Abstract
During meiosis, high levels of recombination initiated by DNA double-strand breaks (DSBs) occur only after DNA replication. However, how DSB formation is coupled to DNA replication is unknown. We examined several DNA replication proteins for a role in this coupling in Schizosaccharomyces pombe, and we show that ribonucleotide reductase, the rate-limiting enzyme of deoxyribonucleotide synthesis and the target of the DNA synthesis inhibitor hydroxyurea (HU) is indirectly required for DSB formation linked to DNA replication. However, in cells in which the function of the DNA-replication-checkpoint proteins Rad1p, Rad3p, Rad9p, Rad17p, Rad26p, Hus1p, or Cds1p was compromised, DSB formation occurred at similar frequencies in the absence or presence of HU. The DSBs in the HU-treated mutant cells occurred at normal sites and were associated with recombination. In addition, Cdc2p is apparently not involved in this process. We propose that the sequence of meiotic S phase and initiation of recombination is coordinated by DNA-replication-checkpoint proteins.
Keywords: cell cycle, DNA replication, meiosis, double-strand break formation
During meiosis, DNA replication occurs, followed by two rounds of chromosome segregation (1, 2). A high level of homologous recombination occurs after meiotic DNA replication but before the first meiotic nuclear division (1, 3–7). Meiotic recombination is required for the proper segregation of homologs at meiosis I, because a chromatid of one homolog recombines with a chromatid of the other homolog, thereby joining homologous chromosomes.
It is well established that meiotic recombination in both budding and fission yeasts is initiated by DNA double-strand breaks (DSBs), catalyzed by the DNA topoisomerase type-2-related enzyme Spo11 in budding yeast (8) and Rec12p in fission yeast (6). Both Spo11 in budding yeast and Rec12p in fission yeast are also essential for meiotic recombination. In addition to Rec12p, meiotic DSB requires multiple gene products that are also essential for meiotic recombination in two yeasts (1, 2).
The DNA-replication checkpoint operates during both the mitotic cell cycle and meiosis to ensure that chromosome segregation is blocked if DNA replication is incomplete (9–12). The proteins Rad1p, Rad3p, Rad9p, Rad17p, Rad26p, Hus1p, and Cds1p are required for operation of the DNA-replication checkpoint in fission yeast (9–11, 13). The DNA-replication-checkpoint proteins send a signal to block mitosis via the cyclin-dependent kinase Cdc2p complexed with the B-type cyclin Cdc13p (9). The Cdc2p-Cdc13p protein kinase is fully activated at the onset of mitosis by dephosphorylation of Cdc2p Tyr-15 (9). Phosphorylation of Cdc2p on Tyr-15 is catalyzed by both the Wee1p and Mik1p Tyr kinases, and dephosphorylation is carried out mainly by the Cdc25p Tyr phosphatase (14).
It has been proposed that DNA replication is directly coupled to initiation of DSB formation in budding yeast, because inhibition of the completion of DNA replication either by hydroxyurea (HU) treatment or by mutation of both the clb5 and clb6 genes (which encode B-type cyclin partners of Cdc28, a homolog of the cyclin-dependent kinase Cdc2p of fission yeast) blocks the formation of DSBs (4, 5). In addition, it has been reported that the removal of active replication origin delays both replication and DSB appearance in budding yeast (5). However, in fission yeast, DSB formation occurs when completion of DNA replication is prevented by the inactivation of initiators of DNA replication (15).
To investigate whether this discrepancy between budding and fission yeasts might result from different ways of preventing meiotic DNA replication, we searched for DNA replication mutants of fission yeast that manifest a reduced level of meiotic DSB formation. Here, we show that ribonucleotide reductase (RNR), the rate-limiting enzyme of deoxyribonucleotide synthesis and the target of the DNA synthesis inhibitor HU, is indirectly required for DSB formation linked to DNA replication. In addition, we present evidence that the sequence of meiotic S phase and initiation of recombination is coordinated by DNA-replication-checkpoint proteins.
Materials and Methods
Fission Yeast Strains and Methods. All strains were constructed by standard procedures (16). The original rad50S strain was kindly provided by E. Hartsuiker (University of Sussex, Brighton, United Kingdom). The strains used for this study are given in Table 1.
