Abstract
P85α, which acts as a tumour suppressor, is frequently found to be downregulated in various human cancers. However, the role of p85α in the tumour microenvironment is unknown. Here, we report that aberrantly low expression of p85α in breast cancer stroma is clinically relevant to breast cancer disease progression. Stromal fibroblasts can acquire the hallmarks of cancer-associated fibroblasts (CAFs) as a result of the loss of p85α expression. Paracrine Wnt10b from p85α-deficient fibroblasts can promote cancer progression via epithelial-to-mesenchymal transition (EMT) induced by the canonical Wnt pathway. Moreover, exosomes have a key role in paracrine Wnt10b transport from fibroblasts to breast cancer epithelial cells. Our results reveal that p85α expression in stromal fibroblasts haves a crucial role in regulating breast cancer tumourigenesis and progression by modifying stromal–epithelial crosstalk and remodelling the tumour microenvironment. Therefore, p85α can function as a tumour suppressor and represent a new candidate for diagnosis, prognosis and targeted therapy.
Introduction
Breast tumours are highly complex and are composed of neoplastic epithelial cells within a tumour-associated microenvironment. The peritumoral environment consists of an extracellular matrix and numerous stromal components, including cancer-associated fibroblasts (CAFs), inflammatory immune cells, mesenchymal stem cells and endothelial cells of the blood and lymphatic systems.1, 2 Stromal–epithelial interactions have a critical role in tumourigenesis by regulating cell proliferation, survival, polarity, differentiation and angiogenesis as well as by modifying cellular compartments, which results in the coevolution of tumour cells and their microenvironment.3, 4 Fibroblasts are the major cell type found in the stromal compartment, and these cells tightly maintain homeostasis by preventing neoplastic transformation of epithelial cells in normal tissues through networks of cytokines and growth factors.4 CAFs promote tumour progression, but the emergence of this cellular feature and the regulation of stromal–epithelial interactions are poorly understood.
Although overwhelming evidence indicates that the behaviour of tumourigenic cells is highly regulated by the complex tumour microenvironment,1 genome instability has classically been considered the underlying mechanism behind tumour initiation and progression. Chromosomal instability can account for the generation of a variety of genetically transformed cancer cells. Breast cancer is a heterogeneous disease with abnormalities in the function and expression of a variety of proteins that contribute to progression, including BRCA1, RB1, TP53, PTEN, AKT1, PIK3CA, KRAS and GATA3.5, 6 Most of these genes are involved in cell cycle regulation, apoptosis, gene transcription and other cell signalling cascades.7 Hence, most research has focused on mutations in the tumour epithelial cells themselves and the subsequent neoplastic progression and diversity of histological subtypes. However, actually, the tumour stroma can also be inappropriately activated in cancer progression. Some reports have revealed that specific signalling pathways in CAFs are involved in suppressing tumour formation. Loss or downregulation of the Notch effector CSL in fibroblasts is sufficient for CAF activation and promotes tumourigenesis.8 Loss of tumour growth factor-β (TGF-β) type II receptor in fibroblasts can promote mammary carcinoma growth and invasion in mouse model.9 Timp-deficient fibroblasts can augment human tumour xenografts and spontaneous metastasis.10 Loss of Pten in stromal fibroblasts can accelerate the initiation, progression and malignance of breast cancer.11 In general, it remains unclear whether stromal cells harbouring mutations or alterations in the expression of these genes can orchestrate the progression of mammary malignancies by crucially modifying stromal–epithelial crosstalk and remodelling the tumour microenvironment. Additionally, the molecular mechanisms underlying the potential role of the abnormally expressed stromal genes in cancer signal transduction require further elucidation.
Members of the phosphoinositide 3-kinase (PI3K) family of proteins, consisting of three classes of PI3K isoforms, are critical intracellular signal nodules. PI3K isoforms regulate tumourigenesis and malignant progression by activating multiple downstream effector proteins that control various cellular processes, such as growth, survival and motility.12, 13, 14 Thus far, only the class IA PI3Ks, which are heterodimers composed of a regulatory subunit (p85α, p55α, p50α, p85β or p55γ) in complex with a catalytic subunit (p110α, p110β or p110δ), have been implicated in human cancer.15, 16 Oncogenic mutations of PI3K subunits are frequent in various human cancers, including breast cancer;16 PIK3CA is mutated in ~25–40% of breast cancers, especially in ER-positive breast cancer.14, 17 Although the role of PI3K pathway in cancer cells had been elucidated, the potential mechanism of PI3K pathway in tumour stroma needs to be explored. Actually, alterations in the PI3K pathway are involved in the activation of tumour stroma.18, 19 Recently Pten, a key regulator of PI3K signalling, have proved to be a suppressor in tumour stroma. Deletion of Pten in stromal fibroblasts can accelerate the initiation, progression and malignant transformation of breast tumours.11 P85α is also a crucial regulatory subunit that mediates the activation of class IA PI3Ks through receptor tyrosine kinases. Somatic mutations in p85α can promote cancer cell survival, anchorage-independent cell growth and oncogenesis through class IA PI3Ks activation.20 However, the effects and functional role of p85α in peritumoral stromal fibroblasts in breast carcinoma initiation and progression remain to be studied.
In this study, we demonstrate that aberrantly low expression of p85α in the stroma is associated with breast tumourigenesis. Specifically, loss of p85α expression stimulates the conversion of fibroblasts into activated myofibroblasts and results in the acquisition of characteristic CAF functions that promote breast cancer epithelial cell proliferation and metastasis through exosome-mediated paracrine Wnt10b signalling. Our results reveal that p85α expression in fibroblasts have a crucial role in modifying stromal–epithelial crosstalk and disrupting homeostasis in the tumour microenvironment, which subsequently regulates breast cancer initiation and malignant progression.
Results
Aberrant low expression of p85α in the stroma is associated with breast tumourigenesis
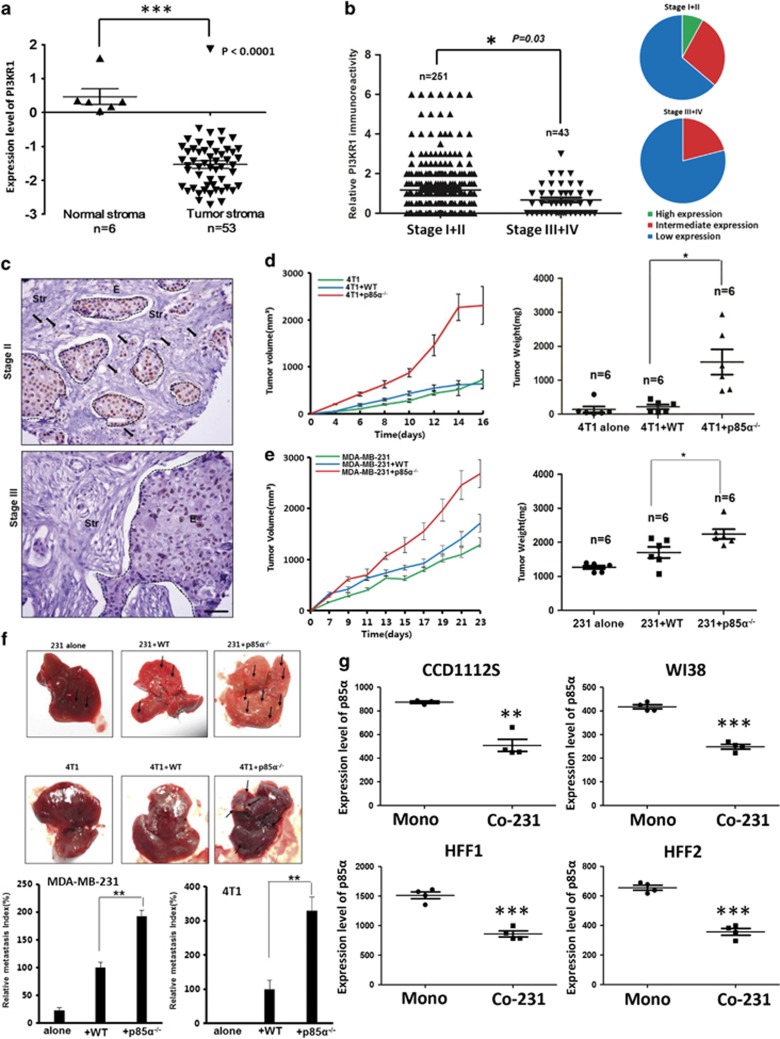
To investigate the role of stromal expression of p85α in breast cancer, we analysed the expression levels of p85α in human breast carcinoma stroma microarrays based on GEO databases. Interestingly, breast cancer stroma samples exhibited lower expression compared with normal breast stroma, which indicates that the p85α expression was significantly decreased in the breast cancer stromal compartment (Figure 1a).21 Moreover, we found that the p85α expression in human breast carcinoma stroma is lower in patients of recurrence within 3 years than that in no-recurrence group and the patients with relatively low level of p85α expression showed a significant increase in recurrence rate compared with the relatively high level of p85α expression group (Supplementary Figure S1A). To further assess the relationship between p85α expression in breast cancer stroma and cancer progression, we collected hundreds of primary breast cancer tumours, including multiple tumour node metastasis (TNM) stages, which were examined by immunohistochemical analysis of p85α expression in a tissue microarray using a digital image analysis algorithm. The results indicated that the protein level of p85α in breast cancer stroma is inversely correlated with breast carcinoma TNM stage, where reduced expression of p85α was found in late TNM stage breast cancer stroma compared with its expression in early TNM stage tissues (Figure 1b). However, the expression of p85α was seemingly unrelated to patient age or grade of cellular differentiation (Supplementary Table S1). Immunohistochemical staining of p85α in representative samples of TNM stage II and III tumours is shown in Figure 1c.
