design with directed evolution and involves stepwise site saturation mutagenesis of individual ligand-contacting residues, accompanied by phenotypic screening for variants with enhanced target ligand selectivity at each stage of mutagenesis, followed by random point mutagenesis and phenotypic screening for further selectivity-enhanced mutants. As our model receptor system, we use the LBD of the hERα protein, and as our target ligand, we use the synthetic nonsteroidal compound, 4,4′-dihydroxybenzil (DHB). The choice of DHB as the target ligand was based on two considerations: (i) demonstrated lack of toxicity in a MCF-7 cell proliferation assay (19) and (ii) structural similarity to diethylstilbestrol, a drug of known pharmacokinetic properties. Our approach has enabled us to effect a >107-fold specificity shift in receptor response to DHB versus E2, resulting in hERα mutants that interact with DHB in a manner almost completely orthogonal to the wild-type hERα–E2 interaction and respond to subnanomolar concentrations of DHB in mammalian cells.
Materials and Methods
Plasmids, Strains, Reagents, and Growth Media. The pGAD424-SRC1 “prey” plasmid containing the full-length steroid receptor coactivator 1 (SRC-1) coactivator was constructed as described in ref. 20 and was a kind gift from Benita S. Katzenellenbogen (University of Illinois). Amino acids 312–595 containing the LBD and F domain of hERα were inserted downstream of the Gal4 DNA binding domain in the pBD-Gal4-Cam “bait” plasmid (Stratagene) as described in ref. 21. The yeast two-hybrid (Y2H) strain YRG2 (Stratagene) was used for this work. The cloning of hERα LBD mutant constructs into the mammalian expression vector pCMV5 has been described in ref. 21. Rich media used for growth of yeast cells was yeast extract-peptonedextrose medium plus adenine (22), whereas minimal media was synthetic complete (SC) dropout media (23) lacking the appropriate amino acids. Taq DNA polymerase was obtained from Promega, and PfuTurbo DNA polymerase was purchased from Stratagene. 4,4′-Dihydroxybenzil was synthesized as described in ref. 24. Unless otherwise specified, all other reagents were obtained from Sigma-Aldrich.
Y2H System-Based Screening. Transformants from individual site saturation mutagenesis library plates and error-prone PCR library plates were picked with sterile toothpicks and incubated overnight (≈16–20 h) at 30°C in round-bottom 96-well plates (Evergreen Scientific, Los Angeles) containing 50 μl of SC-Leu-Trp minimal liquid media in each well. As a control, one well in every microtiter plate was inoculated with a yeast colony expressing the parental hERα LBD construct. After this overnight incubation, 250 μl of sterile double-distilled H2O was added to every well, and 5 μl of each diluted culture was then transferred to the corresponding wells of two sterile flat-bottom 96-well microtiter plates (Rainin Instruments) containing 200 μl of SC-Leu-Trp-His media with an appropriate concentration of either target ligand (DHB) or E2. Appropriate ligand concentrations for this screening were chosen based on the response of the parental hERα LBD construct. For each round of screening, a DHB concentration was selected at which the parental hERα LBD construct responds weakly or not at all, whereas the concentration of E2 for screening was selected such that the parental construct responds moderately. These ligand-containing microtiter plates were incubated at 30°C for 24 h, after which they were visually inspected for identification of mutants with strengthened response toward the target ligand (higher cell density than parental mutant control) and weakened response toward E2 (lower cell density than parent). One hundred ninety mutants were screened per saturation mutagenesis library by using this approach, with 95 library variants and one parental construct-expressing yeast being used as a control per microtiter plate.
Ligand Dose–Response Assay. Overnight cultures of the appropriate yeast cells were diluted in SC-Leu-Trp-His minimal media to a final OD600 of 0.002. Aliquots (190 μl) of this diluted culture were added into the wells of a sterile flat bottom 96-well microtiter plate (Rainin Instruments), followed by the addition of 10 μl of appropriately concentrated ligand consisting of a 50-fold dilution of ethanol stock solution in SC-Leu-Trp-His minimal media. These microtiter plates were incubated at 30°C for 24 h, after which cultures were mixed by pipetting, and OD600 readings were taken by using a Spectramax 340PC plate reader (Molecular Devices).