Table 1. Strains used in this study.
| Strain | Genotype |
|---|---|
| HM34 | h- pat1-114 leu1-32 (pat1) |
| HM113 | h+ pat1-114 cdc2-L7 (pat1 cdc2-L7) |
| HM336 | h-/h- pat1-114/pat1-114 rec12::LEU2/rec12::LEU2 leu1-32/leu1-32 ade6-M210/ade6-M216 (pat1 rec12) |
| HM338 | h+ pat1-114 orp1-4 (pat1 orp1) |
| HM605 | h-/h- pat1-114/pat1-114 mik1::ura4/mik1::ura4 wee1-50/wee1-50 ura4-D18/ura4-D18 leu1-32/leu1-32 ade6-M210/ade6-M216 (pat1 mik1 wee1) |
| HM713 | h- pat1-114 rad9-192 (pat1 rad9) |
| HM731 | h- pat1-114 rad17-h21 leu1-32 (pat1 rad17) |
| HM783 | h- pat1-114 rad26::ura4 ura4-D18 leu1-32 (pat1 rad26) |
| HM798 | h- pat1-114 hus1-14 leu1-32 (pat1 hus1) |
| HM1307 | h+/h+ pat1-114/pat1-114 ade6-M210/ade6-M216 (pat1) |
| HM1482 | h+ pat1-114 rad1::ura4 ura4-D18 leu1-32 (pat1 rad1) |
| HM1691 | h- pat1-114 rad50S-kanr leu1-32 (pat1 rad50S) |
| HM1801 | h- pat1-114 cds1::ura4 rad50S-kanr ura4-D18 leu1-32 (pat1 cds1 rad50S) |
| HM1804 | h+ pat1-114 rad50S-kanr mik1::ura4 wee1-50 ura4-D18 leu1-32 (pat1 mik1 wee1 rad50S) |
| HM1805 | h+ pat1-114 rad50S-kanr rec12::LEU2 leu1-32 (pat1 rec12 rad50S) |
| HM1824 | h- pat1-114 rad50S-kanr mik1::ura4 wee1-50 rec12::LEU2 ura4-D18 leu1-32 (pat1 mik1 wee1 rec12 rad50S) |
| HM1842 | h- pat1-114 cds1::ura4 rad50S-kanr rec12::LEU2 ura4-D18 leu1-32 (pat1cds1 rec12 rad50S) |
| HM1936 | h-/h- pat1-114/pat1-114 cdc22-M45/cdc22-M45 ura4-D18/+ leu1-32/leu1-32 ade6-M210/ade6-M216 (pat1 cdc22) |
| HM2690 | h- pat1-114 rad3-ts rad50S-kanr leu1-32 (pat1 rad3 rad50S) |
| HM2699 | h- pat1-114 rad3-ts rec12::LEU2 rad50S-kanr leu1-32 (pat1 rad3 rec12 rad50S) |
| HM2849 | h+/h+ pat1-114/pat1-114 rad3-ts-kanr/rad3-ts-kanr rec12::LEU2/rec12::LEU2 leu1-32/leu1-32 ade6-M210/ade6-M216(pat1 rad3 rec12) |
| HM2877 | h+/h+ pat1-114/pat1-114 rad3-ts/rad3-ts leu1-32/leu1-32 ade6-M210/ade6-M216(pat1 rad3) |
| HM3696 | h+/h+ pat1-114/pat1-114 rad50S-kanr/rad50S-kanr leu1-32/+ ade6-M210/ade6-M216(pat1 rad50S) |
| HM4065 | h+ pat1-114 rad3-136 cdc2-22 leu1-32(pat1 rad3 cdc2-22) |
| HM4066 | h+ pat1-114 cdc2-22 leu1-32 (pat1 cdc2-22) |
| HM4082 | h+ pat1-114 rad3-136 rec12::LEU2 cdc2-22 leu1-32 (pat1 rad3 rec12 cdc2-22) |
| HM4088 | h+ pat1-114 rad3-ts cdc13::ura4+-pREP41-cdc13+ rad50S-kanr ura4-D18 leu1-32(pat1 rad3 cdc13 rad50S) |
| HM4090 | h+ pat1-114 cdc13::ura4+-pREP41-cdc13+ rad50S-kanr ura4-D18 leu1-32(pat1 cdc13 rad50S) |
| HM4130 | h+ pat1-114 rad3-ts cdc13::ura4+-pREP41-cdc13+ rec12::LEU2 rad50S-kanr ura4-D18 leu1-32 (pat1 rad3 rec12 cdc13 rad50S) |
| HM4132 | h+ pat1-114 cdc13::ura4+-pREP41-cdc13+ rec12::LEU2 rad50S-kanr ura4-D18 leu1-32 (pat1 rec12 cdc13 rad50S) |
| HM4133 | h+ pat1-114 cdc2-22 rec12::LEU2 leu1-32 (pat1 rec12 cdc2-22) |
Pulsed-Field Gel Electrophoresis (PFGE) and Southern Blot Analysis. Samples were prepared for PFGE as described (15). PFGE was performed in 0.8% agarose (chromosomal grade) with a CHEF DR II apparatus (Bio-Rad) for 43–48 h at 2 V/cm and 8–12°C in TAE buffer (Tris·acetate/EDTA) buffer with a switch time of 30 min. For Southern blot analysis, agarose plugs were prepared, and the DNA was digested with NotI and fractionated by PFGE before hybridization (17). The 32P-labeled probes (≈1 kb) were prepared by the PCR with Schizosaccharomyces