Figure 1.
Aberrant low expression of p85α in the stroma is associated with breast tumourigenesis. (a) Gene expression levels of p85α (PIK3R1) in normal and breast cancer stroma from GSE9014. Data are the mean±s.e.m. ***P-value⩽0.001. (b) Left panel: P85α expression levels in breast cancer stroma samples. Right panel: The proportion of different extent of p85α expression in various stage breast cancer stroma. Data are the mean±s.e.m. *P-value⩽0.05. (c) Representative immunohistochemical images from various breast cancer stages. Scale bar, 100 μm. Dotted lines circled structures, cancer epithelium, the arrows, significant expression of p85α in cancer stroma. (d) Tumour volume and weight measurements of 4T1 cells grown subcutaneously alone, with WT or p85α−/− fibroblasts. Each group have six mice (n=6). Data are the mean±s.e.m. *P-value⩽0.05. (e) Tumour volume and weight measurements of MDA-MB-231 cells similar to (d). Each group have six mice (n=6). Data are the mean±s.e.m. *P-value⩽0.05. (f) Quantification of liver metastases in mice described in (d and e) using H&E staining. Scale bar, 100 μm. Data are the mean±s.e.m. **P-value⩽0.01. (g) P85α expression in fibroblasts cocultured with breast cancer cells from GSE41678. Data are the mean±s.e.m. Mono, monoculture of cancer cells; Co-231, fibroblasts cocultured with MDA-MB-231. **P-value⩽0.01, ***P-value⩽0.001.
Fibroblasts are the major component of breast cancer stroma.22 Hence, to confirm these clinical observations, we establish breast tumours implantation model by subcutaneous injection of 4T1 or MDA-MB-231 cells mixed with Wild-type (WT) or p85α−/− fibroblasts. At 16 or 23 days after implantation, 4T1 and MDA-MB-231 cells mixed with p85α−/− fibroblasts both generated tumours of greater volume and weight than 4T1 and MDA-MB-231 cells mixed with WT fibroblasts or alone, respectively (Figures 1d and e). WT or p85α−/− fibroblasts alone failed to form tumours in mice (data not shown). Moreover, evaluation of organs collected from mice that were killed revealed notable liver metastases present in the mice injected with 4T1 or MDA-MB-231 cells mixed with p85α−/− fibroblasts as measured by quantifying the number of metastases in the liver (Figure 1f). These data revealed that p85α−/− fibroblasts can promote breast cancer metastasis compared with effects of WT fibroblasts. Consistent with the anatomical results, haematoxylin and eosin staining of liver tissue also confirmed these results (Supplementary Figure S1B).
Notably, some reports have demonstrated that cancer epithelial cells can also regulate tumour stroma.23, 24 Interestingly, when various human fibroblast cell lines were cocultured with MDA-MB-231 breast cancer cells, they all exhibited downregulation of p85α expression (Figure 1g).25 We also verified these data from public database by western blot (Supplementary Figure S1C). Therefore, according to a theory of stromal coevolution,26 we propose that the origin of abnormal p85α expression in CAFs not only differentiates themselves but also leads to coevolution of stromal–epithelial interactions.
Therefore, these data suggest that aberrantly low or loss of p85α expression in tumour stroma could be correlated with breast carcinoma initiation and progression.
P85α deletion converts fibroblasts into activated myofibroblasts exhibiting features of CAFs
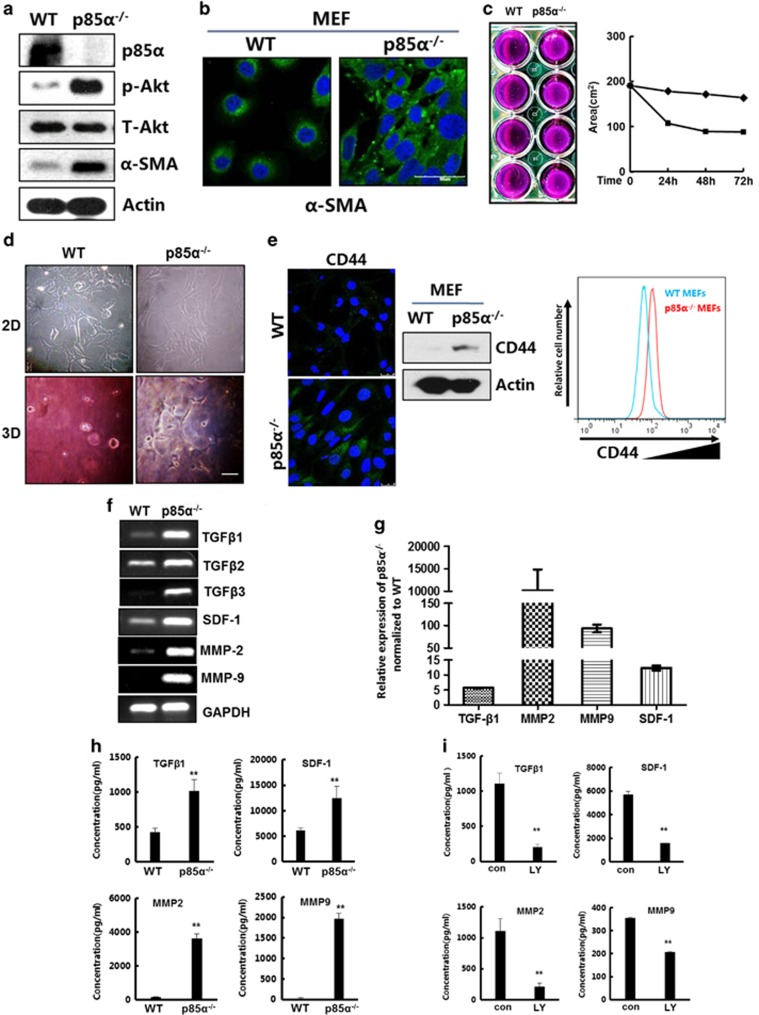
CAFs include a large number of myofibroblasts, which are activated fibroblasts that have an ability to substantially promote tumourigenesis.27, 28 Immunoblot and immunofluorescence data indicated that p85α deletion in fibroblasts significantly promoted the expression of α-smooth muscle actin, which is a marker of myofibroblasts (Figures 2a and b). Furthermore, we found that the loss of p85α expression markedly activated Akt, a critical kinase downstream of PI3K, which is consistent with previous reports regarding the function of p85α as a key regulatory subunit of PI3K (Figure 2a).29 Other characteristics of myofibroblasts include actomyosin fibre formation and stronger contractility. Collagen gel contraction assays indicated that p85α−/− fibroblasts exhibited an increased ability to induce gel contraction compared with WT fibroblasts (Figure 2c). Moreover, p85α−/− fibroblasts exhibited a stretched morphology in dish (two-dimensional culture) and collagen gels (three-dimensional culture; Figure 2d). Recently others have reported that CD44 expression contributes to the acquisition of an activated fibroblast phenotype in the tumour microenvironment.30, 31 Consistently, we found enhanced CD44 expression and accumulation on the surface of p85α−/− fibroblasts compared with that of WT fibroblasts (Figure 2e).
Figure 2.
P85α deletion converts fibroblasts into activated myofibroblasts exhibiting features of CAFs. (a) Western blot analysis of p85α, phospho-Akt, Akt and α-smooth muscle actin (α-SMA) expression in WT and p85α−/− fibroblasts. p-Akt, Phospho-Akt (Ser473); T-Akt, Akt (pan). (b) Immunofluorescence analysis of α-SMA expression in WT and p85α−/− fibroblasts. Scale bar, 50μm. (c) The area of the contracted gels induced by WT or p85α−/− fibroblasts. The appearance of the contracted gels that have four replications at 72 h is also shown. (d) The morphology of WT or p85α−/− fibroblasts in a dish or collagen lattice described in (c). (e) Immunofluorescence, western blot and fluorescence-activated cell sorting (FACS) analysis of CD44 expression in WT and p85α−/− fibroblasts. Scale bar, 20 μm. (f) Reverse transcription–PCR analysis of TGF-β family proteins and SDF-1. MMP2 and MMP9 expression in WT and p85α−/− fibroblasts. (g) Quantitative PCR analysis of TGF-β1, SDF-1, MMP2 and MMP9 expression in WT and p85α−/− fibroblasts. Data are the mean±s.e.m. (h) Enzyme-linked immunosorbent assay (ELISA) analysis of TGF-β1, SDF-1, MMP2 and MMP9 expression in conditioned medium of WT and p85α−/− fibroblasts. Data are the mean±s.e.m. **P-value⩽0.01. (i) ELISA analysis of TGF-β1, SDF-1, MMP2 and MMP9 expression in p85α−/− fibroblast-conditioned medium of when treated with or without the PI3K inhibitor LY294002 (50 μM) for 24 h and the DMSO solution as a control. Con, p85α−/− fibroblasts treated with DMSO, LY, p85α−/− fibroblasts treated with LY294002. Data are the mean±s.e.m. **P-value⩽0.01.