Mammalian Transfection and Luciferase Assay. Methods used for cell culture, transfection, and performance of luciferase assay have been described in ref. 25.
Supporting Materials and Methods. Library creation, library cloning and transformation, and molecular modeling are described in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Results
Library Creation Strategy. Library creation used the following steps: (i) identify all ligand-contacting residues in the receptor structure, (ii) perform individual site saturation mutagenesis of all or a subset of these selected residues, (iii) screen each library in 96-well plates, (iv) select the mutant most selective for the target ligand relative to the natural ligand, (v) perform another round of individual site saturation mutagenesis at the remaining unmutated ligand-contacting residues, (vi) repeat steps iii–v until no further improvement can be achieved, and (vii) perform random mutagenesis on the whole receptor, followed by library screening to isolate mutants with mutations that are not within the ligand binding pocket and yet affect ligand selectivity. These steps have been summarized in the form of a flow chart in Fig. 5, which is published as supporting information on the PNAS web site.
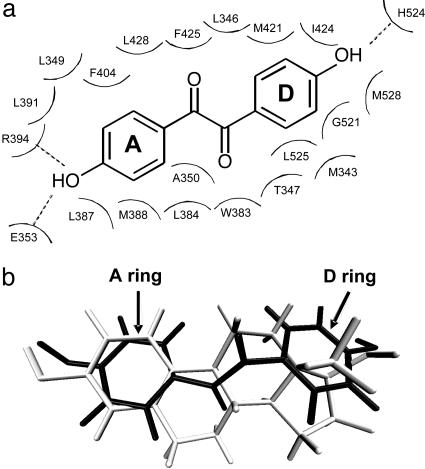
In this work, 21 residues were identified to be in direct contact (within 4.6 Å) with the docked DHB ligand (Fig. 1a). To reduce the load for screening, seven of these residues were not subjected to randomization: R394, E353, and H524 were left unchanged because of their known role in hydrogen bonding with the terminal hydroxyl groups of the ligand; residues L349, L387, F404, and L392, which contact the A ring portion of the ligand, form a tightly maintained ligand-binding subpocket restricting the conformational flexibility of the A ring (Fig. 1b; see also ref. 26) and were similarly left unchanged. Thus, 14 residues in total were selected for individual site saturation mutagenesis. For each site, only 32 distinct library variant possibilities exist (32 possible codon substitutions). The screening of 95 library transformants per randomized site in a convenient 96-well plate format (or 190 transformants per site, as done here) should therefore provide comprehensive coverage of the created variants.
Fig. 1.
Selection of residues for randomization. (a) Two-dimensional depiction of DHB and its surrounding residues when docked into the hERα ligand binding pocket. Twenty-one residues were identified to be within 4.6 Å of DHB. The A ring and D ring analogues of DHB are indicated. Dashed lines denote hydrogen bonds. (b) Superposition of docked DHB (black) and E2 (gray) from crystal structure (PDB code 1GWR).
Library Screening/Selection. Phenotypic screening of library variants was carried out based on a Y2H system consisting of two constructs: (i) the hERα LBD construct fused to the DNA binding domain of the yeast Gal4 transactivator, and (ii) the common mammalian transcriptional coactivator SRC-1 fused to the yeast Gal4 transcriptional activation domain. The hERα–SRC-1 interaction, which is elemental in the role of hERα as a transcriptional activator, is strengthened by the binding of agonist ligands to hERα. The hERα–SRC-1 interaction is also necessary for the host YRG2 yeast cells to effect histidine biosynthesis (21). Thus, this system couples the strength of ligand–receptor interaction within host yeast cells to their growth on media lacking histidine, and it can be applied in either a selection or screening mode.