pombe genomic DNA as the template and 20-nt primers. The probe for the telomere-proximal part of the NotI J fragment of chromosome 1 spanned nucleotides 24,636–25,586 on cosmid c227 (GenBank accession no. AL133156). The probe for rae1+ on chromosome 2 spanned nucleotides 30,966–32,220 on cosmid c16A3 (GenBank accession no. AL021748). Sizes of DNA fragments were estimated from Saccharomyces cerevisiae chromosomal DNA (CHEF DNA Size Standard, Bio-Rad) and Low-Range PFG Markers (New England Biolabs) stained with ethidium bromide.
Quantitation of Meiotic DSB Frequency. The analysis of Southern blots of rad50S cell DNA was performed with a bioimage analyzer bas1800 (Fuji Photo Film). A background level of radioactivity from the lightest part of each lane was subtracted from each value, and the total remaining level of radioactivity for each lane was normalized relative to the highest value.
Other Techniques. Meiosis was induced as described in ref. 12. Samples for flow-cytometric analysis were prepared and DNA content was determined with a FACScan (Becton Dickinson) also as described (12).
Results and Discussion
To investigate which DNA replication protein is required for DSB formation in fission yeast, we constructed double temperature-sensitive (ts) mutants of various DNA replication protein genes and pat1, which encodes a negative regulator of meiosis whose inactivation by a ts mutation synchronously induces meiosis regardless of its ploidy and high levels of recombination (18). In addition, the frequency of meiotic DSB formation in haploid strains is similar to that in diploid strains in pat1-induced meiosis (ref. 17 and this study). Cells were synchronized in G1 phase of the cell cycle by nitrogen deprivation and shifted to the restrictive temperature of 36°C to induce meiosis. Meiotic DNA replication was largely complete 3 h after the temperature shift in pat1 cells, as judged by flow-cytometric analysis (Fig. 6, which is published as supporting information on the PNAS web site). Meiotic DSBs were detected in these cells by Southern blot analysis with a telomere-proximal part of the NotI J fragment of chromosome 1 as a probe (Fig. 1 A and B) (17). Bands corresponding to the prominent meiotic break site (mbs)1 and mbs2 were detected in pat1 cells in addition to several minor bands (Fig. 1B) (17). One of the DNA replication mutants, cdc22, manifested a defect in DSB formation (Fig. 1B), and most meiotic DNA replication was blocked in these cells (Fig. 6). Cdc22p is the large subunit of RNR in fission yeast (19). A relatively high temperature (36°C) was required to inactivate cdc22 in the ts mutant. Given that DNA replication is delayed and the efficiency of sporulation is reduced at this temperature (15), we used HU to inhibit RNR and a temperature of 34°C to induce meiosis efficiently. Treatment with HU inhibited DNA replication (Fig. 6) and reduced the intensity of bands corresponding to mbs1 and mbs2 in pat1 cells to a level similar to that observed in pat1 rec12 cells (Figs. 1C and 6) (17). Analysis with the rae1+ fragment of chromosome 2 as a probe also revealed that DSB formation was blocked efficiently by HU (Fig. 7, which is published as supporting information on the PNAS web site).