Additionally, CAFs have the potential to produce high levels of secreted factors.32, 33 We found that p85α−/− fibroblasts can express and secrete increased levels of TGF-β, stromal cell-derived factor 1 (SDF-1), matrix metalloproteinase-2 (MMP2) and MMP9 compared with that of WT fibroblasts (Figures 2f–h). However, LY294002, an inhibitor of the PI3K/Akt signalling pathway, markedly decreased the ability of p85α−/− fibroblasts to express and secrete cytokines (Figure 2i and Supplementary Figure S2).
These results indicate that p85α crucially dominates stromal fibroblast activation and promotes the expression and secretion of high levels of cytokines in an Akt activity-dependent manner.
Fibroblast p85α deficiency promotes the proliferation and metastasis of cancerous mammary epithelial cells
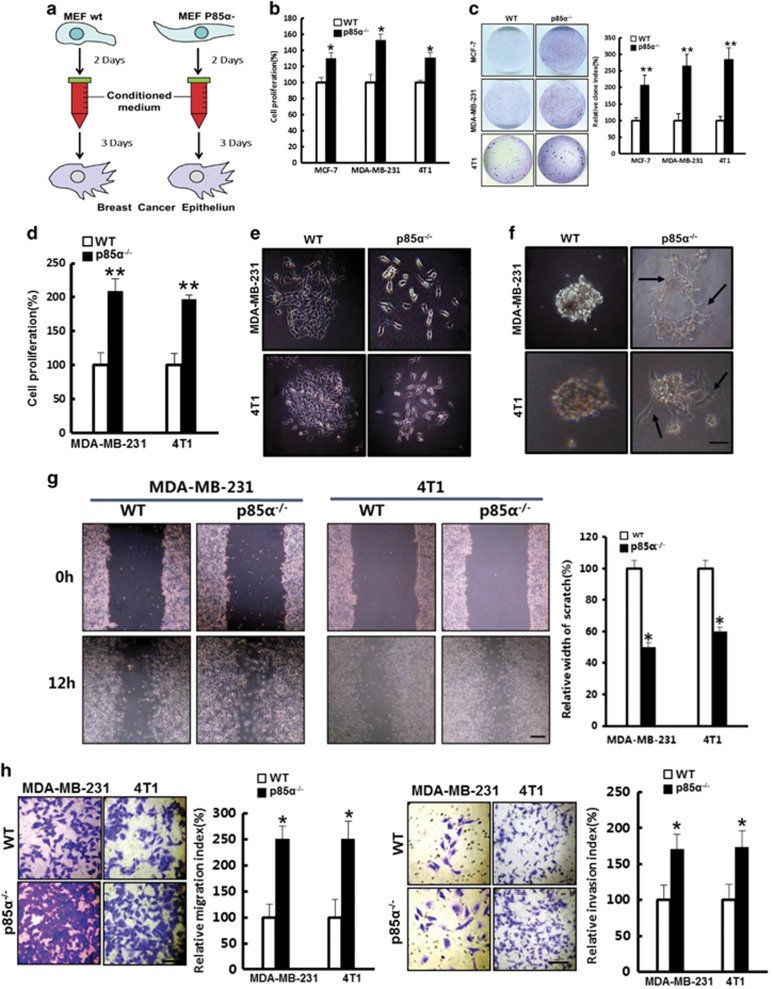
The elevated expression of soluble factors in p85α−/− fibroblasts has been suggested to act as a possible regulator of adjacent cancer cell proliferation and migration.32 To test this hypothesis, we treated MDA-MB-231, MCF-7 and 4T1 cells with prepared conditioned medium (Figure 3a) from WT and p85α−/− fibroblasts, respectively, or cocultured them with WT or p85α−/− fibroblasts, respectively, and then examined cell proliferation. As shown in Figures 3b–d, p85α−/− fibroblasts promoted proliferation and plate colony formation of these cancer cells compared with WT fibroblasts.
Figure 3.
Fibroblast p85α deficiency promotes the proliferation and metastasis of cancerous mammary epithelial cells. (a) Schematic of the experimental design. (b) Cell proliferation of MCF-7, MDA-MB-231 and 4T1 cells in WT or p85α−/− fibroblast-conditioned medium. Data are the mean±s.e.m. *P-value⩽0.05. (c) Plate colony formation assay of MCF-7, MDA-MB-231 and 4T1 cells in WT or p85α−/− fibroblast-conditioned medium. Data are the mean±s.e.m. **P-value⩽0.01. (d) Cell proliferation of MDA-MB-231 and 4T1 cells in coculture with WT or p85α−/− fibroblasts separately by transwell chambers (0.4-μm pore). Data are the mean±s.e.m. **P-value⩽0.01. (e) The morphology of MDA-MB-231 and 4T1 cells in WT or p85α−/− fibroblast-conditioned medium. Scale bar, 100 μm. (f) The morphology of MDA-MB-231 and 4T1 cells cultured in Matrigel treated with WT or p85α−/− fibroblast-conditioned medium. Scale bar, 100 μm. (g) The relative width of the scratch assay in MDA-MB-231 and 4T1 cells in different conditioned medium from WT or p85α−/− fibroblasts. Scale bar, 1 cm. Data are the mean±s.e.m. *P-value⩽0.05. (h) The migration and invasion assay of MDA-MB-231 and 4T1 cells cultured in transwell with fibroblasts. Scale bar, 50 μm. Data are the mean±s.e.m. *P-value⩽0.05.
Interestingly, we found that MDA-MB-231 and 4T1 cells treated with conditioned medium from p85α−/− fibroblasts present an elongated spindle-like morphology and lose their intercellular tight junctions (Figure 3e). Moreover, the cells treated with p85α−/− fibroblast-conditioned medium exhibited distinct stretching features and enhanced motility in three-dimensional cultures in Matrigel (Figure 3f). These observations suggested that loss of p85α expression in fibroblasts can render cancer cells capable of enhanced motility and invasive properties.34 Further investigation using scratch assay and transwell migration/invasion assays confirmed that conditioned medium from p85α−/− fibroblasts potently stimulated breast cancer epithelial cell migration and invasion (Figures 3g and h).
P85α-deficient fibroblasts promote the induction of EMT in breast cancer epithelial cells via a paracrine Wnt/β-catenin pathway
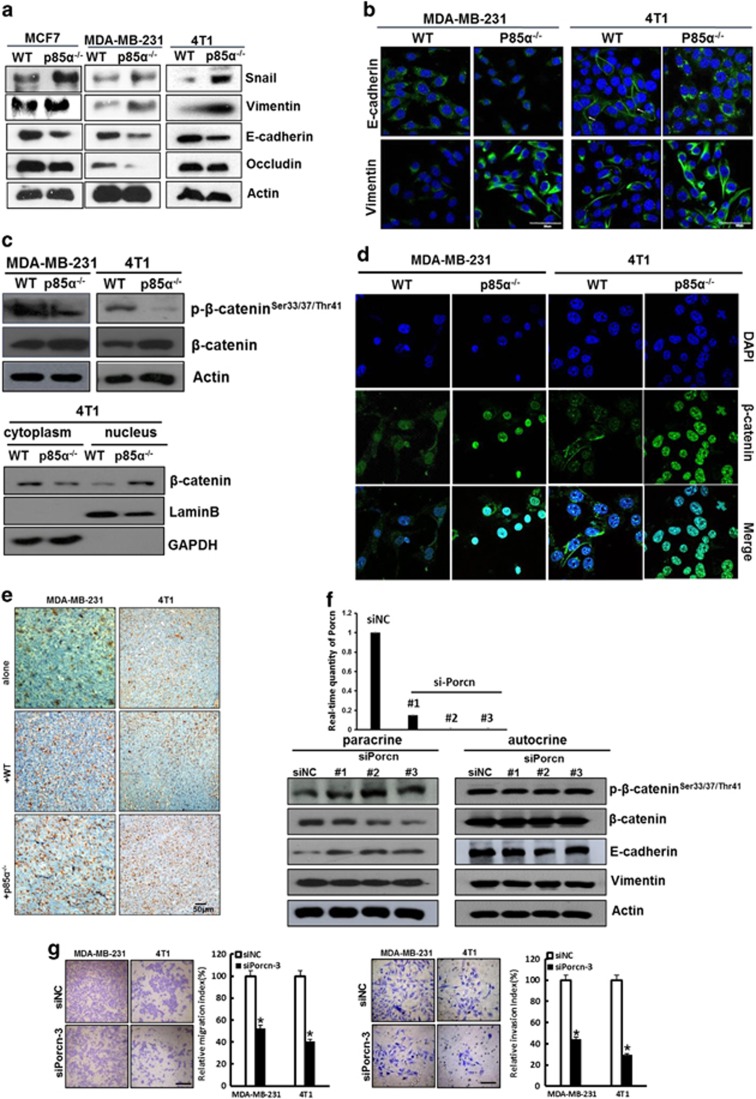
Cancer epithelial cells lose cell–cell junctions and apical–basal polarity leading to a change in cell shape and induction of EMT, which enables stationary epithelial cells to acquire traits consistent with enhanced motility, invasiveness and self-renewal.35, 36 As shown in Figure 3e, MDA-MB-231 and 4T1 cells seemingly underwent an EMT-like phenotypic change when treated with p85α−/− fibroblast-conditioned medium. Therefore, we measured the expression of EMT markers and found that the mesenchymal markers vimentin and Snail were upregulated, and the epithelial markers E-cadherin and occludin were downregulated when MDA-MB-231 and 4T1 cells were treated with p85α−/− fibroblast-conditioned medium compared with those treated with WT fibroblast-conditioned medium, respectively (Figures 4a and b). These findings indicated that the p85α−/− fibroblasts could promote the induction of an EMT event in breast cancer cells.