In the selection approach, variants with strengthened response to DHB relative to the parental construct were selected based on growth of the host yeast cells on agar plates lacking histidine and containing an appropriate concentration of DHB. The selected mutants were subsequently assayed against both DHB and the natural hERα ligand, E2, in a cell growth-based 96-well plate assay to ensure sufficient selectivity. In the screening approach, transformants were individually picked from nonselective (with histidine, without DHB) growth media plates and assayed for cell growth-based response to both target ligand (look for strengthened response) and natural ligand (look for weakened response) in 96-well plates. The screening approach was applied to the libraries created by individual site saturation mutagenesis, whereas the selection approach was used to screen large libraries of variants, which was the case with error-prone PCR-based random point mutagenesis.
Yeast Transactivation Profiles. Mutants leading to increased or unchanged growth in DHB-containing media while simultaneously showing decreased growth in E2-containing media relative to the parental mutant were visually identified and subjected to a growth-based ligand dose–response assay in yeast cells. The plasmids from promising mutants based on this ligand response assay were isolated and retransformed into fresh yeast cells, and the ligand response assay was carried out again to eliminate possible false positives.
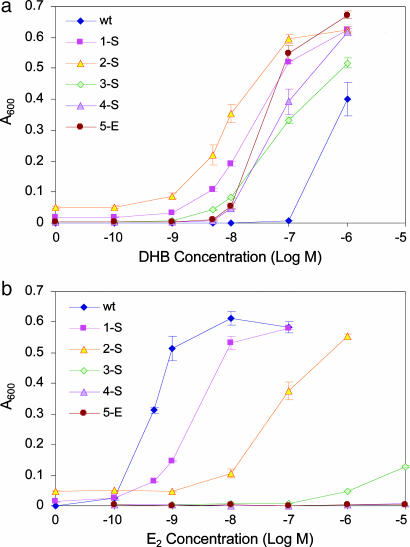
In total, four rounds of individual site saturation mutagenesis and one round of error-prone PCR-based random point mutagenesis were performed. One hundred and ninety transformants were picked from each saturation mutagenesis library and assayed in 96-well plates. For the random mutagenesis library, 3.3 × 106 transformants were subjected to selection, and 1,900 colonies appearing on selective agar growth plates were picked and assayed in 96-well plates. In each round, a number (ranging from 1 to 6) of DHB-selective mutants were identified, the most selective of which was picked and carried forth to the next round of mutagenesis and screening. It should be noted that in cases where more than one DHB-selective mutant was found in a given round of mutagenesis, these mutants appeared in libraries for different randomized sites. The Y2H dose responses and corresponding ligand concentrations leading to half-maximal response (EC50) of the best mutants identified at each round of screening are presented in Fig. 2 and Table 1.
Fig. 2.
Transactivation profiles in Y2H cells for the wild-type hERα construct (wt) and the most DHB-selective mutants identified at each round of screening. (a) Dose responses to DHB. (b) Dose responses to E2. See Table 1 for identity of the mutants. Values represent mean ± SE or the range of two or more experiments.
Table 1. Summary of results of five rounds of mutagenesis and screening, based on Y2H and HEC-1 transactivation profiles.