Fig. 1.
RNR is required for DSB formation. (A) Position of the probe corresponding to the telomere-proximal portion of the NotI J fragment of chromosome 1. (B) pat1 (HM1307) and pat1 cdc22 (HM1936) cells were grown and then transferred to nitrogen-free medium for 16 h at 24°C to arrest cells in G1 phase. Meiosis was induced by shifting the temperature to 36°C at time 0. Samples were subjected to digestion of DNA with NotI and then analyzed by PFGE and Southern blot hybridization with the probe described above. (C and D) pat1 (HM1307) (C) and pat1 rad50S (HM1691) (D) cells were arrested in G1 phase, and meiosis was induced at 34°C in the absence (–) or presence (+) of 24 mM HU. Samples collected at the indicated times were analyzed as in B.(E) Quantitation of meiotic DSB frequency at mbs1 and mbs2 determined from experiments similar to that shown in D. Data are means ± SE of values from three independent experiments.
We next examined a rad50S mutant in which the repair of DSBs is prevented and with which it is possible to obtain a quantitative estimate of a low level of DSBs (17). HU treatment effectively blocked DNA replication and reduced DNA fragments including those corresponding to breakage at mbs1 and mbs2 to virtually undetectable levels in pat1 rad50S cells (Figs. 1D and 6). The frequency of DSB formation at mbs1 or mbs2 at 6 h in pat1 rad50S cells was ≈7% and 4%, respectively, in the absence of HU and <0.5% at both sites in the presence of HU, the latter frequency being similar to that observed in pat1 rec12 rad50S cells (data not shown) or the background level (Figs. 1E and 6) (17). These results were confirmed with the rae1+ probe (Fig. 7). Together, our findings suggested that RNR is directly or indirectly required for DSB formation, consistent with observations in budding yeast (5).
If HU directly inhibited meiotic DSB formation, the latter would be expected to be blocked by HU treatment even after meiotic DNA replication. We added HU to the culture medium of pat1 rad50S cells before, during, or after meiotic DNA replication. The formation of DSBs was blocked almost completely only when HU was added before meiotic DNA replication at 0 or 1.5 h after the induction of meiosis (Fig. 2 A and B). After quantitative analysis of DSB formation (Fig. 2C), we found that the proportion of DSB formation was correlated with DNA replication. Thus, these results indicate that HU does not inhibit DSB formation directly but that HU achieves this effect through inhibition of meiotic DNA replication.
Fig. 2.
HU treatment inhibits DSB formation by acting in meiotic S phase. (A) pat1 rad50S (HM3696) cells were arrested in G1 phase, and meiosis was induced in the absence (–) or presence (+) of HU, with the exception that HU was added at the indicated times after the shift to 34°C. Samples were subjected to PFGE, as described for Fig. 1B. The probe used was the one as shown in Fig. 1A.(B) DNA content of the cells in A was determined by flow cytometry. (C) Quantitation of meiotic DSB frequency at mbs1 and mbs2 determined from experiments similar to that shown in A. Data are means ± SE of values from four independent experiments.
Because the DNA-replication checkpoint couples DNA replication and nuclear division, it is possible that this checkpoint also operates to coordinate meiotic DNA replication and DSB formation. We investigated whether Rad3p is required for prevention of DSB formation in cells in which meiotic DNA replication is incomplete by examining pat1 rad3 double-mutant cells induced to undergo meiosis in the absence or presence of HU (Fig. 3). Mutant cells in which meiotic DNA replication was prevented by HU treatment (Fig. 8, which is published as supporting information on the PNAS web site) progressed through meiotic nuclear division (12) and manifested DSBs, although the appearance of the DSBs was delayed by 1 h and did not disappear (Figs. 3A and 8). To examine whether the DSBs observed in the presence of HU were generated by the normal meiotic DSB machinery, we constructed a pat1 rad3 rec12 mutant. No specific DNA fragments were detected in this mutant after treatment with HU (Figs. 3B and 8), suggesting that most of the DSBs depend on Rec12p. These results also suggest that Rad3p negatively regulates DSB formation in a Rec12p-dependent manner. In addition, the same Rec12p-dependent prominent bands were detected with the rae1+ probe in pat1 rad3 cells that had been incubated in the absence or presence of HU (Fig. 8). Thus, most of the DSBs in the pat1 rad3 cells treated with HU appeared not to be attributable to replication fork stalling, given that the collapse of stalled replication forks would produce DSBs at sites other than those normally targeted and would be independent of Rec12p. We confirmed the above results by using the rad50S mutation (Figs. 3 C and D and 8) and quantitated the frequency of DSB formation in these cells (Fig. 3E). The frequencies of DSB formation at mbs1 and mbs2 were ≈7% and 4%, respectively, in the absence of HU treatment, and ≈6% and 4% in HU-treated cells. Together, these findings suggest that Rad3p plays a major role in preventing DSB formation in HU-treated cells that are not mediated through replication fork stabilization whose function is found in the DNA-replication-checkpoint proteins (20).