Figure 4.
P85α−/− fibroblasts promote the induction of EMT in breast cancer epithelial cells via a paracrine Wnt/β-catenin pathway. (a) Western blot analysis of EMT markers in MCF-7, MDA-MB-231 and 4T1 cells cultured in WT or p85α−/− fibroblast-conditioned medium. (b) Immunofluorescence analysis of the expression of EMT markers in MDA-MB-231 and 4T1 cells cultured in WT or p85α−/− fibroblast-conditioned medium. Scale bar, 50 μm. (c) Western blot analysis of Wnt/β-catenin activation by the expression and localization of phospho-β-catenin and β-catenin in MDA-MB-231 and 4T1 cells cultured in WT or p85α−/− fibroblast-conditioned medium. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a loading control for cytoplasmic proteins, and laminin B was used as a loading control for nuclear proteins. (d) Immunofluorescence analysis of the location of β-catenin in MDA-MB-231 and 4T1 cells cultured in WT or p85α−/− fibroblast-conditioned medium. Scale bar, 20 μm. (e) β-Catenin immunostaining in tumour tissue described in Figures 1d and e. Scale bar, 50 μm. (f) Western blot analysis of Wnt pathway and EMT markers in paracrine and autocrine signalling conditions. The effects of siPorcn are shown on the upper panel. (g) The migration and invasion assay of MDA-MB-231 and 4T1 cells cultured in negative control or Porcn knockdown p85α−/− fibroblast-conditioned medium. Data are the mean±s.e.m. *P-value⩽0.05. Scale bar, 100 μm.
Reports indicate that Wnt signalling can regulate gene expression during EMT in breast cancer.37 Therefore, we analysed Wnt/β-catenin signalling pathway activity in our research model. Compared with conditioned medium from WT fibroblasts, p85α−/− fibroblast-conditioned medium markedly decreased the levels of phosphorylated β-catenin at Ser33/37/Thr41 and promoted β-catenin translocation into the nucleus (Figures 4c and d), indicating that the Wnt/β-catenin signalling pathway was activated. Consistent with the in vitro results, p85α−/− fibroblasts also activated tumour Wnt signalling in vivo (Figure 4e).
Here, we speculated that the probable source of secreted Wnt ligands acting on epithelial cells was either fibroblasts (as a paracrine factor) or breast cancer cells (as an autocrine factor). To explore this issue, we knocked down Porcupine (Porcn), which encodes for an acyltransferase essential for Wnt lipidation and secretion,38 in 4T1 cells or p85α−/− fibroblastst to block Wnt autocrine or paracrine signalling, respectively. Interestingly, as shown in Figure 4f, knockdown of Porcn in 4T1 cells did not change the activation of the Wnt pathway or the levels of EMT markers. In contrast, knockdown of Porcn in p85α−/− fibroblasts inhibited the activation of Wnt and EMT of 4T1 cells. Further analysis of cell migration and invasion also suggested that paracrine Wnt was related to metastasis of MDA-MB-231 and 4T1 cells (Figure 4g). Therefore, we concluded that Wnt ligands that induced epithelial cell EMT represented a paracrine factor originating from p85α−/− fibroblasts.
Wnt10b acts as a paracrine factor and has a crucial role in inducing breast cancer epithelial cell EMT and facilitating metastasis
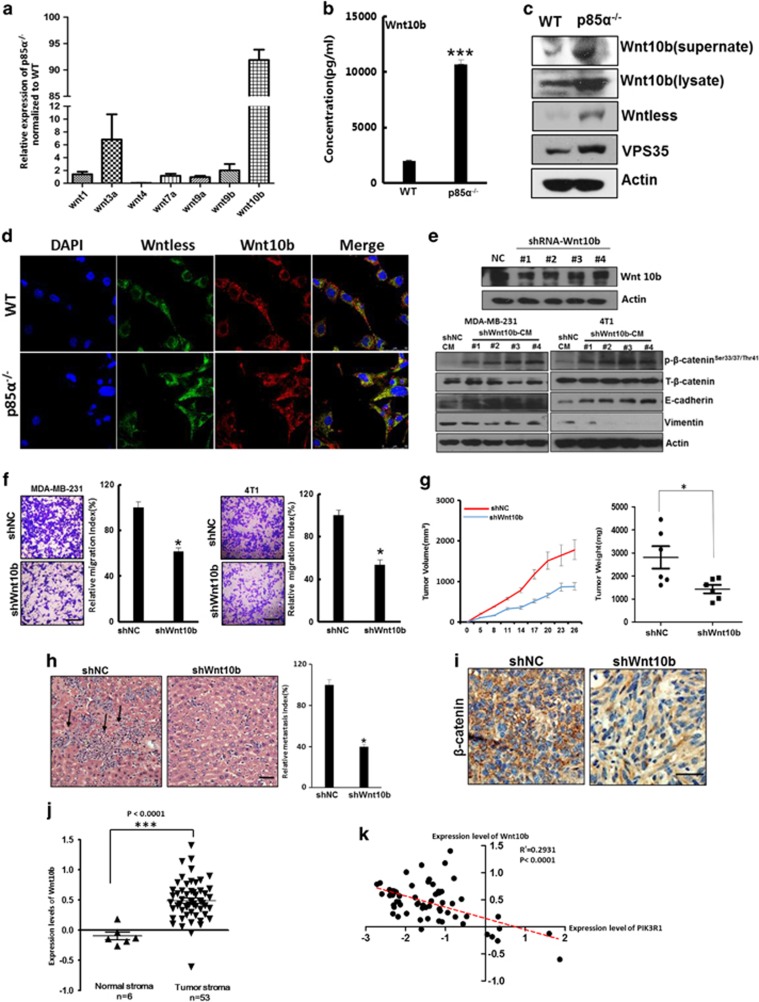
Next, we measured the expression of Wnt family proteins involved in canonical pathways in p85α−/− fibroblasts and found that the expression of Wnt10b were significantly increased in p85α−/− fibroblasts compared with WT fibroblasts (Figures 5a and b). Furthermore, western blot analyses indicated that p85α−/− fibroblasts expressed distinctly high levels of Wnt10b both in supernatants and cell lysates. In addition, Wntless and Vps35, which are specifically required for Wnt secretion,39, 40 were also upregulated in p85α−/− fibroblasts (Figures 5c and d). To examine the role of Wnt10b in breast cancer cell EMT and metastasis, 4T1 cells were treated with conditioned medium from p85α−/− fibroblasts in which Wnt10b was knocked down. As shown in Figures 5e and f, Wnt10b knockdown significantly reduced the ability of p85α−/− fibroblasts to promote activation of the Wnt pathway, EMT of the cancer cells and cell motility. In addition, Wnt10b knockdown markedly impaired the ability of p85α−/− fibroblasts to promote tumour growth, metastasis and activation of the Wnt pathway in vivo (Figures 5g and i). Moreover, in stroma associated with human breast cancer, Wnt10b expression was significantly upregulated (Figure 5j).21 Correlation analysis also demonstrated the inverse relationship between p85α and Wnt10b (Figure 5k).21 These data suggest that Wnt10b from p85α−/− fibroblasts has a crucial role in inducing breast cancer EMT and promoting tumourigenesis.
Figure 5.
Wnt10b as a paracrine factor has a crucial role in inducing breast cancer epithelial cell EMT and facilitates metastasis. (a) Quantitative PCR analysis of Wnt family expression in p85α−/− fibroblasts normalized to WT fibroblasts. (b) ELISA analysis of Wnt10b expression in WT and p85α−/− fibroblast-conditioned medium. Data are the mean±s.e.m. ***P-value⩽0.001. (c) Western blot analysis of Wnt10b expression in WT and p85α−/− fibroblast-conditioned medium, and Wnt10b, Wntless, VPS35 in cell lysates. (d) Immunofluorescence analysis of Wnt10b and Wntless expression and localization in WT and p85α−/− fibroblasts. Scale bar, 20 μm. (e) Western blot analysis of Wnt pathway and EMT markers in MDA-MB-231 cells and 4T1 cells cultured in Wnt10b knockdown and negative control p85α−/− fibroblast-conditioned medium. The effect of shWnt10b are shown on the upper panel. (f) The migration assay of MDA-MB-231 and 4T1 cells cultured in negative control or Wnt10b knockdown p85α−/− fibroblast-conditioned medium. Data are the mean±s.e.m. *P-value⩽0.05. Scale bar, 100 μm. (g) Tumour volume and weight measurements of 4T1 cell grown subcutaneously with negative control vector or with Wnt10b knockdown p85α−/− fibroblasts. Each group have six mice (n=6). Data are the mean±s.e.m. *P-value⩽0.05. (h) Quantification of liver metastasis in mice described in (g). Scale bar, 50 μm. Data are the mean±s.e.m. *P-value⩽0.05. (i) Representative images of β-catenin immunostaining of tumours in mice described in (g and h). Scale bar, 50 μm. (j) Gene expression level of Wnt10b in human breast cancer stroma versus normal breast stroma from GSE9014. Data are the mean±s.e.m. ***P-value⩽0.001. (k) Correlation analysis between Wnt10b and p85α (PIK3R1) in breast stroma from GSE9014. The line with linear regression is also shown as a red line. The P-values and R2 are also shown.