| Yeast
|
HEC-1
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Round | Best variant | EC50, DHB, nm | EC50, E2, nm | Selectivity | Fold improvement | EC50, DHB, nm | EC50, E2, nm | Selectivity | Fold improvement |
| 0 | wt | 500 ± 200 | 0.5 ± 0.3 | 1.0 × 10-3 | 1.0 | 66 ± 19 | 0.012 | 1.8 × 10-4 | 1.0 |
| 1-S | A350M | 25 ± 20 | 3.0 ± 2.2 | 0.1 | 1.0 ± 102 | ND | ND | ND | ND |
| 2-S | A350M, L346I | 10 ± 5 | 70 ± 30 | 7.0 | 7.0 × 103 | ND | ND | ND | ND |
| 3-S | A350M, L346I, M388Q | 100 ± 80 | ≥5,000 | ≥50 | ≥5.0 × 104 | ND | ND | ND | ND |
| 4-S | A350M, L346I, M388Q, G521S, Y526D | 65 ± 40 | ≥65,000* | ≥1.0 × 103† | ≥1.0 × 106† | 0.37 ± 0.02 | ≥1.0 × 104 | ≥2.7 × 104 | ≥1.5 × 108 |
| 5-E | A350M, L346I, M388Q, G521S, Y526D, F461L, V560M | 100 ± 40 | ≥106* | ≥1.0 × 104† | ≥1.0 × 107† | 0.38 ± 0.17 | ≥1.0 × 104 | ≥2.6 × 104 | ≥1.4 × 108 |
ND, not determined.
Calculated from the estimated selectivity (†) and EC50 values for DHB.
Estimated based on incubation of yeast two-hybrid ligand response microtiter plates at room temperature for 3-4 days, after which time mutants responded to high concentrations (≥1 μM) of E2.
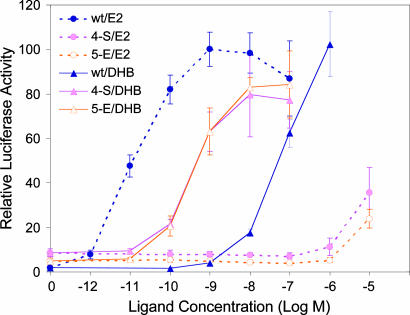
Mammalian Cell Transactivation Profiles. Mammalian cell transactivation profiles for the wild-type hERα and the two best mutants, 4-S and 5-E, were carried out in ER-negative human endometrial cancer (HEC-1) cells after cloning the hERα LBD from the chimeric Y2H construct into the full-length estrogen receptor construct. Dose responses from this analysis are presented in Fig. 3, and the corresponding EC50 values are presented in Table 1.
Fig. 3.
Transactivation profiles in HEC-1 cells for the wild-type hERα construct (wt) and the most DHB-selective engineered mutants (4-S and 5-E) in response to DHB and E2. Luciferase activity was corrected for effects of ligands with β-galactosidase activity driven by a constitutive promoter as described in ref. 25. Values represent mean ± SE or the range of two experiments conducted in duplicate.
Discussion
In this report, we describe the development of a general protein engineering approach that we have applied to reengineer the ligand specificity of hERα. In particular, by combining stepwise targeted site saturation mutagenesis of ligand-contacting protein residues and random point mutagenesis with phenotypic screening or selection in a Y2H system, we have been able to shift the hERα specificity for the synthetic ligand (DHB) versus the natural ligand (E2) by >107-fold. The resulting ligand–receptor pair is highly sensitive to DHB in mammalian cells and is almost fully orthogonal to the natural ligand–receptor pair.