Fig. 3.
Rad3p is required for coupling of meiotic DNA replication to DSB formation. (A–D) pat1 rad3 (HM2877) (A), pat1 rad3 rec12 (HM2849) (B), pat1 rad3 rad50S (HM2690) (C), or pat1 rad3 rec12 rad50S (HM2699) (D) cells were arrested in G1 phase, and meiosis was induced at 34°C in the absence (–) or presence of HU (+), with HU added at time 0. Samples collected at the indicated times thereafter were analyzed as described for Fig. 1B. The same probe was used as in Fig. 1 A. (E) Quantitation of meiotic DSB frequency at mbs1 and mbs2 for pat1 rad3 rad50S (HM2690) cells determined from experiments similar to that shown in C. Data are means ± SE of values from three independent experiments.
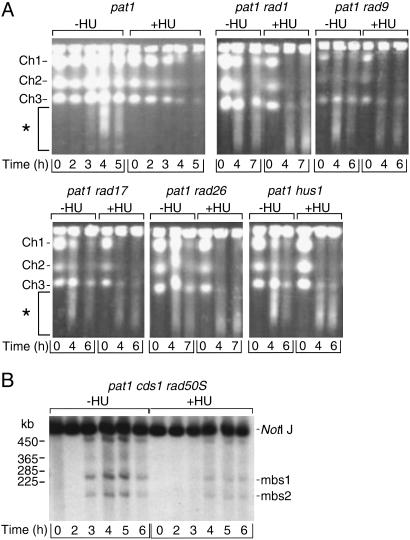
To determine whether other DNA-replication-checkpoint proteins, including Rad1p, Rad9p, Rad17p, Rad26p, and Hus1p, are required for the coupling of DNA replication to DSB formation, we monitored the formation of DSBs in the corresponding DNA-replication-checkpoint mutants with or without HU. DSB formation in the presence of HU was observed in these mutants (Fig. 4A), indicating that all of these proteins are required for prevention of DSB formation when DNA replication is reduced by HU. We next monitored DSB formation and DNA replication in pat1 cds1 rad50S cells without or with HU (Fig. 4B and Fig. 9, which is published as supporting information on the PNAS web site). Formation of DSBs at both mbs1 and mbs2 was observed in pat1 cds1 rad50S cells in the presence of HU in a Rec12-dependent manner, indicating that Cds1p is also required for coupling of DNA replication to DSB formation (Figs. 4B and 9). This result was confirmed by using the rae1+ probe (Fig. 9).
Fig. 4.
The DNA-replication-checkpoint proteins are required for coupling of meiotic DNA replication to DSB formation. (A) pat1 (HM34), pat1 rad1 (HM1482), pat1 rad9 (HM713), pat1 rad17 (HM731), pat1 rad26 (HM783), and pat1 hus1 (HM798) cells were arrested in G1 phase, and meiosis was induced at 34°C in the absence (–) or presence (+) of HU, with HU added at time 0. Samples collected at the indicated times thereafter were analyzed by PFGE, followed by ethidium bromide staining. Ch1, Ch2, and Ch3 indicated the positions of chromosomes 1 (5.7 Mb), 2 (4.6 Mb), and 3 (3.5 Mb), respectively. The smear (*) shows DSBs generated during meiosis. (B) pat1 cds1 rad50S (HM1801) cells were arrested in G1 phase, and meiosis was induced at 34°C. Samples were analyzed as described for Fig. 1B. The probe used in B was the one shown in Fig. 1 A.