Exosomes from p85α-deficient fibroblasts regulate stromal–epithelial crosstalk in breast cancer
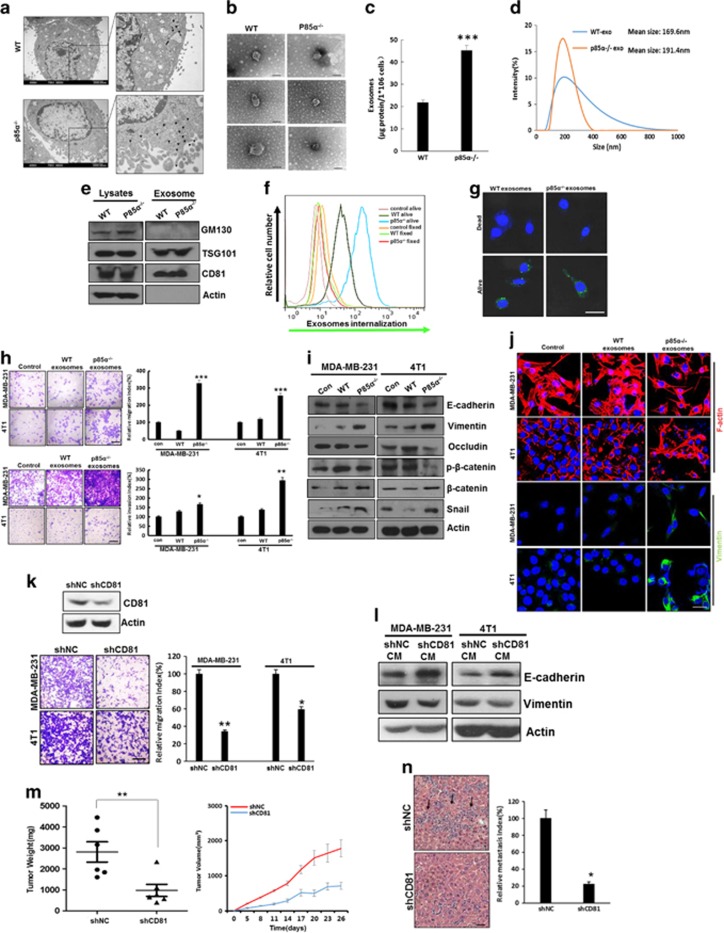
Recently reports demonstrated that exosomes, which are nanosize vesicles secreted by cells, from the tumour microenvironment could promote traits of cancer progression.10, 41, 42 Therefore, we investigated the effects of p85α loss in fibroblasts on the function of exosomes. Electron microscopy observations indicated that there are more exosome-like structures located close to the cell membrane in p85α−/− fibroblasts than in WT fibroblasts (Figure 6a). Moreover, the purified exosomes from WT and p85α−/− fibroblast-conditioned medium showed comparable size and appearance (Figure 6b). We also found by bicinchoninic acid protein assay that p85α−/− fibroblasts can produce more exosomes than WT fibroblasts (Figure 6c). The particle size analysis of isolated exosomes showed that particle size distribution primarily ranged from 80 to 300 nm (Figure 6d). Western blots showed that expression of the exosome markers CD81 and TSG101, but not GM130, in exosome lysates confirmed that there was no cellular contamination in our isolated exosomes (Figure 6e). To further examine exosome transfer, purified exosomes from various fibroblasts were labelled with the fluorescent lipophilic dye DiD. Fluorescence-activated cell sorting and microscopic analysis of exosomes transfer indicated that the exosomes collected from various fibroblasts transferred into living but not dead (fixed) cancer cells via active transport (Figures 6f and g). Fluorescence-activated cell sorting also revealed that exosomes from p85α−/− fibroblasts had an increased transfer capacity compared with those from WT fibroblasts.
Figure 6.
Exosomes from p85α-deficient fibroblasts regulate stromal–epithelial crosstalk in breast cancer. (a) Representative electron microscopy image of WT and p85α−/− fibroblasts. Scale bar, 2000 nm. (b) Representative electron microscopy image of exosomes purified from WT and p85α−/− fibroblast-conditioned medium, each group have three replications. Scale bar, 100 nm. (c) Quantification of the exosome protein yield from WT and p85α−/− fibroblasts by BCA protein quantitation. Data are the mean±s.e.m. ***P-value⩽0.001. (d) Exosome size distribution of WT and p85α−/− fibroblasts measured by a Zetasizer analyser. (e) Western blot analysis of exosomes from WT and p85α−/− fibroblasts. TSG101 and CD81 are markers of exosomes. GM130 is a check for cellular contamination. (f) Flow cytometry of DiD dye transfer from WT or p85α−/− fibroblasts to breast cancer cells (4T1). (g) Image of DiD dye transfer from various fibroblasts to breast cancer cells (4T1) as described in (f). Scale bar, 50 μm. (h) The migration and invasion assay of MDA-MB-231 and 4T1 cells treated with exosomes purified from WT or p85α−/− fibroblast-conditioned medium. Data are the mean±s.e.m. *P-value⩽0.05. **P-value⩽0.01. ***P-value⩽0.001. Scale bar, 100 μm. (i) Western blot analysis of Wnt pathway and EMT markers in MDA-MB-231 cells and 4T1 cells treated with exosomes purified from WT or p85α−/− fibroblasts. (j) Representative immunofluorescence images of cytoskeleton and vimentin in cancer cells treated with exosomes from WT or p85α−/− fibroblasts. Scale bar, 25 μm. (k) Migration assay of MDA-MB-231 and 4T1 cells cultured in shNC or shCD81 p85α−/− fibroblast-conditioned medium. Data are the mean±s.e.m. *P-value⩽0.05. **P-value⩽0.01. Scale bar, 100 μm. (l) Western blot analysis of EMT markers in MDA-MB-231 cells and 4T1 cells cultured in shNC or shCD81 p85α−/− fibroblast-conditioned medium. (m) Tumour volume and weight measurements of 4T1 cell grown subcutaneously with shNC or shCD81 p85α−/− fibroblasts. Each group have six mice (n=6). Data are the mean±s.e.m. **P-value⩽0.01. (n) Quantification of liver metastasis in mice described in (m). Data are the mean±s.e.m. *P-value⩽0.05. Scale bar, 50 μm.
To determine whether exosomes produced by fibroblasts are sufficient to induce protrusive activity and metastasis of breast cancer cells, we exposed MDA-MB-231 or 4T1 cells to exosomes isolated from WT or p85α−/− fibroblasts, respectively, and evaluated cell motility. As shown in Figure 6h, exosomes derived from p85α−/− fibroblasts notably promoted MDA-MB-231 and 4T1 migration and invasion compared with exosomes from WT fibroblasts. Accordingly, exosomes from p85α−/− fibroblasts also more potently induced EMT and cytoskeletal remodelling in breast cancer epithelial cells, which is closely related to tumour metastasis (Figures 6i and j). Tetraspanin CD81, an exosome marker, is critically involved in exosome function, secretion and cellular uptake during intercellular communication.43 To further explore the importance of exosomes in stromal–epithelial crosstalk and their effects on cancer epithelial cell motility, we generated p85α−/− fibroblasts in which CD81 was constitutively knocked down by short hairpin RNAs (shRNAs), thereby creating fibroblasts with aberrant exosome function. MDA-MB-231 or 4T1 cells were treated with the conditioned medium from negative control or CD81 knockdown fibroblasts, respectively. As shown in Figures 6k and l, the conditioned medium contained reduced amount of CD81 and significantly decreased the ability of p85α−/− fibroblasts to stimulate cell migration and EMT. Similarly, downregulation of CD81 expression inhibited the tumour-promoting effects of p85α−/− fibroblasts in vivo (Figures 6m and n). Taken together, these results reveal that exosomes from p85α−/− fibroblasts can induce EMT, promote cancer cell motility and increase tumour metastasis.