The stepwise individual site saturation mutagenesis/random point mutagenesis strategy used here stands in contrast with other approaches that have been proposed for creating new specific receptor–ligand pairs in a number of ways. These contrasting features can be summarized as follows: (i) Universality: the current library creation strategy can be readily generalized to other receptor–ligand systems, given the availability of sufficient structural information about the receptor, without having to choose specific allowable amino acid substitutions for randomized ligand-contacting receptor sites. Currently, we are applying this approach to create a specific receptor–ligand pair based on hERα and the synthetic thiazole compound 2,4-di(4-hydroxyphenyl)-5-ethylthiazole (27). So far, our application of this approach has enabled us to obtain a hERα variant that exhibits an ≈2.5 × 104-fold improved selectivity for the thiazole compound over the natural ligand E2 relative to the wild-type hERα (unpublished data) (ii) Screenability: there are only 32 possible codon substitutions or 19 possible amino acid substitutions per site for the saturation mutagenesis libraries. Subjecting 96 transformants to screening in a convenient 96-well plate format should therefore be sufficient to represent most, if not all, the possible library variants. For random point mutagenesis of the entire LBD, on average approximately six amino acid substitutions can be accessed per residue by a single base-pair substitution, thus creating a potential library size of ≈1,800 single mutants, which should be screened comprehensively by using a selection system such as the one used here. In contrast, the combinatorial randomization strategy presented by Doyle and coworkers (18) relies on the dominant presence of selective variants within a large library (note that despite the ≈3 × 106 possible codon combinations, only ≈3.8 × 105 transformants were subjected to selection) (iii) Sensitivity: because the potential library size of protein variants is very small for saturation mutagenesis, essentially all randomized variants can be subjected to simultaneous positive screening (for strengthened target ligand response) and negative screening (for weakened natural ligand response) in a sensitive manner. This two-pronged screening is essential for the efficient generation of ligand–receptor pairs with shifted specificity. It should be noted that mutants were identified by using this approach that reproducibly show a 2- to 3-fold enhanced selectivity for DHB compared with the parental construct in yeast (data not shown), although these mutants were not picked for further engineering because others with higher selectivity were identified. (iv) Accessibility to amino acid substitutions: methods for creating a library of protein variants based on single base-pair substitutions on a DNA level can access only a limited number (approximately six on average) of amino acid substitutions per residue. This fact may at least partially explain the inability of Miller and Whelan (16, 17) to identify hERα variants with significantly altered ligand selectivity by using an error-prone PCR-based random mutagenesis strategy. In contrast, a stepwise site saturation mutagenesis approach allows every site in the ligand-binding pocket to be randomized to all 20 possible amino acids, ensuring that important amino acid substitutions at critical receptor positions are not overlooked. Notably, three of four substitutions created in the ligand-binding pocket (A350M, L346I, and M388Q), contributing a combined target ligand selectivity improvement of ≥5 × 104-fold relative to the wild-type hERα construct (Table 1), could not have been obtained through single base-pair substitutions.
It should be noted that the mutants identified by using the receptor reengineering strategy we have described need not represent the only variants with enhanced selectivity for the target ligand. Here, the best, or most selective, receptor variant identified at each round of screening was held fixed and carried forth to the next round of mutagenesis and screening. Had another mutant (not necessarily the best) been chosen as a starting point for subsequent rounds of screening, the resulting final selective variant might well be different.
In contrast to our initial expectation that a predominantly polar binding pocket would be required to complement the polar α-dicarbonyl core of DHB, much of the engineered selectivity derived from variations in hydrophobicity. This observation underlines the potential drawbacks of limiting the amino acids available for substitution at particular receptor sites based on rational considerations.
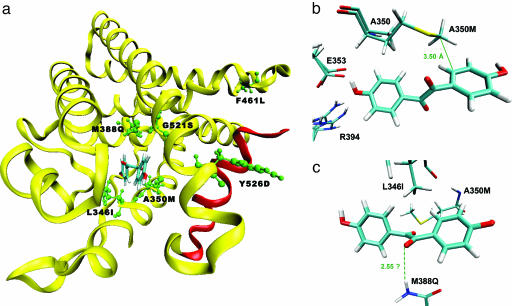
The roles played by individual mutations (Fig. 4) in determining selectivity toward DHB are not immediately clear. To understand the potential role played by the A350M mutation by modeling, this substitution (after energy minimization of all binding pocket and surrounding residues) was made to the docked DHB–hERα complex (Fig. 4b). This cursory analysis reveals that the extended hydrophobic side chain of methionine might make a favorable hydrophobic contact with the D ring analogue of DHB, whereas the short side chain of alanine cannot make this contact. In addition to this favorable hydrophobic interaction, the sulfur atom of the methionine is within 6 Å of carbon atoms in both the A ring and D ring of DHB, resulting in potentially favorable sulfur–aromatic dispersion interactions (28). Although not obvious from the same analysis, we suspect that the long side chain of methionine might clash with the bulky hydrophobic core of E2, leading to a weakened E2 response. A similar analysis to gauge the effect of the M388Q mutation (Fig. 4c) revealed that the glutamine might donate a hydrogen bond to one of the ketone moieties of DHB. The accompanying unfavorable interaction with E2 is presumably due to the introduction of a polar side group into direct contact with the hydrophobic core of E2. Thus, both of these substitutions appear to make dual contributions to the shift in ligand-binding selectivity, enhancing the stability of DHB binding while disabling E2 binding.