In HU-treated cells in which DNA replication is incomplete, the DNA-replication-checkpoint proteins inhibit the kinase activity of Cdc2p by mediating its phosphorylation on Tyr-15, and thereby, they prevent entry into mitosis or the first meiotic nuclear division (9–11). If the block of DSB formation by HU were mediated through Cdc2p, then inactivation of Cdc2p would be expected to prevent DSB formation even in rad3 cells. We used a cdc2–22 allele whose inactivation gives rise to cell-cycle arrest in G2 only during the mitotic cell cycle (21). As expected, meiotic DNA replication occurred normally in pat1cdc2–22 and pat1 rad3 cdc2–22 cells (Fig. 10, which is published as supporting information on the PNAS web site). However, meiotic nuclear division was severely delayed in both of these cells. At 8 h, >60% of pat1 or pat1 rad3 cells entered meiosis II whereas <9% of pat1 cdc2–22 or pat1 rad3 cdc2–22 cells did. Rec12-dependent DSB formation occurred in pat1 rad3 cdc2–22 cells without or with HU, whereas it was severely blocked in pat1 cdc2–22 cells (Figs. 5 and 10). These results suggest that Cdc2p is not involved in coupling DNA replication and DSB formation. To confirm this hypothesis, we next examined a cdc13+ switch-off strain in which cdc13+ expression is inhibited by thiamine (22). Meiotic DNA replication occurred normally in pat1 cdc13 rad50S and pat1 rad3 cdc13 rad50S cells (Fig. 10), and at 8 h, >94% of these cells failed to enter the first meiotic nuclear division, whereas <8% of pat1 rad50S or pat1 rad3 rad50S cells did. DSB formation occurred in pat1 rad3 cdc13 rad50S cells depending on Rec12p without or with HU, whereas it was severely blocked in pat1 cdc13 rad50S cells (Figs. 5 C and D and 10). These results suggest that the major B-type cyclin Cdc13p is not required for DSB formation. If the block of DSB formation by HU was mediated through phosphorylation of Cdc2p on Tyr-15, then prevention of this phosphorylation would be expected to allow DSB formation even in the presence of HU. DSB formation occurred (although at a low level) in pat1 mik1 wee1 and pat1 mik1 wee1 rad50S cells in the absence of HU (Fig. 11, which is published as supporting information on the PNAS web site). Treatment with HU prevented DNA replication and DSB formation in both of these cells (Fig. 11), even though Tyr-15 of Cdc2p is dephosphorylated and the kinase activity of Cdc2p is high in pat1 mik1 wee1 cells (12). All of these findings suggest that the target of the DNA-replication checkpoint in this situation is something other than Cdc2p.
Fig. 5.
Cdc2p/Cdc13p is not involved in coupling meiotic DNA replication to DSB formation. (A and B) pat1 rad3 cdc2–22 (HM4065) (A) and pat1 cdc2–22 (HM4066) (B) cells were arrested in G1 phase, and meiosis was induced at 36.5°C in the absence (–) or presence (+) of HU, with HU added at time 0. Samples collected at the indicated times thereafter were analyzed by PFGE as in described for Fig. 1B. The probe used was the one shown in Fig. 1 A.(C and D) pat1 rad3 cdc13 rad50S (HM4088) (C) and pat1 cdc13 rad50S (HM4090) (D) cells were arrested in G1 phase, and meiosis was induced at 34°C in the absence or presence of HU, with HU added at time 0. Thiamine was added at time 0 to switch off cdc13+ expression. Samples collected at the indicated times thereafter were analyzed by PFGE and Southern blotting, as described above.
Our data lead us to propose the following model for the coupling of meiotic DNA replication and recombination initiation. Treatment of cells with HU results in the inactivation of RNR, leading to the inhibition of the bulk of DNA replication in meiotic S phase. The resulting change in DNA structure likely results in the activation of DNA-replication-checkpoint proteins, including Rad1p, Rad3p, Rad9p, Rad17p, Rad26p, and Hus1p, and this signal is then transmitted to Cds1p, which prevents the initiation of recombination through regulation of unidentified targets (possibly including Rec12p).