Exosomes act as a cargo delivery system that transports Wnt10b from stroma to cancer cells
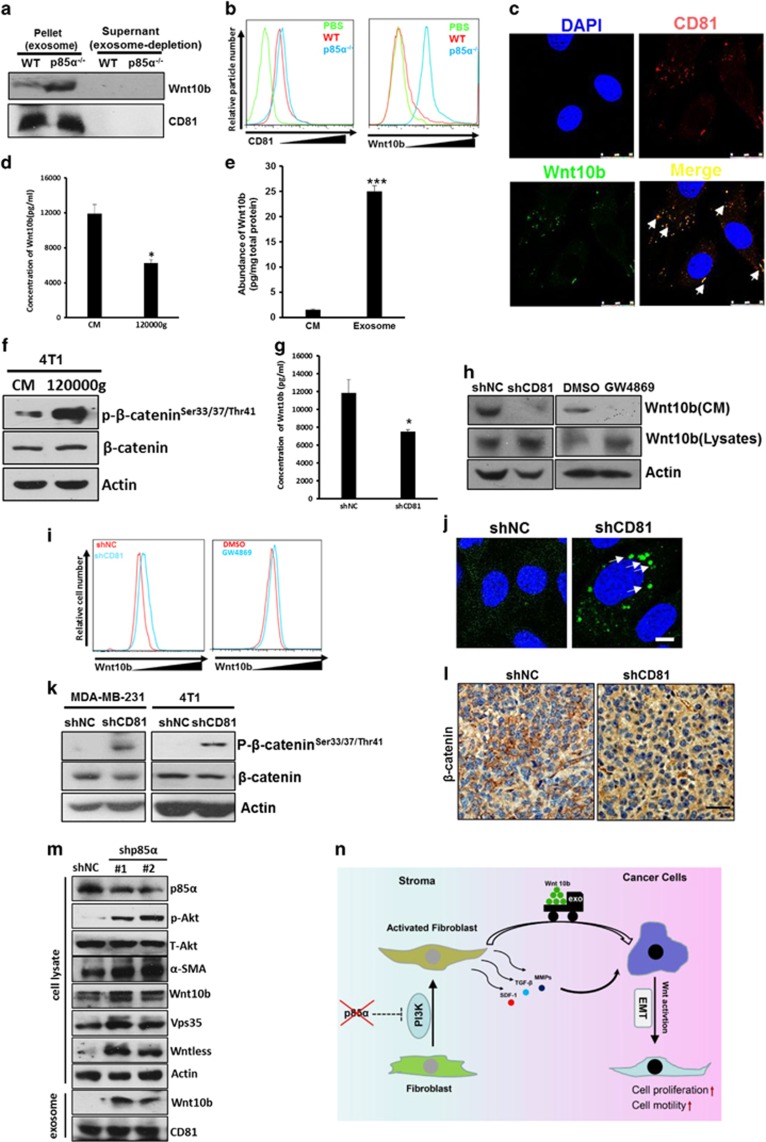
Considering the fact that some reports have also demonstrated that active Wnt proteins could be secreted in exosomes,44 we speculated that stromal-derived exosomes might carry Wnt10b into cancer cells to induce Wnt/β-catenin signalling and regulate stromal–epithelial dialogue. Western blots indicated that Wnt10b is more abundant in p85α−/− fibroblast-derived exosomes compared with WT fibroblast exosomes. On the other hand, the expression of Wnt10b in exosomes is far higher than in exosome-depleted conditioned medium when the total loading protein contents were equal (Figure 7a). Flow cytometric analysis of Wnt10b expression in exosomes captured by aldehyde latex beads confirmed this trend (Figure 7b). Furthermore, we found that Wnt10b colocalized with the exosome marker CD81 in p85α−/− fibroblasts via immunofluorescence (Figure 7c). Additionally, ELISA analyses showed that the Wnt10b concentration in p85α−/− fibroblast-conditioned medium was obviously reduced after ultracentrifugation overnight (Figure 7d). In addition, we found that the abundance of Wnt10b in exosomes was far higher (~16-fold) than in conditioned medium from p85α−/− fibroblasts (Figure 7e). Moreover, the ability of the conditioned medium to activate the Wnt pathway was also markedly reduced after ultracentrifugation (Figure 7f). Knockdown of CD81 in p85α−/− fibroblasts or treatment of p85α−/− fibroblasts with GW4869, an N-SMase inhibitor that blocks exosome generation,45 reduced Wnt10b secretion in conditioned medium (Figures 7g and h). Consistently, disturbing exosomes resulted in increased intracellular accumulation of Wnt10b (Figures 7i and j). Moreover, we found that blocking exosomes via CD81 knockdown in p85α−/− fibroblasts abrogated the activation of Wnt/β-catenin signalling in vitro and in vivo (Figures 7k and l). We also knockdown p85α in WT fibroblasts to mimic p85α−/−, and the result also confirmed our conclusions (Figure 7m). These results indicated that exosomes acted as a cargo delivery system that transported Wnt10b from stroma to cancer cells (Figure 7n).
Figure 7.
Exosomes act as a cargo delivery system that transports Wnt10b from stroma to cancer cells. (a) Western blot analysis of Wnt10b in exosomes purified from various fibroblasts. (b) Flow cytometry of CD81 and Wnt10b expression in exosomes purified from various fibroblasts. (c) Immunofluorescence analysis of Wnt10b and CD81 co-location in p85α−/− fibroblasts. Scale bar, 25μm. (d) ELISA analysis of Wnt10b in conditioned medium from p85α−/− fibroblasts before (conditioned medium (CM)) and after (120 000 g) ultracentrifugation overnight. Data are the mean±s.e.m. *P-value⩽0.05. (e) The protein abundance of Wnt10b in conditioned medium and exosomes from p85α−/− fibroblasts. Data are the mean±s.e.m. ***P-value⩽0.001. (f) Western blot analysis of Wnt activity in 4T1 cells cultured in original CM or after (120 000 g) ultracentrifugation overnight. (g) ELISA analysis of Wnt10b in CM from shNC or shCD81 p85α−/− fibroblasts. Data are the mean±s.e.m. *P-value⩽0.05. (h) Western blot analysis of Wnt10b in cell lysates and concentrated conditioned medium from shNC or shCD81 p85α−/− fibroblasts and p85α−/− fibroblasts treated with DMSO or GW4869 (10 μM) for 2 days. (i) Flow cytometric analysis of Wnt10b intracellular accumulation in shNC or shCD81 p85α−/− fibroblasts and p85α−/− fibroblasts treated with DMSO or GW4869 as described in (I). (j) Immunofluorescence analysis of intracellular Wnt10b accumulation in shNC or shCD81 p85α−/− fibroblasts. Scale bar, 10 μm. (k) Western blot analysis of Wnt pathway activation in MDA-MB-231 cells and 4T1 cells cultured in shNC or shCD81 p85α−/− fibroblast-conditioned medium. (l) Representative images of β-catenin immunostaining in tumours from mice described in Figure 6m. Scale bar, 50 μm. (m) Western blot analysis of PI3K activity and Wnt10b in negative control vector (NC) and in p85α knockdown (shp85α) WT fibroblasts and exosomes. (n) Schematic representation of tumour-promoting effects provoked by fibroblasts instigated by loss expression of p85α.
Discussion
Mutations in or abnormal expression of the p85α regulatory subunit of PI3K are reported in various human cancers,46, 47, 48 but the role of these mutations in tumour microenvironment remains unclear. In this study, we have determined that reduced expression of p85α occurs in the stromal compartment in breast cancer and has a critical role in stromal–epithelial crosstalk, as well as in tumour growth and metastasis. With respect to promoting tumourigenesis, p85α−/− fibroblasts exhibit similar functions to CAFs.
Cancer-related deaths are primarily attributed to metastasis. It is established that crosstalk between the neoplastic stroma and the epithelium is essential to metastasis, yet the master regulators of metastasis are largely unknown. Wnt pathway, including canonical (Wnt/β-catenin) and noncanonical (Wnt-PCP) Wnt signaling, has a key role in these progressions. Jeff Wrana's group had reported that stromal fibroblasts could regulate breast cancer cell migration via autocrine Wnt-PCP signaling.41 However, in our research, we found that p85α−/− fibroblasts could promote breast cancer cell motility mainly via paracrine Wnt/β-catenin signaling. Although Wnt-PCP signaling and Wnt/β-catenin signaling are two different kinds of pathways, there is ample evidence that there is a close link between these pathways. Moreover, our findings also demonstrated that Wnt10b secreted from fibroblasts acted as a potent mediator of tumour–stroma communication to promote metastasis.
Interestingly, we also observed that considerable levels of Wnt10b were located in exosomes from p85α-deficient fibroblasts. Moreover, we observed that p85α−/− fibroblasts expressed increased levels of Wntless and Vps35 proteins, indicating that the loss of p85α comprehensively regulates the Wnt signalling axis not only with respect to production but also with respect to transportation. EMT is also involved in both Wnt/β-catenin signalling and cancer cell metastasis. To further verify the function of p85α in CAFs, we developed stable and ectopic expression of p85α in p85α−/− fibroblasts for rescue experiments in WT fibroblasts. As expected, the rescue experiments confirmed our conclusions (Supplementary Figures S3A–E). Therefore, we propose the following model to explain our findings: loss of p85α stimulates fibroblasts to express and secrete more Wnt10b. Then, these Wnt10b ligands are transported to adjacent epithelial cancer cells, where they activate Wnt/β-catenin signalling and then induce EMT, eventually leading to breast cancer cell metastasis.
Notably, we found that active Wnt10b is also present in exosomes from p85α-deficient fibroblasts that exhibit characteristics of CAFs. The exosomal Wnt derived from fibroblasts could activate canonical Wnt signalling and promote the mobility of breast cancer cells both in vitro and in vivo. Exosomes are nano-sized extracellular microvesicles that have an endosomal origin, are secreted by various cells and have important roles in intercellular communication.45 Exosomes contain many bioactive molecules, such as nucleic acids and proteins that can mediate cell–cell communication associated with development and cancer progression.49, 50 When we disturbed exosome function via knockdown of the exosomal marker CD81 in fibroblasts, Wnt10b secretion was inhibited, leading to accumulation of Wnt10b in fibroblasts. Hence, the CD81 knockdown in CAF-like fibroblasts failed to activate canonical Wnt signalling and could not promote tumourigenesis. Thus, we conclude that exosomes from fibroblasts in the tumour microenvironment transport Wnt proteins to cancer epithelial cells, which may be an indispensable and efficient approach to activate Wnt signalling and cellular functions involved in cancer progression.
Taken together, our data demonstrate that p85α expression is frequently reduced in breast cancer stroma and is functionally associated with the regulation of breast tumourigenesis and malignant progression through its ability to modify the tumour microenvironment. Thus, p85α acts as a tumour suppressor in the tumour microenvironment and may represent a new candidate for diagnosis, prognosis and targeted therapy.