Fig. 4.
Analysis of mutations. (a) Location of the mutations in the most selective mutant for DHB, 5-E. Note that residue 560 is located in the C-terminal F domain of hERα, of which structural information is not available. Helix 12, which contributes to a ligand-dependent activation function (AF-2), is colored in red. (b) Effect of mutation A350M. Shown is the methionine residue superimposed on the original alanine. The distance between the terminal carbon atom of the methionine and the D ring analogue of DHB is indicated. (c) Effect of mutation M388Q. The dashed line represents a potential hydrogen bonding interaction. All graphics were made with vmd (29).
It should be noted that in rounds 4 and 5 of mutagenesis and screening, two mutations were introduced into the best identified mutants (mutants 4-S and 5-E). In the fourth round, the nonbinding pocket mutation (Y526D) was the result of a point mutation introduced during polymerase amplification. Site-directed mutagenesis to separate the contributions of G521S and Y526D in mutant 4-S revealed that G521S is primarily responsible for the observed selectivity enhancement relative to mutant 3-S (data not shown). However, interestingly, it was found that in the absence of the Y526D mutation, a significant amount of basal level (ligand-independent) response is present (data not shown). This result may indicate that the Y526D mutation (positioned on helix 11) directly or indirectly influences the conformation of helix 12 (red helix in Fig. 4a), which comprises a ligand-dependent activation function (AF-2) in hERα. In mutant 5-E, site-directed mutagenesis experiments revealed that the observed selectivity enhancement in yeast cells (Table 1) relative to mutant 4-S is entirely due to the F461L mutation, and that V560M had no detectable effect (data not shown). Note that residue 461 is distant from the ligand-binding pocket (Fig. 4a).
For the most part, the ligand selectivity displayed by the chimeric hERα mutants in yeast cells is reproduced well by the full-length hERα constructs in mammalian cells (Table 1). The EC50 values in mammalian cells are, in fact, lower than the corresponding values in yeast cells; this phenomenon has been observed in refs. 18 and 21 and could be attributed to an increased permeability of the ligands for entry into mammalian cells. Another factor that might contribute to the observed difference in ligand response potencies in yeast and mammalian cells is the differing receptor constructs (chimeric versus full-length hERα) used in the yeast and mammalian cell systems. Overall, the ligand selectivities of the mutants in yeast and mammalian cells correlate well, with the mutants being actually more selective for DHB compared with E2 in HEC-1 cells than in Y2H cells (Table 1).
One note should be made regarding the fifth round mutant (5-E). This mutant seems to show no selectivity enhancement toward DHB relative to the fourth-round mutant (4-S), both in yeast (according to Fig. 2) and in mammalian cells (according to Fig. 3). In yeast, the estimated selectivity difference (Table 1) arises primarily from a weakened E2 response compared with the 4-S construct, observed after extended incubation of the ligand-response assay plates. In mammalian cells, this weakened E2 response is not apparent. This disparity between the yeast and mammalian cell systems might be related to the presence of numerous interacting coactivators in mammalian cells compared with the single SRC-1 coactivator that was introduced for the assays in yeast. These additional coactivators, unlike SRC-1, might not be able to distinguish between the E2-bound mutants 4-S and 5-E.