Mec1 (a homolog of fission yeast Rad3p) is not required for coupling of meiotic DNA replication to recombination initiation in budding yeast (5), although Mec1 is required for the linkage between meiotic DNA replication and the first meiotic nuclear division (23). This discrepancy might be due to the different roles of the checkpoint proteins in the two yeasts. Budding yeast Mec1 is essential for viability, whereas fission yeast Rad3p is not (11, 24). The hypomorphic mutant mec1–1 is deficient only in coupling of DNA replication and meiotic nuclear division, the link between DNA replication and DSB formation being intact (23). It is also possible that these two yeasts rely on completely different systems for the coordination between meiotic S phase and recombination initiation. DSBs were detected in fission yeast cells deficient in the B-type cyclins Cig2p (data not shown) or Cdc13p (this study) but not in budding yeast clb5 and clb6 mutants (4). However, B-type cyclins are required for meiotic S phase in both yeasts (23, 25, 26). These observations suggest that the roles of B-type cyclins and this cyclin-dependent kinase during meiosis differ between the two yeast species (4, 15, 18, 23, 26).
Compared with the results in ref. 15, Cdc22p inactivation is different from inactivation of other DNA replication initiation proteins that include Orp1p, Cdc18p, Cdc19p, and Cdc21p. Cdc22p inactivation or HU treatment inhibits DSB formation, whereas inactivation of these DNA replication proteins does not. We investigated the relation between Cdc22p and other DNA replication proteins. HU treatment significantly inhibited the DSB formation observed in pat1 orp1 and pat1 cdc2-L7 cells, whereas DNA replication was effectively blocked in both of these cells without or with HU as judged by flow-cytometric analysis (Fig. 12, which is published as supporting information on the PNAS web site) (15). There are at least three possibilities that cause these results. First, the degree of DNA replication may be important; that is, little replication is evident in the cdc22 or HU treated cells, whereas a small amount of DNA replication has occurred in orp1 and other DNA replication mutants (15). It is possible that a subtle difference of the degree of DNA replication may be important. Second, the absence of dNTPs in the HU-treated cells may have a physiological effect that is independent of the replication fork stalling. In mitotic cells, Chk1p is not required for an arrest in HU, but it is required for arrest in the replication mutants (27). Third, the DNA structure that activates the checkpoint signal may play a role. The DNA structure may be different depending on the way of blocking DNA replication. The checkpoint signal that activates Cds1p is generated in the cdc22 or HU treated cells but not in orp1 cdc18 cells during the mitotic cell cycle (28). Therefore, if meiotic S phase is not initiated, DSB formation can, nonetheless, take place. The parallel conclusion for this study would then be that if meiotic S phase is initiated, but blocked, then DSB formation is delayed until replication is complete. Perhaps then, the corollary to this conclusion is that DSB formation occurs in the DNA replication initiation mutants because there is nothing (i.e., no unreplicated DNA) to prevent it. Furthermore, DSB formation is blocked only if an aberrant DNA structure arises that activates the checkpoint.
Supplementary Material
Acknowledgments
We thank P. Nurse for helpful discussions; E. Hartsuiker for the rad50S strain; G. Smith for critical reading of the manuscript and providing strains and the protocol of Southern blot analysis; and C. Yamada, H. Niida, and S. Mori for technical assistance. This work was supported in part by grants-in-aid for Scientific Research from the Ministry of Education, Science, Sports, and Culture of Japan (to H.M.).
Author contributions: H.M. designed research; Y.T. and H.M. performed research; H.M., K.S., and M.N. analyzed data; and H.M. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: DSB, double-strand break; RNR, ribonucleotide reductase; HU, hydroxyurea; PFGE, pulsed-field gel electrophoresis; mbs, meiotic break site.