Materials and methods
Chemicals and antibodies
LY294001 was purchased from Cell Signalling Technology (Beverly, MA, USA), GW4869 was purchased from Santa Cruz (Santa Cruz, CA, USA). The antibodies used in this research were listed in Supplementary Table S4.
Cell culture
The immortalized WT mouse embryonic fibroblasts (MEFs) and the p85α−/− MEFs were a gift from Professor Lewis C Cantley.51 The human breast cancer cell line MDA-MB-231, the mouse breast cancer cell line 4T1, the immortalized WT MEFs and the p85α−/− MEFs were cultured in Dulbecco’s modified Eagle’s medium (HyClone, Logan, UT, USA) supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin and 10% fetal bovine serum (HyClone) at 37 °C and 5% CO2. Cell lines were acquired from the China Center For Type Culture Collection, where it was tested and authenticated, and were cultured continuously for no more than 2 months.
Conditioned medium preparation
Various MEFs were incubated with complete medium for 3 days. The conditioned medium was collected, centrifuged at 300 g for 10 min, 2000 g for 10 min and 10 000 g for 30 min, and then filtered with a 0.22-μm filter.
Cell migration and invasion assays
The cell migration and invasion assay was performed as described by Hu et al.52
Subcutaneous tumourigenicity assays
A total of 1 × 106 4T1 or MDA-MB-231 cells mixed with 3 × 106 MEFs of various types in Matrigel (BD Biosciences, San Jose, CA, USA) and coinjected subcutaneously into 4–6-week-old female BALB/c mice or nude mice. Each group had six mice in the animal experiments. Tumourigenicity assays were performed as described previously.32 The animal care and experimental protocols were approved by the Experimental Animal Center of Wuhan University.
Exosome isolation
Exosomes were purified from cells cultured in exosome-depleted fetal bovine serum (ultracentrifuged at 120 000 g overnight) as described previously.53 For western blot analysis, the exosome pellet was resuspended in 1% sodium dodecyl sulphate lysis buffer, and analysed with loadings of exosomes that amounted to equal content of proteins, respectively. For examination by electron microscopy, the exosome pellet was resuspended in phosphate-buffered saline, and analyzed again with loadings of exosomes that amounted to equal content of proteins, respectively. For cell treatment, the exosome pellets were resuspended in Dulbecco’s modified Eagle’s medium, respectively, and the exosomes isolated from equal volume of conditioned mediums were used. For quantification by the BCA assay, exosome size distribution measurement by Zetasizerand flow cytometry, the exosome pellets were resuspended in phosphate-buffered saline and exosomes isolated from equal volume of conditioned mediums were analyzed.
Analysis of human stroma gene expression data
The data regarding human breast cancer stroma (GSE9014) and human fibroblast cocultured with breast cancer cells (GSE41678) were downloaded from the NCBI GEO datasets database.
Tissue microarray and immunohistochemistry
The human breast tissue microarray chip was purchased from US Biomax Inc. (Rockville, MD, USA). Immunohistochemistry was performed as described by Hu et al.52
Immunohistochemical score
For the immunohistochemical semiquantitative assessment of p85α expression, the product of the staining intensity scores and the quantity of immunoreactive cells were calculated based on the following scoring system: the optical stain intensity was graded as 0=negative, 1=weak, 2=moderate or 3=strong staining; the quantity of cells was graded as 0=no expression, 1=⩽10% positive cells, 2=10–50% positive cells, 3=51–80% positive cells and 4=⩾80% positive cells. The number of positive cells was assessed by counting three random fields at × 100 magnification. The final immunohistochemical score was obtained by multiplying the intensity score and the quantity score. According to the total Immunohistochemical score, p85α expression levels in tissues were categorized as low expression when the score was <1, as intermediate expression when the score was 1–3 and as high expression when the score was >3. The staining intensity was scored by two investigators in a blinded procedure.
Cell extracts and western blot analysis
The cell extracts were performed as described previously.54 The supernatants were centrifuged to remove the cells and debris and concentrated with Amicon Ultra columns (Millipore, Billerica, MA, USA). Proteins were extracted from the nucleus and cytoplasm using Nuclear and Cytoplasmic Protein Extraction Kits (Beyotime, Jiangsu, China), respectively, according to the manufacturer’s instructions. Western blotting was performed as described previously.54
Immunofluorescence
The assay was performed as described by Hu et al.52 Images were acquired using a confocal microscope (Leica, Wetzlar, Germany).
Measurement of collagen gel contraction
The assay was performed as described previously.55
Fluorescence-activated cell sorting analysis
Fibroblasts of different genotypes were stained with antibodies against CD44 (BD Pharmingen, San Diego, CA, USA) and assayed with flow cytometry (Beckman Coulter, Miami, FL, USA).
Flow cytometry analysis of purified exosomes was performed as described previously.56
RNA extraction, PCR and real-time quantitative PCR
The primers are listed in Supplementary Table S2. All quantitations were normalized to endogenous glyceraldehyde 3-phosphate dehydrogenase as a loading control. Relative changes in gene expression were quantified using the 2−ΔΔCT method.57
Immunoassay for conditioned medium
Approximately 1.5 × 106 fibroblasts were cultured in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum on a 10-cm plate for 2–3 days. The supernatants were measured using a commercially available SDF-1 ELISA Kit (R&D Systems, Minneapolis, MN, USA), a TGF-β1 ELISA Kit (Boster Bio-Tech, Wuhan, China), an MMP2 ELISA Kit (Boster Bio-Tech), an MMP9 ELISA Kit (Boster Bio-Tech) and a Wnt10b ELISA Kit (Cusabio Biotech, Wuhan, China).
Cell proliferation assay
The assay was performed as described by Hu et al.52
Colony formation assays
The assay was performed as described by Hu et al.52
Matrigel three-dimensional cultures
Cells were suspended in conditioned media containing 2% growth factor reduced Matrigel (BD Biosciences) and plated on 100% Matrigel as described previously.58
Scratch assay
The assay was performed as described by Hu et al.52
shRNA/siRNA preparation and cell transfection
shRNA sequences against Wnt10b and p85α were designed and synthesized by GenePharma Co. Ltd (Shanghai, China). The fragments were digested with BbsI and BamHI and were inserted into the pGPU/GFP/Neo vector (GenePharma Co. Ltd). shRNA sequences against CD81 were designed according to information given by Sigma-Aldrich (St Louis, MO, USA), which has been validated. The fragments were inserted into the retroviral vector pSuper. The full-length cDNA encoding p85α was cloned into the lentiviral vector pHAGE-CMV-MCS-PGK puro+3tag. An empty pHAGE-CMV-MCS-PGK puro+3tag plasmid was used similarly to establish control clones. Transient transfection was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Small interfering RNA (siRNA) against Porcn was designed and synthesized by RiboBio Co. Ltd (Guangzhou, China). Transient transfection of siRNA was also performed using Lipofectamine 2000. The siRNA and shRNA sequences are listed in Supplementary Table S3.
Electron microscopy
WT or p85α MEFs were fixed in 2.5% glutaraldehyde. The next day, the cells were washed in phosphate-buffered saline and fixed with 1% osmium tetroxide, followed by washing in phosphate-buffered saline and dehydration through an alcohol gradient. After that, the cells were embedded in epoxide resin, sliced by an LKB-V ultramicrotome (LKB Bromma, Sollentuna, Sweden) and observed with a transmission electron microscope (Hitachi Ltd, Tokyo, Japan).
Purified exosomes were left to settle on carbon-coated grids. After the grids were stained with 3% uranyl acetate, they were air-dried and visualized using a transmission electron microscope.
Statistical analysis
Statistical significance was calculated using Prism (GraphPad Software, La Jolla, CA, USA). Data are reported as the mean±s.e.m. The results between the two independent groups were determined by Student's t-test, unless otherwise stated. In the case of different variances, a two-tailed unpaired t-test with Welch's correction was also performed. When data did not pass the normality test, a Mann–Whitney test was also used for the statistical analysis as indicated. Values of P<0.05 were considered statistically significant. All experiments were repeated at least three times.
Acknowledgments
We thank Professor Lewis C Cantley for the gift of the knockout cell lines. This study was supported by National Basic Research Program of China (2014CB910603), the National Natural Science Foundation of China (81472684, 81072151) and Program for New Century Excellent Talents in University of Ministry of Education of China (NCET-13-0436).
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
The authors declare no conflict of interest.