As evident from Table 1, the best receptor variant obtained after four rounds of individual site saturation mutagenesis and one round of error-prone PCR (5-E), despite being highly selective for DHB compared to E2, does not respond to DHB with a potency fully equivalent to that of the very high potency of the wild-type hERα-E2 response (although it comes within ≈30-fold in HEC-1 cells). To enhance the ligand response potency for DHB, further rounds of error-prone PCR mutagenesis and selection based on mutant 5-E were attempted. Despite subjecting a library of 2.4 × 106 transformants to Y2H selection, no variants with significantly improved potency or selectivity for DHB were found. We suspect that the inability to identify mutants more sensitive for DHB may be due to the inability of error-prone PCR to access important amino acid substitutions from single base-pair changes. We expect that other mutagenesis methods could be used to introduce amino acid substitutions otherwise inaccessible by error-prone PCR, so as to further enhance potency of response toward DHB.
In summary, by combining straightforward selection of target protein residues with the power of directed evolution, we have altered the selectivity of a natural nuclear hormone receptor, hERα, for a synthetic ligand DHB by >107-fold. The resulting hERα mutant responds to subnanomolar concentrations of DHB in mammalian cells and is essentially unresponsive to E2, thus being essentially orthogonal to the wild-type hERα-E2 combination. The demonstrated protein engineering approach constitutes a conceptually simple and readily generalizable method for significantly altering the selectivity of nuclear hormone receptors for a target ligand. This approach involves screening very manageably sized protein variant libraries and is sensitive to the detection of variants enhanced in target ligand selectivity. We envision that the described technology could provide a powerful, broadly applicable tool for engineering receptors/enzymes with improved or novel ligand/substrate specificity.
Supplementary Material
Acknowledgments
We thank Dr. Benita S. Katzenellenbogen for providing HEC-1 cells and culture media and for assistance with the HEC-1 cell luciferase assay and Ka Chun Lai for performing yeast-based screening experiments for the target ligand 2,4-di(4-hydroxyphenyl)-5-ethylthiazole. This work was supported by National Science Foundation CAREER Award BES-0348107.
Author contributions: K.C. and H.Z. designed research; K.C. and Z.C. performed research; K.C., Z.C., and J.A.K. analyzed data; and K.C. and H.Z. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: DHB, 4,4′-dihydroxybenzil; E2, 17β-estradiol; HEC-1, human endometrial cancer; hER, human estrogen receptor; LBD, ligand binding domain; SRC-1, steroid receptor coactivator 1; Y2H, yeast two-hybrid; SC, synthetic complete.
References
- 1.Koh, J. T. (2002) Chem. Biol. 9, 17–23. [DOI] [PubMed] [Google Scholar]
- 2.Harvey, D. M. & Caskey, C. T. (1998) Curr. Opin. Chem. Biol. 2, 512–518. [DOI] [PubMed] [Google Scholar]
- 3.Fussenegger, M. (2001) Biotechnol. Progr. 17, 1–51. [DOI] [PubMed] [Google Scholar]
- 4.Bishop, A., Buzko, O., Heyeck-Dumas, S., Jung, I., Kraybill, B., Liu, Y., Shah, K., Ulrich, S., Witucki, L., Yang, F., et al. (2000) Annu. Rev. Biophys. Biomol. Struct. 29, 577–606. [DOI] [PubMed] [Google Scholar]