References
- 1.Roeder, G. S. (1997) Genes Dev. 11, 2600–2621. [DOI] [PubMed] [Google Scholar]
- 2.Davis, L. & Smith, G. R. (2001) Proc. Natl. Acad. Sci. USA 98, 8395–8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Padmore, R., Cao, L. & Kleckner, N. (1991) Cell 66, 1239–1256. [DOI] [PubMed] [Google Scholar]
- 4.Smith, K. N., Penkner, A., Ohta, K. & Nicols, A. (2001) Curr. Biol. 11, 88–97. [DOI] [PubMed] [Google Scholar]
- 5.Borde, V., Goldman, A. S. & Lichten, M. (2000) Science 290, 806–809. [DOI] [PubMed] [Google Scholar]
- 6.Cervantes, M. D., Farah, J. A. & Smith, G. R. (2000) Mol. Cell 5, 883–888. [DOI] [PubMed] [Google Scholar]
- 7.Lee, B. & Amon, A. (2001) Curr. Opin. Cell Biol. 13, 770–777. [DOI] [PubMed] [Google Scholar]
- 8.Keeney, S., Giroux, C. N. & Kleckner, N. (1997) Cell 88, 375–384. [DOI] [PubMed] [Google Scholar]
- 9.Russell, P. (1998) Trends Biochem. Sci. 23, 399–402. [DOI] [PubMed] [Google Scholar]
- 10.O`Connell, M. J., Walworth, N. C. & Carr, A. M. (2000) Trends Cell Biol. 10, 296–303. [DOI] [PubMed] [Google Scholar]
- 11.Murakami, H. & Nurse, P. (2000) Biochem. J. 349, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murakami, H. & Nurse, P. (1999) Genes Dev. 13, 2581–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murakami, H. & Okayama, H. (1995) Nature 374, 817–819. [DOI] [PubMed] [Google Scholar]
- 14.MacNeill, S. A. & Nurse, P. (1997) in The Molecular and Cellular Biology of the Yeast Saccharomyces: Life Cycle and Cell Biology, eds. Pringle, J. R., Broach, J. R. & Jones, E. W. (Cold Spring Harbor Lab. Press, Woodbury, NY), pp. 697–763.
- 15.Murakami, H. & Nurse, P. (2001) Nat. Genet. 28, 290–293. [DOI] [PubMed] [Google Scholar]
- 16.Moreno, S., Klar, A. & Nurse, P. (1991) Methods Enzymol. 194, 795–823. [DOI] [PubMed] [Google Scholar]
- 17.Young, J. A., Schreckhise, R. W., Steiner, W. W. & Smith, G. R. (2002) Mol. Cell 9, 253–263. [DOI] [PubMed] [Google Scholar]
- 18.Yamamoto, M., Imai, Y. & Watanabe, Y. (1997) in The Molecular and Cellular Biology of the Yeast Saccharomyces: Life Cycle and Cell Biology, eds. Pringle, J. R., Broach, J. R. & Jones, E. W. (Cold Spring Harbor Lab. Press, Woodbury, NY), pp. 1037–1106.
- 19.Fernandez Sarabia, M. J., McInerny, C., Harris, P., Gordon, C. & Fantes, P. (1993) Mol. Gen. Genet. 238, 241–251. [DOI] [PubMed] [Google Scholar]
- 20.Lopes, M., Cotta-Ramusino, C., Pellicioli, A., Liberi, G., Plevani, P., Muzi-Falconi, M., Newlon, C. S. & Foiani, M. (2001) Nature 412, 557–561. [DOI] [PubMed] [Google Scholar]
- 21.MacNeill, S. A., Creanor, J. & Nurse, P. (1991) Mol. Gen. Genet. 229, 109–118. [DOI] [PubMed] [Google Scholar]
- 22.Fisher, D. L. & Nurse, P. (1996) EMBO J. 15, 850–860. [PMC free article] [PubMed] [Google Scholar]
- 23.Stuart, D. & Wittenberg, C. (1998) Genes Dev. 12, 2698–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Desany, B. A., Alcasabas, A. A., Bachant, J. B. & Elledge, S. J. (1998) Genes Dev. 12, 2956–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iino, Y., Hiramine, Y. & Yamamoto, M. (1995) Genetics 140, 1235–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borgne, A., Murakami, H., Ayte, J. & Nurse, P. (2002) Mol. Biol. Cell 13, 2080–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walworth, N., Davey, S. & Beach, D. (1993) Nature 363, 368–371. [DOI] [PubMed] [Google Scholar]
- 28.Murakami, H., Yanow, S. K., Griffiths, D., Nakanishi, M. & Nurse, P. (2002) Nat. Cell Biol. 4, 384–388. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.