Supplementary Material
References
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674. [DOI] [PubMed] [Google Scholar]
- Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature 2007; 449: 557–563. [DOI] [PubMed] [Google Scholar]
- Bhowmick NA, Neilson EG, Moses HL. Stromal fibroblasts in cancer initiation and progression. Nature 2004; 432: 332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med 2013; 19: 1423–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchor L, Benitez J. The complex genetic landscape of familial breast cancer. Hum Genet 2013; 132: 845–863. [DOI] [PubMed] [Google Scholar]
- Shannon KM, Chittenden A. Genetic testing by cancer site: breast. Cancer J 2012; 18: 310–319. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature 2012; 490: 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procopio MG, Laszlo C, Al Labban D, Kim DE, Bordignon P, Jo SH et al. Combined CSL and p53 downregulation promotes cancer-associated fibroblast activation. Nat Cell Biol 2015; 17: 1193–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng N, Bhowmick NA, Chytil A, Gorksa AE, Brown KA, Muraoka R et al. Loss of TGF-beta type II receptor in fibroblasts promotes mammary carcinoma growth and invasion through upregulation of TGF-alpha-, MSP- and HGF-mediated signaling networks. Oncogene 2005; 24: 5053–5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoda M, Principe S, Jackson HW, Luga V, Fang H, Molyneux SD et al. Loss of the Timp gene family is sufficient for the acquisition of the CAF-like cell state. Nat Cell Biol 2014; 16: 889–901. [DOI] [PubMed] [Google Scholar]
- Trimboli AJ, Cantemir-Stone CZ, Li F, Wallace JA, Merchant A, Creasap N et al. Pten in stromal fibroblasts suppresses mammary epithelial tumours. Nature 2009; 461: 1084–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Leevers SJ, Panayotou G, Waterfield MD. Phosphoinositide 3-kinases: a conserved family of signal transducers. Trends Biochem Sci 1997; 22: 267–272. [DOI] [PubMed] [Google Scholar]
- Katso R, Okkenhaug K, Ahmadi K, White S, Timms J, Waterfield MD. Cellular function of phosphoinositide 3-kinases: implications for development, homeostasis, and cancer. Annu Rev Cell Dev Biol 2001; 17: 615–675. [DOI] [PubMed] [Google Scholar]
- Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene 2008; 27: 5497–5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantley LC. The phosphoinositide 3-kinase pathway. Science 2002; 296: 1655–1657. [DOI] [PubMed] [Google Scholar]
- Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer 2002; 2: 489–501. [DOI] [PubMed] [Google Scholar]
- Klarenbeek S, van Miltenburg MH, Jonkers J. Genetically engineered mouse models of PI3K signaling in breast cancer. Mol Oncol 2013; 7: 146–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cully M, You H, Levine AJ, Mak TW. Beyond PTEN mutations: the PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat Rev Cancer 2006; 6: 184–192. [DOI] [PubMed] [Google Scholar]
- Bergamaschi A, Tagliabue E, Sorlie T, Naume B, Triulzi T, Orlandi R et al. Extracellular matrix signature identifies breast cancer subgroups with different clinical outcome. J Pathol 2008; 214: 357–367. [DOI] [PubMed] [Google Scholar]
- Jaiswal BS, Janakiraman V, Kljavin NM, Chaudhuri S, Stern HM, Wang W et al. Somatic mutations in p85alpha promote tumorigenesis through class IA PI3K activation. Cancer Cell 2009; 16: 463–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med 2008; 14: 518–527. [DOI] [PubMed] [Google Scholar]
- Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer 2006; 6: 392–401. [DOI] [PubMed] [Google Scholar]
- Albrengues J, Bourget I, Pons C, Butet V, Hofman P, Tartare-Deckert S et al. LIF mediates proinvasive activation of stromal fibroblasts in cancer. Cell Rep 2014; 7: 1664–1678. [DOI] [PubMed] [Google Scholar]
- Webber J, Steadman R, Mason MD, Tabi Z, Clayton A. Cancer exosomes trigger fibroblast to myofibroblast differentiation. Cancer Res 2010; 70: 9621–9630. [DOI] [PubMed] [Google Scholar]
- Rajaram M, Li J, Egeblad M, Powers RS. System-wide analysis reveals a complex network of tumor–fibroblast interactions involved in tumorigenicity. PLoS Genet 2013; 9: e1003789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg RA. Coevolution in the tumor microenvironment. Nat Genet 2008; 40: 494–495. [DOI] [PubMed] [Google Scholar]
- Franco OE, Shaw AK, Strand DW, Hayward SW. Cancer associated fibroblasts in cancer pathogenesis. Semin Cell Dev Biol 2010; 21: 33–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Phan SH. Myofibroblasts. Curr Opin Rheumatol 2013; 25: 71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi CM, Winnay J, Kondo T, Bronson RT, Guimaraes AR, Aleman JO et al. The Phosphoinositide 3-kinase regulatory subunit p85 alpha can exert tumor suppressor properties through negative regulation of growth factor signaling. Cancer Res 2010; 70: 5305–5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature 2007; 449: U557–U554. [DOI] [PubMed] [Google Scholar]
- Spaeth EL, Labaff AM, Toole BP, Klopp A, Andreeff M, Marini FC. Mesenchymal CD44 expression contributes to the acquisition of an activated fibroblast phenotype via TWIST activation in the tumor microenvironment. Cancer Res 2013; 73: 5347–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell 2005; 121: 335–348. [DOI] [PubMed] [Google Scholar]
- Kojima Y, Acar A, Eaton EN, Mellody KT, Scheel C, Ben-Porath I et al. Autocrine TGF-beta and stromal cell-derived factor-1 (SDF-1) signaling drives the evolution of tumor-promoting mammary stromal myofibroblasts. Proc Natl Acad Sci USA 2010; 107: 20009–20014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond N, d'Agua BB, Ridley AJ. Crossing the endothelial barrier during metastasis. Nat Rev Cancer 2013; 13: 858–870. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial–mesenchymal transitions in development and disease. Cell 2009; 139: 871–890. [DOI] [PubMed] [Google Scholar]
- Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer 2009; 9: 265–273. [DOI] [PubMed] [Google Scholar]
- Scheel C, Eaton EN, Li SH, Chaffer CL, Reinhardt F, Kah KJ et al. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell 2011; 145: 926–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willert K, Nusse R. Wnt proteins. Cold Spring Harbor Perspect Biol 2012; 4: a007864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang PT, Lorenowicz MJ, Silhankova M, Coudreuse DY, Betist MC, Korswagen HC. Wnt signaling requires retromer-dependent recycling of MIG-14/Wntless in Wnt-producing cells. Dev Cell 2008; 14: 140–147. [DOI] [PubMed] [Google Scholar]
- Franch-Marro X, Wendler F, Guidato S, Griffith J, Baena-Lopez A, Itasaki N et al. Wingless secretion requires endosome-to-Golgi retrieval of Wntless/Evi/Sprinter by the retromer complex. Nat Cell Biol 2008; 10: 170–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luga V, Zhang L, Viloria-Petit AM, Ogunjimi AA, Inanlou MR, Chiu E et al. Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell 2012; 151: 1542–1556. [DOI] [PubMed] [Google Scholar]
- Boelens MC, Wu TJ, Nabet BY, Xu B, Qiu Y, Yoon T et al. Exosome transfer from stromal to breast cancer cells regulates therapy resistance pathways. Cell 2014; 159: 499–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoller M. Tetraspanins: push and pull in suppressing and promoting metastasis. Nat Rev Cancer 2009; 9: 40–55. [DOI] [PubMed] [Google Scholar]
- Gross JC, Chaudhary V, Bartscherer K, Boutros M. Active Wnt proteins are secreted on exosomes. Nat Cell Biol 2012; 14: 1036–1045. [DOI] [PubMed] [Google Scholar]
- Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F et al. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 2008; 319: 1244–1247. [DOI] [PubMed] [Google Scholar]
- Taniguchi CM, Winnay J, Kondo T, Bronson RT, Guimaraes AR, Aleman JO et al. The phosphoinositide 3-kinase regulatory subunit p85alpha can exert tumor suppressor properties through negative regulation of growth factor signaling. Cancer Res 2010; 70: 5305–5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K, Wang J, Jayasinghe R, Lameijer EW, McMichael JF, Ning J et al. Systematic discovery of complex insertions and deletions in human cancers. Nat Med 2016; 22: 97–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer 2009; 9: 550–562. [DOI] [PubMed] [Google Scholar]
- Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Exosomes: proteomic insights and diagnostic potential. Expert Rev Proteomics 2009; 6: 267–283. [DOI] [PubMed] [Google Scholar]
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007; 9: 654–659. [DOI] [PubMed] [Google Scholar]
- Chiu YH, Lee JY, Cantley LC. BRD7, a tumor suppressor, interacts with p85alpha and regulates PI3K activity. Mol Cell 2014; 54: 193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu HM, Chen Y, Liu L, Zhang CG, Wang W, Gong K et al. C1orf61 acts as a tumor activator in human hepatocellular carcinoma and is associated with tumorigenesis and metastasis. FASEB J 2013; 27: 163–173. [DOI] [PubMed] [Google Scholar]
- Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protocols Cell Biol 2006; 30: 3.22, 3.22.1–3.22.29. [DOI] [PubMed] [Google Scholar]
- Gong K, Chen C, Zhan Y, Chen Y, Huang Z, Li W. Autophagy-related gene 7 (ATG7) and reactive oxygen species/extracellular signal-regulated kinase regulate tetrandrine-induced autophagy in human hepatocellular carcinoma. J Biol Chem 2012; 287: 35576–35588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogatkevich GS, Tourkina E, Silver RM, Ludwicka-Bradley A. Thrombin differentiates normal lung fibroblasts to a myofibroblast phenotype via the proteolytically activated receptor-1 and a protein kinase C-dependent pathway. J Biol Chem 2001; 276: 45184–45192. [DOI] [PubMed] [Google Scholar]
- Stoeck A, Keller S, Riedle S, Sanderson MP, Runz S, Le Naour F et al. A role for exosomes in the constitutive and stimulus-induced ectodomain cleavage of L1 and CD44. Biochem J 2006; 393: 609–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 2001; 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Viloria-Petit AM, David L, Jia JY, Erdemir T, Bane AL, Pinnaduwage D et al. A role for the TGFbeta-Par6 polarity pathway in breast cancer progression. Proc Natl Acad Sci USA 2009; 106: 14028–14033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.