- 5.Nagy, L. & Schwabe, J. W. (2004) Trends Biochem. Sci. 29, 317–324. [DOI] [PubMed] [Google Scholar]
- 6.Rich, R. L., Hoth, L. R., Geoghegan, K. F., Brown, T. A., LeMotte, P. K., Simons, S. P., Hensley, P. & Myszka, D. G. (2002) Proc. Natl. Acad. Sci. USA 99, 8562–8567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braselmann, S., Graninger, P. & Busslinger, M. (1993) Proc. Natl. Acad. Sci. USA 90, 1657–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang, Y. L., DeMayo, F. J., Tsai, S. Y. & O'Malley, B. W. (1997) Nat. Biotechnol. 15, 239–243. [DOI] [PubMed] [Google Scholar]
- 9.Yaghmai, R. & Cutting, G. R. (2002) Mol. Ther. 5, 685–694. [DOI] [PubMed] [Google Scholar]
- 10.Ansari, A. Z. & Mapp, A. K. (2002) Curr. Opin. Chem. Biol. 6, 765–772. [DOI] [PubMed] [Google Scholar]
- 11.Shi, Y. H. & Koh, J. T. (2001) Chem. Biol. 8, 501–510. [DOI] [PubMed] [Google Scholar]
- 12.Shi, Y. H. & Koh, J. T. (2002) J. Am. Chem. Soc. 124, 6921–6928. [DOI] [PubMed] [Google Scholar]
- 13.Tedesco, R., Thomas, J. A., Katzenellenbogen, B. S. & Katzenellenbogen, J. A. (2001) Chem. Biol. 8, 277–287. [DOI] [PubMed] [Google Scholar]
- 14.Koh, J. T., Putnam, M., Tomic-Canic, M. & McDaniel, C. M. (1999) J. Am. Chem. Soc. 121, 1984–1985. [Google Scholar]
- 15.Doyle, D. F., Braasch, D. A., Jackson, L. K., Weiss, H. E., Boehm, M. F., Mangelsdorf, D. J. & Corey, D. R. (2001) J. Am. Chem. Soc. 123, 11367–11371. [DOI] [PubMed] [Google Scholar]
- 16.Miller, N. & Whelan, J. (1998) J. Steroid Biochem. 64, 129–135. [DOI] [PubMed] [Google Scholar]
- 17.Whelan, J. & Miller, N. (1996) J. Steroid Biochem. 58, 3–12. [DOI] [PubMed] [Google Scholar]
- 18.Schwimmer, L. J., Rohatgi, P., Azizi, B., Seley, K. L. & Doyle, D. F. (2004) Proc. Natl. Acad. Sci. USA 101, 14707–14712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilbert, J., Doré, J. C., Bignon, E., Pons, M. & Ojasoo, T. (1994) Quant. Struct.-Act. Relat. 13, 262–274. [Google Scholar]
- 20.Ding, X. F., Anderson, C. M., Ma, H., Hong, H., Uht, R. M., Kushner, P. J. & Stallcup, M. R. (1998) Mol. Endocrinol. 12, 302–313. [DOI] [PubMed] [Google Scholar]
- 21.Chen, Z. L., Katzenellenbogen, B. S., Katzenellenbogen, J. A. & Zhao, H. M. (2004) J. Biol. Chem. 279, 33855–33864. [DOI] [PubMed] [Google Scholar]
- 22.Woods, R. A. & Gietz, R. D. (2000) Yeast Transformation (Eaton, Natick, MA).
- 23.Rose, M. D. (1987) Methods Enzymol. 152, 481–504. [DOI] [PubMed] [Google Scholar]
- 24.Katzenellenbogen, J. A., Johnson, H. J., Jr., Carlson, K. E. & Myers, H. N. (1974) Biochemistry 13, 2986–2994. [DOI] [PubMed] [Google Scholar]
- 25.Muthyala, R. S., Sheng, S., Carlson, K. E., Katzenellenbogen, B. S. & Katzenellenbogen, J. A. (2003) J. Med. Chem. 46, 1589–1602. [DOI] [PubMed] [Google Scholar]
- 26.Anstead, G. M., Carlson, K. E. & Katzenellenbogen, J. A. (1997) Steroids 62, 268–303. [DOI] [PubMed] [Google Scholar]
- 27.Fink, B. E., Mortensen, D. S., Stauffer, S. R., Aron, Z. D. & Katzenellenbogen, J. A. (1999) Chem. Biol. 6, 205–219. [DOI] [PubMed] [Google Scholar]
- 28.Reid, K. S. C., Lindley, P. F. & Thornton, J. M. (1985) FEBS Lett. 190, 209–213. [Google Scholar]
- 29.Humphrey, W., Dalke, A. & Schulten, K. (1996) J. Mol. Graphics 14, 33–38. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.